Monitoring Antibiotic Resistance in Wastewater: Findings from Three Treatment Plants in Sicily, Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Handling
2.2. Sample Processing and Concentration
2.3. Nucleic Acid Extraction
2.4. DNA Pooling for Composite Samples
2.5. Real-Time PCR
2.6. Statistical Analysis
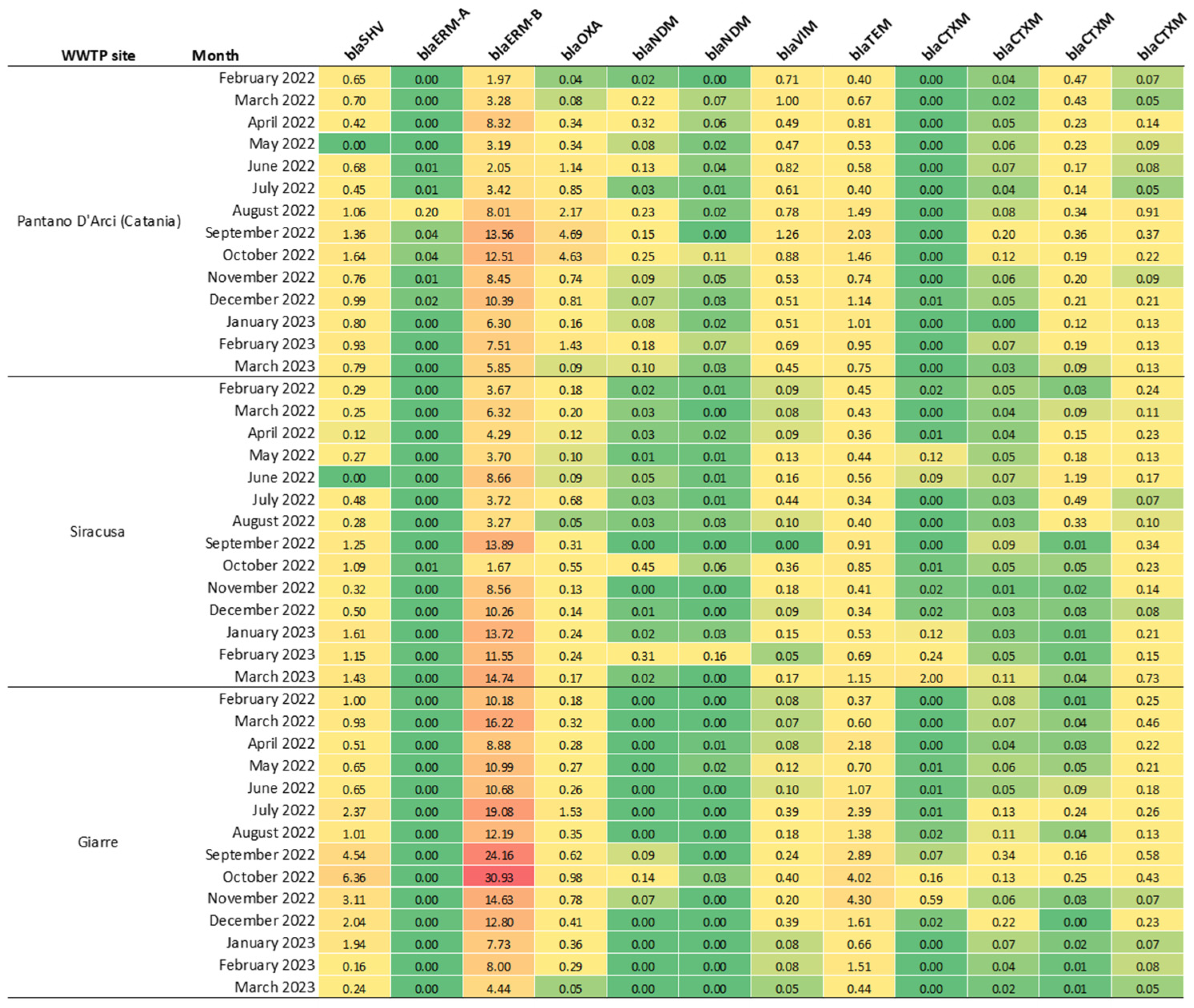
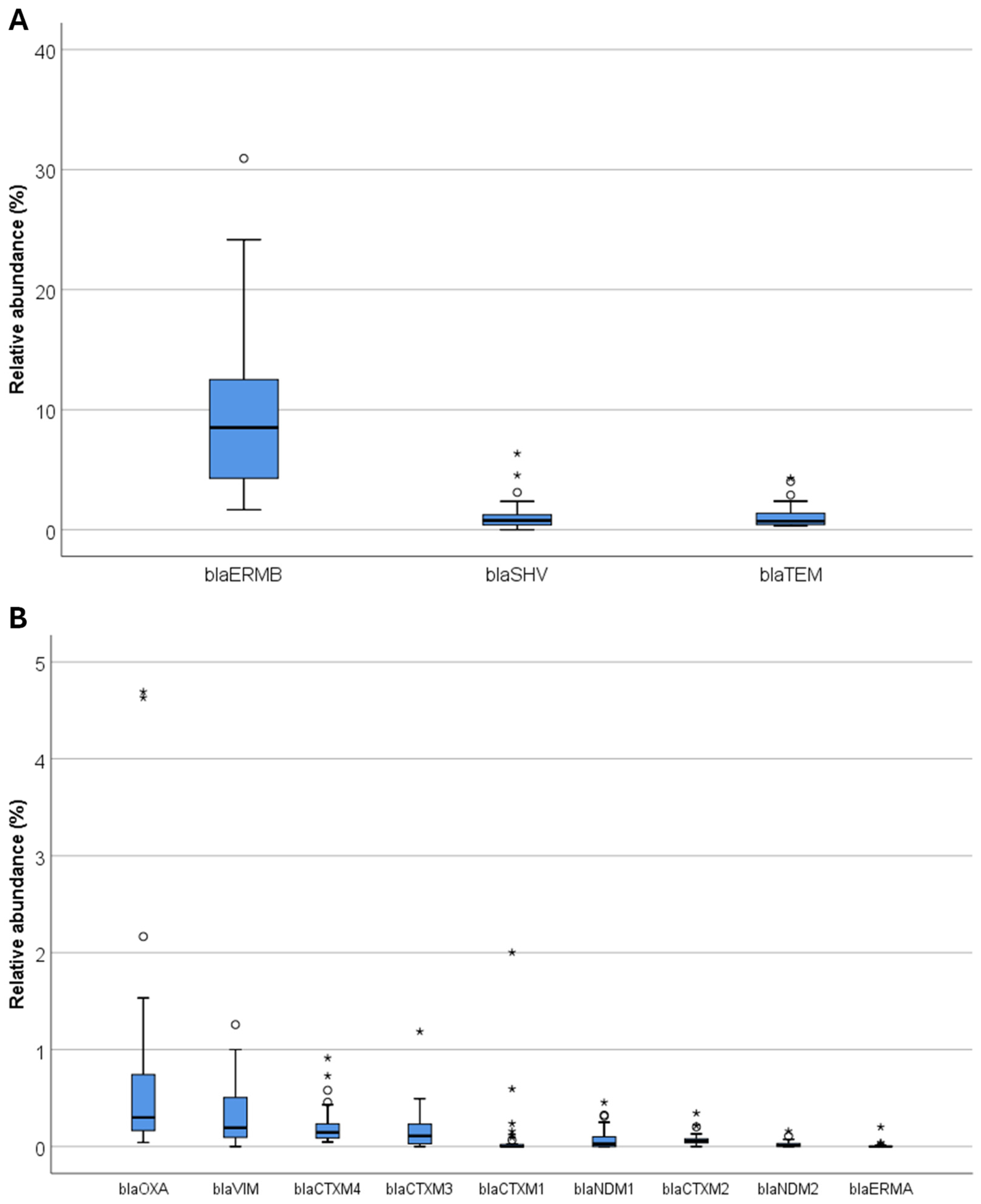
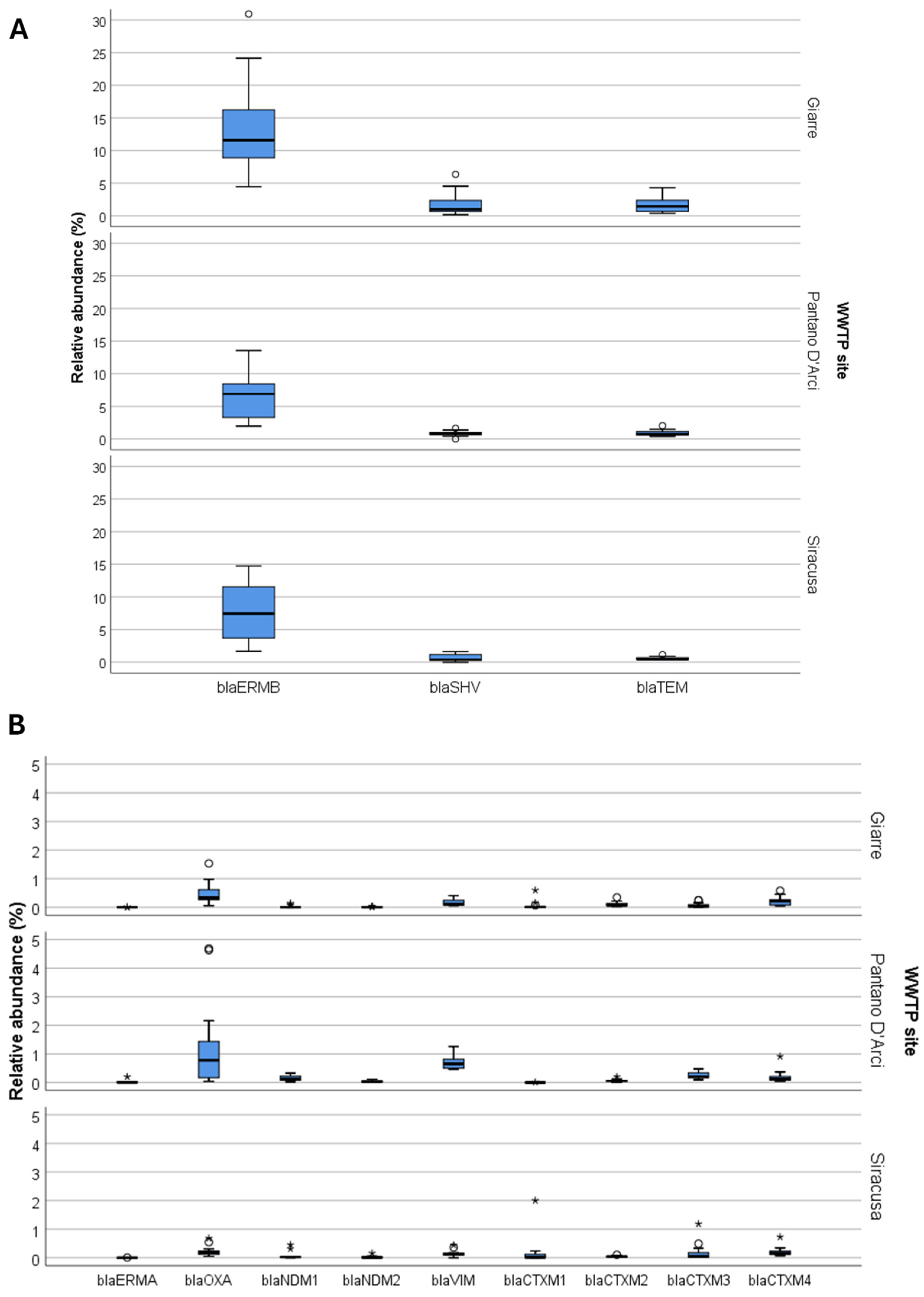
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| ARG | Antibiotic resistance gene |
| WWTP | Wastewater treatment plants |
| WBE | Wastewater-based epidemiology |
References
- O’NEILL, C.B.J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; HM Government: London, UK, 2016.
- World Health Organization, WHO. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: Early Implementation 2020. 2021. Available online: https://www.who.int/publications/i/item/9789240005587 (accessed on 23 January 2025).
- Chau, K.K.; Barker, L.; Budgell, E.P.; Vihta, K.D.; Sims, N.; Kasprzyk-Hordern, B.; Harriss, E.; Crook, D.W.; Read, D.S.; Walker, A.S.; et al. Systematic review of wastewater surveillance of antimicrobial resistance in human populations. Environ. Int. 2022, 162, 107171. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mahfouz, N.; Caucci, S.; Achatz, E.; Semmler, T.; Guenther, S.; Berendonk, T.U.; Schroeder, M. High genomic diversity of multi-drug resistant wastewater Escherichia coli. Sci. Rep. 2018, 8, 8928. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Choi, P.M.; Tscharke, B.J.; Donner, E.; O’Brien, J.W.; Grant, S.C.; Kaserzon, S.L.; Mackie, R.; O’Malley, E.; Crosbie, N.D.; Thomas, K.V.; et al. Wastewater-based epidemiology biomarkers: Past, present and future. TrAC Trends Anal. Chem. 2018, 105, 453–469. [Google Scholar] [CrossRef]
- Gholipour, S.; Shamsizadeh, Z.; Halabowski, D.; Gwenzi, W.; Nikaeen, M. Combating antibiotic resistance using wastewater surveillance: Significance, applications, challenges, and future directions. Sci. Total Environ. 2024, 908, 168056. [Google Scholar] [CrossRef] [PubMed]
- Keenum, I.; Liguori, K.; Calarco, J.; Davis, B.C.; Milligan, E.; Harwood, V.J.; Pruden, A. A framework for standardized qPCR-targets and protocols for quantifying antibiotic resistance in surface water, recycled water and wastewater. Crit. Rev. Environ. Sci. Technol. 2022, 52, 4395–4419. [Google Scholar] [CrossRef]
- Prieto Riquelme, M.V.; Garner, E.; Gupta, S.; Metch, J.; Zhu, N.; Blair, M.F.; Arango-Argoty, G.; Maile-Moskowitz, A.; Li, A.D.; Flach, C.F.; et al. Demonstrating a Comprehensive Wastewater-Based Surveillance Approach That Differentiates Globally Sourced Resistomes. Environ. Sci. Technol. 2022, 56, 14982–14993. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- McArthur Andrew, G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad Marisa, A.; Baylay Alison, J.; Bhullar, K.; Canova Marc, J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Husna, A.; Rahman, M.M.; Badruzzaman, A.T.M.; Sikder, M.H.; Islam, M.R.; Rahman, M.T.; Alam, J.; Ashour, H.M. Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities. Biomedicines 2023, 11, 2937. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lund, D.; Kieffer, N.; Parras-Moltó, M.; Ebmeyer, S.; Berglund, F.; Johnning, A.; Larsson, D.G.J.; Kristiansson, E. Large-scale characterization of the macrolide resistome reveals high diversity and several new pathogen-associated genes. Microb. Genom. 2022, 8, 000770. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meng, M.; Li, Y.; Yao, H. Plasmid-Mediated Transfer of Antibiotic Resistance Genes in Soil. Antibiotics 2022, 11, 525. [Google Scholar] [CrossRef]
- Stokes, H.W.; Gillings, M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011, 35, 790–819. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- La Rosa, G.; Bonadonna, L.; Suffredini, E.; Istituto Superiore di Sanità. Protocollo Della Sorveglianza di SARS-CoV-2 in Reflui urbani (SARI)—Rev. 3. Available online: https://zenodo.org/records/5758725 (accessed on 23 January 2025).
- Environmental Protection Agency—United States. NPDES Compliance Inspection Manual. Available online: https://www.epa.gov/sites/default/files/2017-03/documents/npdesinspect-chapter-05.pdf (accessed on 23 January 2025).
- TaqPath™ qPCR Master Mix, CG. USER GUIDE For Two-Step RT-PCR in Gene Expression Experiments or Quantitative Analysis. Applied Biosystems. Thermofisher Scientific. Available online: https://assets.fishersci.com/TFS-Assets/LSG/manuals/TaqPath_qPCR_MasterMixCG_man.pdf (accessed on 23 January 2025).
- Rawat, D.; Nair, D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010, 2, 263–274. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Poirel, L.; Dortet, L.; Bernabeu, S.; Nordmann, P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 2011, 55, 5403–5407. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumari, M.; Verma, S.; Venkatesh, V.; Gupta, P.; Tripathi, P.; Agarwal, A.; Siddiqui, S.S.; Arshad, Z.; Prakash, V. Emergence of blaNDM-1 and blaVIM producing Gram-negative bacilli in ventilator-associated pneumonia at AMR Surveillance Regional Reference Laboratory in India. PLoS ONE 2021, 16, e0256308. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Effendi, M.H.; Hartadi, E.B.; Witaningrum, A.M.; Permatasari, D.A.; Ugbo, E.N. Molecular identification of blaTEM gene of extended-spectrum beta-lactamase-producing Escherichia coli from healthy pigs in Malang district, East Java, Indonesia. J. Adv. Vet. Anim. Res. 2022, 9, 447–452. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bialvaei, A.Z.; Kafil, H.S.; Asgharzadeh, M.; Aghazadeh, M.; Yousefi, M. CTX-M extended-spectrum β-lactamase-producing Klebsiella spp, Salmonella spp, Shigella spp and Escherichia coli isolates in Iranian hospitals. Braz. J. Microbiol. 2016, 47, 706–711. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Saribas, Z.; Tunckanat, F.; Pinar, A. Prevalence of erm genes encoding macrolide-lincosamide-streptogramin (MLS) resistance among clinical isolates of Staphylococcus aureus in a Turkish university hospital. Clin. Microbiol. Infect. 2006, 12, 797–799. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, M.; Laird, E.; Gentry, T.J.; Brooks, J.P.; Karthikeyan, R. Increased Antimicrobial and Multidrug Resistance Downstream of Wastewater Treatment Plants in an Urban Watershed. Front. Microbiol. 2021, 12, 657353. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Teixeira, A.M.; Vaz-Moreira, I.; Calderón-Franco, D.; Weissbrodt, D.; Purkrtova, S.; Gajdos, S.; Dottorini, G.; Nielsen, P.H.; Khalifa, L.; Cytryn, E.; et al. Candidate biomarkers of antibiotic resistance for the monitoring of wastewater and the downstream environment. Water Res. 2023, 247, 120761. [Google Scholar] [CrossRef] [PubMed]
- Waśko, I.; Kozińska, A.; Kotlarska, E.; Baraniak, A. Clinically Relevant β-Lactam Resistance Genes in Wastewater Treatment Plants. Int. J. Environ. Res. Public Health 2022, 19, 13829. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cheney, C.; Johnson, J.D.; Ste Marie, J.P.; Gacosta, K.Y.M.; Denlinger Drumm, N.B.; Jones, G.D.; Waite-Cusic, J.; Navab-Daneshmand, T. Resolved genomes of wastewater ESBL-producing Escherichia coli and metagenomic analysis of source wastewater samples. Microbiol. Spectr. 2024, 12, e0071724. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Coyte, K.Z.; Stevenson, C.; Knight, C.G.; Harrison, E.; Hall, J.P.J.; Brockhurst, M.A. Horizontal gene transfer and ecological interactions jointly control microbiome stability. PLoS Biol. 2022, 20, e3001847. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Jeannot, K.; Xiao, Y.; Knapp, C.W. Editorial: Horizontal Gene Transfer Mediated Bacterial Antibiotic Resistance. Front. Microbiol. 2019, 10, 1933. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Martinez, E.P.; Cepeda, M.; Jovanoska, M.; Bramer, W.M.; Schoufour, J.; Glisic, M.; Verbon, A.; Franco, O.H. Seasonality of antimicrobial resistance rates in respiratory bacteria: A systematic review and meta-analysis. PLoS ONE 2019, 14, e0221133. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Brown, C.; Bürgmann, H.; Larsson, D.G.J.; Nambi, I.; Zhang, T.; Flach, C.-F.; Pruden, A.; Vikesland, P.J. Long-read metagenomic sequencing reveals shifts in associations of antibiotic resistance genes with mobile genetic elements from sewage to activated sludge. Microbiome 2022, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Robins, K.; Leonard, A.F.C.; Farkas, K.; Graham, D.W.; Jones, D.L.; Kasprzyk-Hordern, B.; Bunce, J.T.; Grimsley, J.M.S.; Wade, M.J.; Zealand, A.M.; et al. Research needs for optimising wastewater-based epidemiology monitoring for public health protection. J. Water Health 2022, 20, 1284–1313. [Google Scholar] [CrossRef] [PubMed]
- Barancheshme, F.; Munir, M. Strategies to Combat Antibiotic Resistance in the Wastewater Treatment Plants. Front. Microbiol. 2017, 8, 2603. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magnano San Lio, R.; Maugeri, A.; Barchitta, M.; Favara, G.; La Rosa, M.C.; La Mastra, C.; Agodi, A. Monitoring Antibiotic Resistance in Wastewater: Findings from Three Treatment Plants in Sicily, Italy. Int. J. Environ. Res. Public Health 2025, 22, 351. https://doi.org/10.3390/ijerph22030351
Magnano San Lio R, Maugeri A, Barchitta M, Favara G, La Rosa MC, La Mastra C, Agodi A. Monitoring Antibiotic Resistance in Wastewater: Findings from Three Treatment Plants in Sicily, Italy. International Journal of Environmental Research and Public Health. 2025; 22(3):351. https://doi.org/10.3390/ijerph22030351
Chicago/Turabian StyleMagnano San Lio, Roberta, Andrea Maugeri, Martina Barchitta, Giuliana Favara, Maria Clara La Rosa, Claudia La Mastra, and Antonella Agodi. 2025. "Monitoring Antibiotic Resistance in Wastewater: Findings from Three Treatment Plants in Sicily, Italy" International Journal of Environmental Research and Public Health 22, no. 3: 351. https://doi.org/10.3390/ijerph22030351
APA StyleMagnano San Lio, R., Maugeri, A., Barchitta, M., Favara, G., La Rosa, M. C., La Mastra, C., & Agodi, A. (2025). Monitoring Antibiotic Resistance in Wastewater: Findings from Three Treatment Plants in Sicily, Italy. International Journal of Environmental Research and Public Health, 22(3), 351. https://doi.org/10.3390/ijerph22030351







