Prevalence of Antimicrobial Resistant Escherichia coli from Sinking Creek in Northeast Tennessee
Abstract
:1. Introduction
2. Materials and Methods
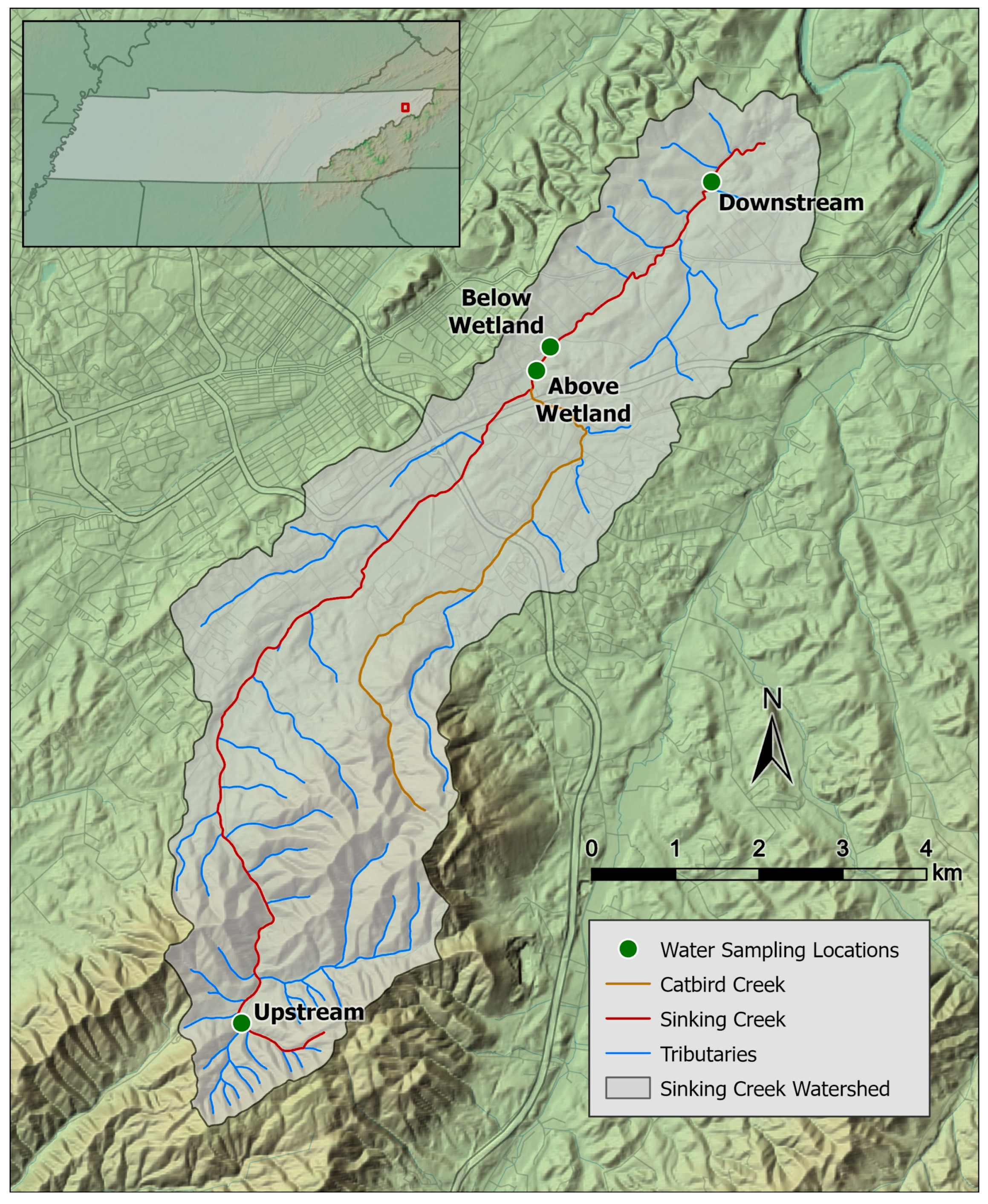
2.1. Study Area and Sample Collection
2.2. Microbiological Analyses
2.2.1. E. coli Isolation
2.2.2. Antibiotic Susceptibility Testing
2.2.3. Polymyxin (Colistin) Susceptibility Testing
2.3. Data Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO (World Health Organization). Antibiotic Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 11 August 2024).
- EclinicalMedicine. Antimicrobial resistance: A top ten global public health threat. eClinicalMedicine 2021, 41, 101221. [Google Scholar] [CrossRef] [PubMed]
- Acar, J.F.; Moulin, G. Antimicrobial resistance: A complex issue. Rev. Sci. Tech. 2012, 31, 23–31. [Google Scholar] [CrossRef]
- Wellington, E.M.; Boxall, A.B.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Kusi, J.; Ojewole, C.O.; Ojewole, A.E.; Nwi-Mozu, I. Antimicrobial Resistance Development Pathways in Surface Waters and Public Health Implications. Antibiotics 2022, 11, 821. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Kaur, R.; Verma, S.; Singh, S. Antimicrobials and Antibiotic Resistance Genes in Water Bodies: Pollution, Risk, and Control. Front. Environ. Sci. 2022, 10, 830861. [Google Scholar] [CrossRef]
- Cho, S.; Jackson, C.R.; Frye, J.G. Freshwater environment as a reservoir of extended-spectrum β-lactamase-producing Enterobacteriaceae. J. Appl. Microbiol. 2023, 134, lxad034. [Google Scholar] [CrossRef]
- Bashir, I.; Lone, F.A.; Bhat, R.A.; Mir, S.A.; Dar, Z.A.; Dar, S.A. Concerns and Threats of Contamination on Aquatic Ecosystems. In Bioremediation and Biotechnology; Springer Nature: Cham, Switzerland, 2020; pp. 1–26. [Google Scholar]
- Nappier, S.P.; Liguori, K.; Ichida, A.M.; Stewart, J.R.; Jones, K.R. Antibiotic Resistance in Recreational Waters: State of the Science. Int. J. Environ. Res. Public Health 2020, 17, 8034. [Google Scholar] [CrossRef]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef]
- Pereira, A.; Santos, A.; Tacão, M.; Alves, A.; Henriques, I.; Correia, A. Genetic diversity and antimicrobial resistance of Escherichia coli from Tagus estuary (Portugal). Sci. Total Environ. 2013, 461–462, 65–71. [Google Scholar] [CrossRef]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-Resistance Genes in Waste Water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef]
- Baquero, F.; Martínez, J.L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Pruden, A.; Virta, M.; Zhang, T. Editorial: Antibiotic Resistance in Aquatic Systems. Front. Microbiol. 2017, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.F.C.; Zhang, L.; Balfour, A.J.; Garside, R.; Hawkey, P.M.; Murray, A.K.; Ukoumunne, O.C.; Gaze, W.H. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: Environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey). Environ. Int. 2018, 114, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef]
- Partridge, S.R. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 2011, 35, 820–855. [Google Scholar] [CrossRef]
- Bennett, P.M. Plasmid encoded antibiotic resistance: Acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 2008, 153 (Suppl. S1), S347–S357. [Google Scholar] [CrossRef]
- Huddleston, J.R. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug Resist. 2014, 7, 167–176. [Google Scholar] [CrossRef]
- Ishii, S.; Ksoll, W.B.; Hicks, R.E.; Sadowsky, M.J. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 2006, 72, 612–621. [Google Scholar] [CrossRef]
- Berthe, T.; Ratajczak, M.; Clermont, O.; Denamur, E.; Petit, F. Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl. Environ. Microbiol. 2013, 79, 4684–4693. [Google Scholar] [CrossRef]
- Djordjevic, S.P.; Stokes, H.W.; Roy Chowdhury, P. Mobile elements, zoonotic pathogens and commensal bacteria: Conduits for the delivery of resistance genes into humans, production animals and soil microbiota. Front. Microbiol. 2013, 4, 86. [Google Scholar] [CrossRef]
- Harwood, V.J.; Staley, C.; Badgley, B.D.; Borges, K.; Korajkic, A. Microbial source tracking markers for detection of fecal contamination in environmental waters: Relationships between pathogens and human health outcomes. FEMS Microbiol. Rev. 2014, 38, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Hiott, L.M.; Barrett, J.B.; McMillan, E.A.; House, S.L.; Humayoun, S.B.; Adams, E.S.; Jackson, C.R.; Frye, J.G. Prevalence and characterization of Escherichia coli isolated from the Upper Oconee Watershed in Northeast Georgia. PLoS ONE 2018, 13, e0197005. [Google Scholar] [CrossRef] [PubMed]
- Amato, H.K.; Wong, N.M.; Pelc, C.; Taylor, K.; Price, L.B.; Altabet, M.; Jordan, T.E.; Graham, J.P. Effects of concentrated poultry operations and cropland manure application on antibiotic resistant Escherichia coli and nutrient pollution in Chesapeake Bay watersheds. Sci. Total Environ. 2020, 735, 139401. [Google Scholar] [CrossRef]
- Haberecht, H.B.; Nealon, N.J.; Gilliland, J.R.; Holder, A.V.; Runyan, C.; Oppel, R.C.; Ibrahim, H.M.; Mueller, L.; Schrupp, F.; Vilchez, S.; et al. Antimicrobial-Resistant Escherichia coli from Environmental Waters in Northern Colorado. J. Environ. Public Health 2019, 2019, 3862949. [Google Scholar] [CrossRef] [PubMed]
- Givens, C.E.; Kolpin, D.W.; Hubbard, L.E.; Meppelink, S.M.; Cwiertny, D.M.; Thompson, D.A.; Lane, R.F.; Wilson, M.C. Simultaneous stream assessment of antibiotics, bacteria, antibiotic resistant bacteria, and antibiotic resistance genes in an agricultural region of the United States. Sci. Total Environ. 2023, 904, 166753. [Google Scholar] [CrossRef]
- Fincher, L.M.; Parker, C.D.; Chauret, C.P. Occurrence and Antibiotic Resistance of Escherichia coli O157:H7 in a Watershed in North-Central Indiana. J. Environ. Qual. 2009, 38, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, D.C.; Chacón, L.M.; Wallace, D.J. Anthropogenic Activities and the Problem of Antibiotic Resistance in Latin America: A Water Issue. Water 2021, 13, 2693. [Google Scholar] [CrossRef]
- Bong, C.W.; Low, K.Y.; Chai, L.C.; Lee, C.W. Prevalence and Diversity of Antibiotic Resistant Escherichia coli from Anthropogenic-Impacted Larut River. Front. Public Health 2022, 10, 794513. [Google Scholar] [CrossRef]
- Truong, T.; Hoang, T.L.; Tran, L.T.; Pham, T.P.T.; Le, T.-H. Prevalence of Antibiotic Resistance Genes in the Saigon River Impacted by Anthropogenic Activities. Water 2021, 13, 2234. [Google Scholar] [CrossRef]
- Gilfillan, D.; Joyner, T.A.; Scheuerman, P. Maxent estimation of aquatic Escherichia coli stream impairment. PeerJ 2018, 6, e5610. [Google Scholar] [CrossRef]
- Hall, K.K.; Evanshen, B.G.; Maier, K.J.; Scheuerman, P.R. Application of multivariate statistical methodology to model factors influencing fate and transport of fecal pollution in surface waters. J. Environ. Qual. 2014, 43, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Hall, K.K.; Scheuerman, P.R. Development of Multiple Regression Models to Predict Sources of Fecal Pollution. Water Environ. Res. 2017, 89, 1961–1969. [Google Scholar] [CrossRef] [PubMed]
- Gilfillan, D.; Hall, K.; Joyner, T.A.; Scheuerman, P. Canonical Variable Selection for Ecological Modeling of Fecal Indicators. J. Environ. Qual. 2018, 47, 974–984. [Google Scholar] [CrossRef] [PubMed]
- McCurdy, P.; Luffman, I.; Joyner, T.A.; Maier, K. Storm sampling to assess inclement weather impacts on water quality in a karst watershed: Sinking Creek, Watauga watershed, East Tennessee. J. Environ. Qual. 2021, 50, 429–440. [Google Scholar] [CrossRef] [PubMed]
- TDEC (Tennessee Department of Environment and Conservation). Water Quality Rules, Reports & Publications. Available online: https://www.tn.gov/environment/program-areas/wr-water-resources/water-quality/water-quality-reports---publications.html (accessed on 29 July 2024).
- USEPA. Method 1603 Escherichia coli (E. coli) in Water by Membrane Filtration Using Modified Membrane-Thermotolerant Escherichia coli Agar (Modified m-TEC), EPA 821-R-02-023; Environmental Protection Agency, Office of Research and Development: Cincinnati, OH, USA, 2002. Available online: https://nepis.epa.gov/Exe/ZyNET.exe/P1008MZV.txt?ZyActionD=ZyDocument&Client=EPA&Index=2000%20Thru%202005&Docs=&Query=&Time=&EndTime=&SearchMethod=1&TocRestrict=n&Toc=&TocEntry=&QField=&QFieldYear=&QFieldMonth=&QFieldDay=&UseQField=&IntQFieldOp=0&ExtQFie (accessed on 10 March 2024).
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Coleman, B.L.; Salvadori, M.I.; McGeer, A.J.; Sibley, K.A.; Neumann, N.F.; Bondy, S.J.; Gutmanis, I.A.; McEwen, S.A.; Lavoie, M.; Strong, D.; et al. The role of drinking water in the transmission of antimicrobial-resistant E. coli. Epidemiol. Infect. 2012, 140, 633–642. [Google Scholar] [CrossRef]
- O‘Flaherty, E.; Solimini, A.; Pantanella, F.; Cummins, E. The potential human exposure to antibiotic resistant-Escherichia coli through recreational water. Sci. Total Environ. 2019, 650, 786–795. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Dorado-García, A.; van Duijkeren, E.; van den Bunt, G.; Dierikx, C.M.; Bonten, M.J.M.; Bootsma, M.C.J.; Schmitt, H.; Hald, T.; Evers, E.G.; et al. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: A population-based modelling study. Lancet Planet. Health 2019, 3, e357–e369. [Google Scholar] [CrossRef]
- Masters, N.; Wiegand, A.; Ahmed, W.; Katouli, M. Escherichia coli virulence genes profile of surface waters as an indicator of water quality. Water Res. 2011, 45, 6321–6333. [Google Scholar] [CrossRef]
- Iverson, G.; Humphrey, C.P.; Postma, M.H.; O’Driscoll, M.A.; Manda, A.K.; Finley, A. Influence of Sewered versus Septic Systems on Watershed Exports of E. coli. Water Air Soil Pollut. 2017, 228, 237. [Google Scholar] [CrossRef]
- USEPA. Septic System Impacts on Water Sources. Available online: https://www.epa.gov/septic/septic-system-impacts-water-sources (accessed on 10 June 2024).
- CDC. All Antibiotic Classes|A.R. & Patient Safety Portal. Available online: https://arpsp.cdc.gov/profile/antibiotic-use/all-classes (accessed on 10 June 2024).
- Karlowsky James, A.; Kelly Laurie, J.; Thornsberry, C.; Jones Mark, E.; Sahm Daniel, F. Trends in Antimicrobial Resistance among Urinary Tract Infection Isolates of Escherichia coli from Female Outpatients in the United States. Antimicrob. Agents Chemother. 2002, 46, 2540–2545. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Thornsberry, C.; Jones, M.E.; Sahm, D.F. Susceptibility of Antimicrobial-Resistant Urinary Escherichia coli Isolates to Fluoroquinolones and Nitrofurantoin. Clin. Infect. Dis. 2003, 36, 183–187. [Google Scholar] [CrossRef]
- Gregory, L.F.; Karthikeyan, R.; Aitkenhead-Peterson, J.A.; Gentry, T.J.; Wagner, K.L.; Harmel, R.D. Nutrient loading impacts on culturable E. coli and other heterotrophic bacteria fate in simulated stream mesocosms. Water Res. 2017, 126, 442–449. [Google Scholar] [CrossRef]
- Ibekwe, A.M.; Murinda, S.E.; Graves, A.K. Genetic diversity and antimicrobial resistance of Escherichia coli from human and animal sources uncovers multiple resistances from human sources. PLoS ONE 2011, 6, e20819. [Google Scholar] [CrossRef] [PubMed]
- Kappell, A.D.; DeNies, M.S.; Ahuja, N.H.; Ledeboer, N.A.; Newton, R.J.; Hristova, K.R. Detection of multi-drug resistant Escherichia coli in the urban waterways of Milwaukee, WI. Front. Microbiol. 2015, 6, 336. [Google Scholar] [CrossRef]
- Webster, L.F.; Thompson, B.C.; Fulton, M.H.; Chestnut, D.E.; Van Dolah, R.F.; Leight, A.K.; Scott, G.I. Identification of sources of Escherichia coli in South Carolina estuaries using antibiotic resistance analysis. J. Exp. Mar. Biol. Ecol. 2004, 298, 179–195. [Google Scholar] [CrossRef]
- Odonkor, S.T.; Addo, K.K. Prevalence of Multidrug-Resistant Escherichia coli Isolated from Drinking Water Sources. Int. J. Microbiol. 2018, 2018, 7204013. [Google Scholar] [CrossRef]
- Thornton, C.N.; Tanner, W.D.; VanDerslice, J.A.; Brazelton, W.J. Localized effect of treated wastewater effluent on the resistome of an urban watershed. GigaScience 2020, 9, giaa125. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, A.S.; Mikolajczyk, F.N.; Fisher, J.C. Antimicrobial Resistance Linked to Septic System Contamination in the Indiana Lake Michigan Watershed. Antibiotics 2023, 12, 569. [Google Scholar] [CrossRef]
- Tabut, P.; Yongyod, R.; Ungcharoen, R.; Kerdsin, A. The Distribution of Mobile Colistin-Resistant Genes, Carbapenemase-Encoding Genes, and Fluoroquinolone-Resistant Genes in Escherichia coli Isolated from Natural Water Sources in Upper Northeast Thailand. Antibiotics 2022, 11, 1760. [Google Scholar] [CrossRef]
- Snyman, Y.; Whitelaw, A.C.; Barnes, J.M.; Maloba, M.R.B.; Newton-Foot, M. Characterisation of mobile colistin resistance genes (mcr-3 and mcr-5) in river and storm water in regions of the Western Cape of South Africa. Antimicrob. Resist. Infect. Control 2021, 10, 96. [Google Scholar] [CrossRef]
- Meinersmann, R.J.; Ladely, S.R.; Plumblee, J.R.; Cook, K.L.; Thacker, E. Prevalence of mcr-1 in the Cecal Contents of Food Animals in the United States. Antimicrob. Agents Chemother. 2017, 61, e02244-16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hou, N.; Johnston, J.; Sarreal, C.; Jarosh, J.; Hughes, A.C.; Gu, Y.; He, X. Low prevalence of mobile colistin-resistance in U.S. meat, catfish, poultry and genomic characterization of a mcr-1 positive Escherichia coli strain. Food Control 2020, 118, 107434. [Google Scholar] [CrossRef]
- Henig, O.; Rojas, L.J.; Bachman, M.A.; Rudin, S.D.; Brennan, B.M.; Soehnlen, M.K.; Jones, K.L.; Mills, J.P.; Dombecki, C.R.; Valyko, A.M.; et al. Identification of four patients with colistin-resistant Escherichia coli containing the mobile colistin resistance mcr-1 gene from a single health system in Michigan. Infect. Control Hosp. Epidemiol. 2019, 40, 1059–1062. [Google Scholar] [CrossRef] [PubMed]
- Monte, D.F.; Nelson, V.; Cerdeira, L.; Keelara, S.; Greene, S.; Griffin, D.; Rath, S.; Hall, R.; Page, N.; Lawson, T.; et al. Multidrug- and colistin-resistant Salmonella enterica 4,[5],12:i:- sequence type 34 carrying the mcr-3.1 gene on the IncHI2 plasmid recovered from a human. J. Med. Microbiol. 2019, 68, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Gelalcha, B.D.; Mohammed, R.I.; Gelgie, A.E.; Kerro Dego, O. Molecular epidemiology and pathogenomics of extended-spectrum beta-lactamase producing- Escherichia coli and - Klebsiella pneumoniae isolates from bulk tank milk in Tennessee, USA. Front. Microbiol. 2023, 14, 1283165. [Google Scholar] [CrossRef]
- Mondal, A.H.; Khare, K.; Saxena, P.; Debnath, P.; Mukhopadhyay, K.; Yadav, D. A Review on Colistin Resistance: An Antibiotic of Last Resort. Microorganisms 2024, 12, 772. [Google Scholar] [CrossRef]

| Drug Class/Antibiotic 1 | Concentration (μg) | Zones of Inhibition Diameter in mm Interpretation 2 | ||
|---|---|---|---|---|
| S | I | R | ||
| Sulfonamides | ||||
| Trimethoprim & Sulfamethoxazole | 1.25/23.75 | ≥16 | 45,245 | ≤10 |
| Penicillin β-lactamase inhibitor combinations | ||||
| Amoxicillin/ Clavulanic acid | 20/10 | ≥18 | 14–17 | ≤13 |
| Tetracyclines | ||||
| Tetracycline | 30 | ≥15 | 45,274 | ≤11 |
| Cephalosporins | ||||
| Ceftriaxone | 30 | ≥23 | 20–22 | ≤19 |
| Fluoroquinolone | ||||
| Ciprofloxacin | 5 | ≥31 | 21–30 | ≤20 |
| Urinary anti-infectives | ||||
| Nitrofurantoin | 300 | ≥17 | 15–16 | ≤14 |
| Phosphonic antibiotics | ||||
| Fosfomycin | 200 | ≥16 | 13–15 | ≤12 |
| No. (%) of Isolates | ||||||
|---|---|---|---|---|---|---|
| Antibiotics/Sampling Site | Upstream (n = 63) | Above Wetland (n = 43) | Below Wetland (n = 29) | Downstream (n = 42) | p-Value 1 | Overall Prevalence (n = 177) |
| Ceftriaxone | 23 (36.5) | 23 (53.5) | 16 (55.2) | 9 (21.4) | 0.005 | 71 (40.1) |
| Tetracycline | 17 (27.0) | 14 (32.6) | 12 (41.4) | 11 (26.2) | 0.482 | 54 (30.5) |
| Ciprofloxacin | 43 (68.3) | 28 (65.1) | 15 (51.7) | 27 (64.3) | 0.488 | 113 (63.8) |
| Amoxicillin/ Clavulanic acid | 18 (28.6) | 19 (44.2) | 10 (34.5) | 5 (11.9) | 0.008 | 52 (29.4) |
| Fosfomycin | 12 (19.1) | 9 (20.9) | 2 (6.9) | 2 (4.8) | 0.064 | 25 (14.1) |
| Trimethoprim-Sulfamethoxazole | 14 (22.2) | 12 (27.9) | 8 (27.6) | 4 (9.5) | 0.126 | 38 (21.5) |
| Nitrofurantoin | 34 (54.0) | 30 (69.8) | 20 (69.0) | 27 (64.3) | 0.333 | 111 (62.7) |
| Colistin 2 | 1 (7.1) | 0 (0) | 2 (18.1) | 0 (0.0) | 0.243 | 3 (6.0) |
| Resistant Phenotype 1 | Frequency | Percentage |
|---|---|---|
| No resistance | 30 | 17.00 |
| Resistant to only one antibiotic | 31 | 17.50 |
| CIP_NIF | 20 | 13.61 |
| CIP | 13 | 8.84 |
| NIF_ | 11 | 7.48 |
| CRO_CIP_NIF | 10 | 6.80 |
| CRO_TET_CIP_NIF | 10 | 6.80 |
| CRO_TET_CIP_AMC_FOS_SXT_NIF | 9 | 6.12 |
| COL | 3 | 6.00 |
| TET_CIP | 6 | 4.08 |
| CRO_CIP_AMC_NIF | 5 | 3.40 |
| CRO_TET_CIP_AMC_NIF | 5 | 3.40 |
| CRO_CIP_AMC_FOS_SXT_NIF | 4 | 2.72 |
| CRO_CIP_AMC_SXT_NIF | 4 | 2.72 |
| CRO_TET_CIP_SXT_NIF | 4 | 2.72 |
| TET | 4 | 2.72 |
| CRO_AMC_FOS_SXT_NIF | 3 | 2.04 |
| CRO_TET_CIP_AMC_SXT_NIF | 3 | 2.04 |
| CIP_AMC_NIF | 2 | 1.36 |
| CIP_SXT | 2 | 1.36 |
| CRO_CIP | 2 | 1.36 |
| CRO_TET_AMC_SXT_NIF | 2 | 1.36 |
| TET_CIP_NIF | 2 | 1.36 |
| AMC | 1 | 0.68 |
| AMC_NIF | 1 | 0.68 |
| AMC_NIF_COL | 1 | 0.68 |
| CIP_AMC | 1 | 0.68 |
| CIP_AMC_FOS_SXT_NIF | 1 | 0.68 |
| CIP_AMC_SXT_NIF | 1 | 0.68 |
| CRO_AMC_NIF | 1 | 0.68 |
| CRO_AMC_SXT_NIF | 1 | 0.68 |
| CRO_CIP_AMC_FOS_NIF | 1 | 0.68 |
| CRO_CIP_FOS_NIF | 1 | 0.68 |
| CRO_CIP_FOS_SXT_NIF | 1 | 0.68 |
| CRO_FOS_NIF | 1 | 0.68 |
| CRO_TET_AMC_FOS_SXT_NIF | 1 | 0.68 |
| CRO_TET_CIP | 1 | 0.68 |
| CRO_TET_CIP_AMC_FOS_NIF | 1 | 0.68 |
| CRO_TET_SXT_NIF | 1 | 0.68 |
| FOS_NIF | 1 | 0.68 |
| TET_AMC_NIF | 1 | 0.68 |
| TET_AMC_NIF_COL | 1 | 0.68 |
| TET_CIP_AMC | 1 | 0.68 |
| TET_CIP_AMC_FOS | 1 | 0.68 |
| TET_CIP_COL | 1 | 0.68 |
| TET_CIP_SXT_NIF | 1 | 0.68 |
| Sampling Site (No. Isolates) | MDR No. (%) | Top Most Frequent MDR (>5%) Phenotype and % | Highest MDR Profile and % |
|---|---|---|---|
| Upstream (n = 63) | 27 (42.9) | CRO_TET_CIP_AMC_FOS_SXT_NIF (5.7%) CRO_CIP_AMC_FOS_SXT_NIF (5.7%) CRO_AMC_FOS_SXT_NIF (5.7%) CRO_CIP_NIF (5.7%) | CRO_TET_CIP_AMC_FOS_SXT_NIF (5.7%) |
| Above Wetland (n = 43) | 26 (60.7) | CRO_CIP_AMC_FOS_SXT_NIF (16.7%) | CRO_CIP_AMC_FOS_SXT_NIF (16.7%) |
| Below Wetland (n = 29) | 15 (51.7) | CRO_TET_CIP_NIF (18.8%) | CRO_TET_CIP_AMC_SXT_NIF (9.1%) |
| Downstream (n = 42) | 15 (35.7) | CRO_CIP_NIF (17.9%) | CRO_CIP_AMC_FOS_SXT_NIF (2.8%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alali, W.Q.; Scheuerman, P.; McClure, C.; Ghimire, A.; Owusu-Mensah, P.; Schultz, J.; Joyner, T.A. Prevalence of Antimicrobial Resistant Escherichia coli from Sinking Creek in Northeast Tennessee. Int. J. Environ. Res. Public Health 2024, 21, 1285. https://doi.org/10.3390/ijerph21101285
Alali WQ, Scheuerman P, McClure C, Ghimire A, Owusu-Mensah P, Schultz J, Joyner TA. Prevalence of Antimicrobial Resistant Escherichia coli from Sinking Creek in Northeast Tennessee. International Journal of Environmental Research and Public Health. 2024; 21(10):1285. https://doi.org/10.3390/ijerph21101285
Chicago/Turabian StyleAlali, Walid Q., Phillip Scheuerman, Clara McClure, Achala Ghimire, Priscilla Owusu-Mensah, Jacob Schultz, and Timothy Andrew Joyner. 2024. "Prevalence of Antimicrobial Resistant Escherichia coli from Sinking Creek in Northeast Tennessee" International Journal of Environmental Research and Public Health 21, no. 10: 1285. https://doi.org/10.3390/ijerph21101285
APA StyleAlali, W. Q., Scheuerman, P., McClure, C., Ghimire, A., Owusu-Mensah, P., Schultz, J., & Joyner, T. A. (2024). Prevalence of Antimicrobial Resistant Escherichia coli from Sinking Creek in Northeast Tennessee. International Journal of Environmental Research and Public Health, 21(10), 1285. https://doi.org/10.3390/ijerph21101285






