Use of Artificial Intelligence in the Classification of Elementary Oral Lesions from Clinical Images
Abstract
1. Introduction
2. Materials and Methods
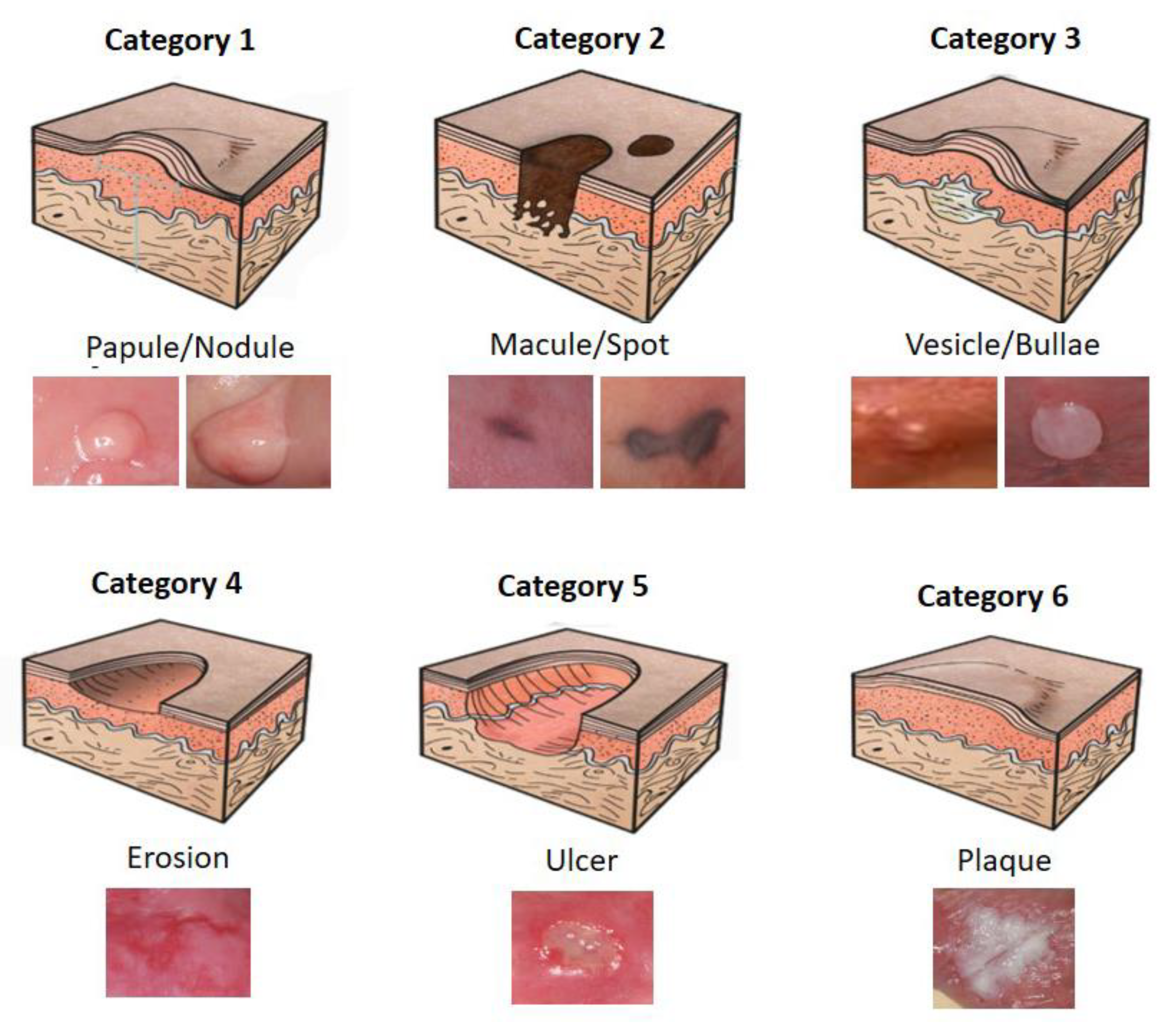
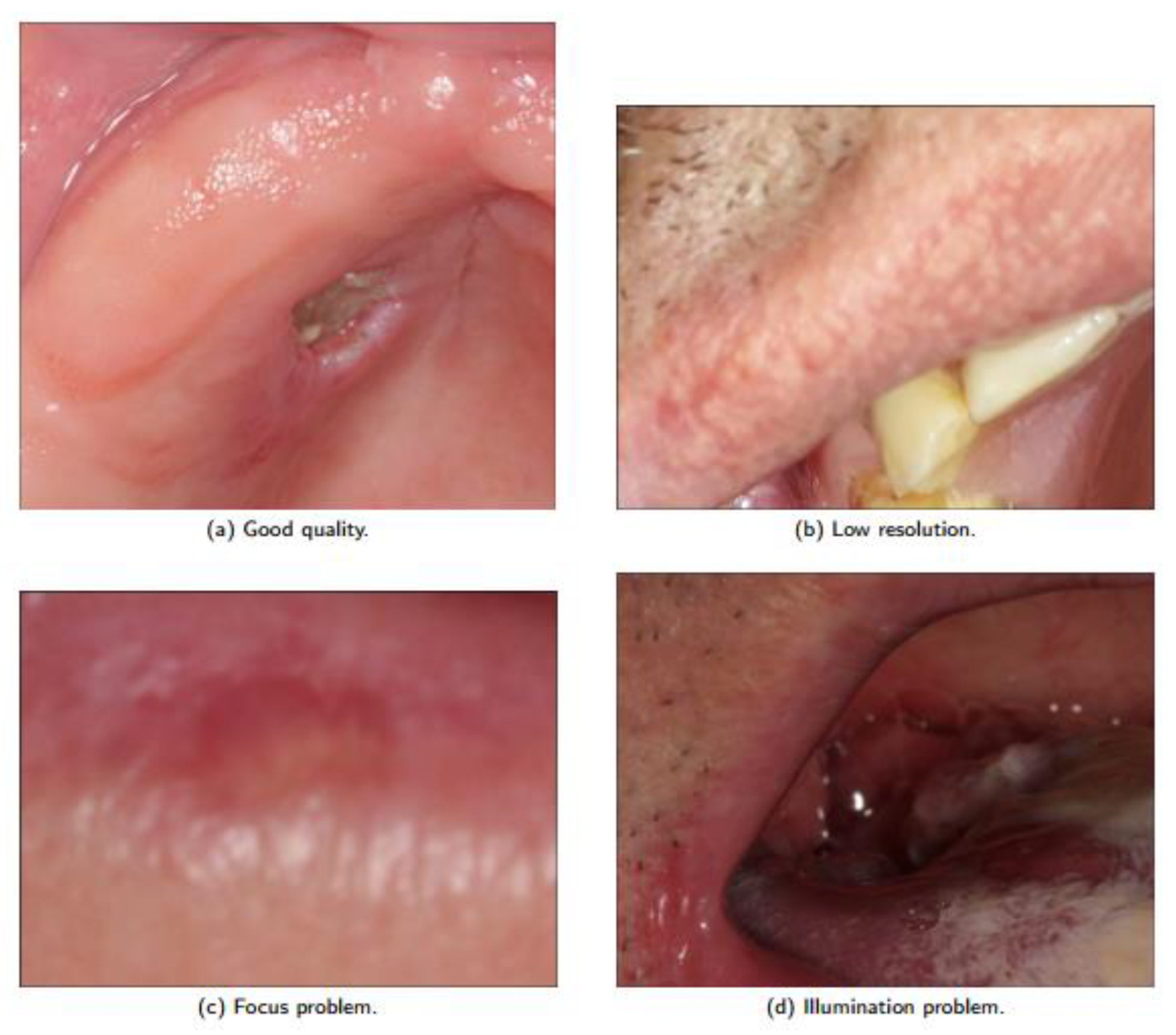
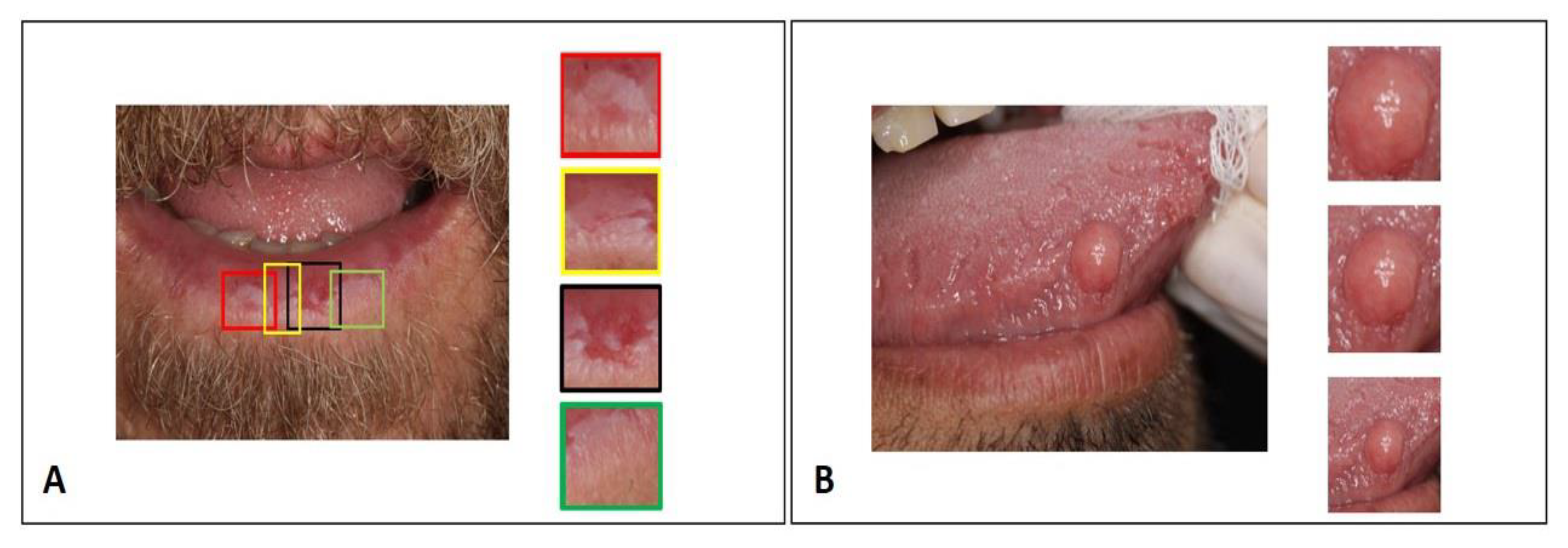
2.1. Dataset
2.2. Convolutional Neural Network
- (1)
- Initialize a model with weights: Usually recommended when the number of images is lower than 1000 per class. During the training, we keep some layers with fixed optimizing parameters in higher-level layers, once the initial layers have similar features regardless of the problem domain.
- (2)
- Retrain an existing model: Usually recommended when the number of images is larger than 1000 per class. An approach that most likely leads to overfitting may need more parameter optimization than the number of images available. Given the similarity with the dataset originally used for training, a large pretrained network can be used as a featurizer.
2.3. Experiments
2.3.1. Data Augmentation
2.3.2. CNN—Architecture Implementation
2.3.3. Hyperparameter Optimization
2.4. Evaluation Metrics
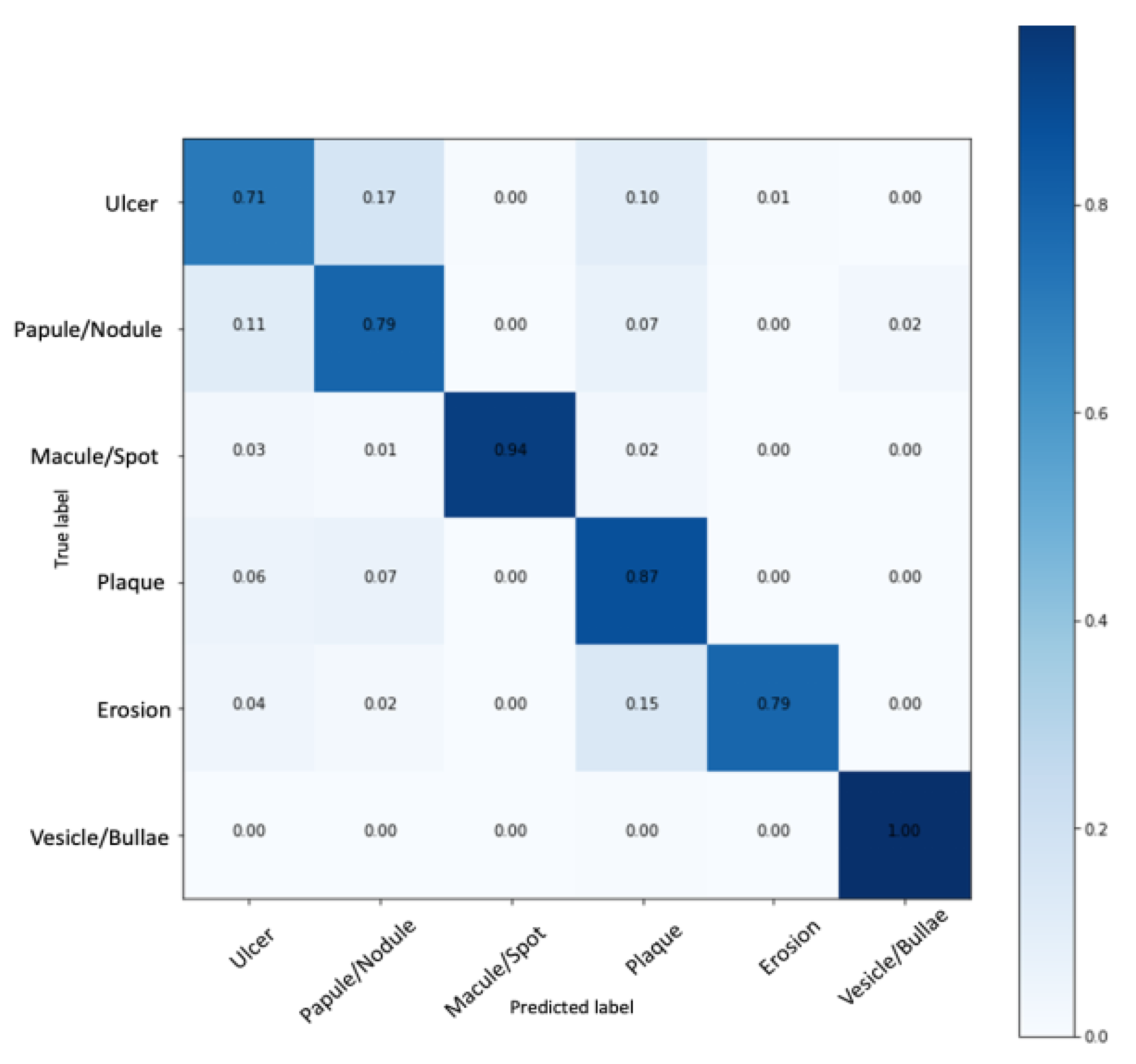
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- García-Pola, M.; Pons-Fuster, E.; Suárez-Fernández, C.; Seoane-Romero, J.; Romero-Méndez, A.; López-Jornet, P. Role of artificial intelligence in the early diagnosis of oral cancer. a scoping review. Cancers 2021, 13, 4600. [Google Scholar] [CrossRef]
- Mahmood, H.; Shaban, M.; Rajpoot, N.; Khurram, S.A. Artificial intelligence-based methods in head and neck cancer diagnosis: An overview. Br. J. Cancer 2021, 124, 1934–1940. [Google Scholar] [CrossRef]
- Khanagar, S.B.; Al-Ehaideb, A.; Maganur, P.C.; Vishwanathaiah, S.; Patil, S.; Baeshen, H.A.; Sarode, S.C.; Bhandi, S. Developments, application, and performance of artificial intelligence in dentistry—A systematic review. J. Dent. Sci. 2021, 16, 508–522. [Google Scholar] [CrossRef]
- Anantharaman, R.; Anantharaman, V.; Lee, Y. Oro vision: Deep learning for classifying orofacial diseases. In Proceedings of the 2017 IEEE International Conference on Healthcare Informatics (ICHI), Park City, UT, USA, 23–26 August 2017; pp. 39–45. [Google Scholar] [CrossRef]
- Seetharam, K.; Shrestha, S.; Sengupta, P.P. Cardiovascular imaging and intervention through the lens of artificial intelligence. Interv. Cardiol. Rev. Res. Resour. 2021, 16, e31. [Google Scholar] [CrossRef]
- Al Turk, L.; Wang, S.; Krause, P.; Wawrzynski, J.; Saleh, G.M.; Alsawadi, H.; Alshamrani, A.Z.; Peto, T.; Bastawrous, A.; Li, J.; et al. Evidence based prediction and progression monitoring on retinal images from three nations. Transl. Vis. Sci. Technol. 2020, 9, 44. [Google Scholar] [CrossRef]
- Aubreville, M.; Knipfer, C.; Oetter, N.; Jaremenko, C.; Rodner, E.; Denzler, J.; Bohr, C.; Neumann, H.; Stelzle, F.; Maier, A. Automatic classification of cancerous tissue in laserendomicroscopy images of the oral cavity using deep learning. Sci. Rep. 2017, 7, 11979. [Google Scholar] [CrossRef]
- Folmsbee, J.; Liu, X.; Brandwein-Weber, M.; Doyle, S. Active deep learning: Improved training efficiency of convolutional neural networks for tissue classification in oral cavity cancer. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; IEEE: New York, NY, USA, 2018; pp. 770–773. [Google Scholar] [CrossRef]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A survey of the recent architectures of deep convolutional neural networks. arXiv 2019, arXiv:1901.06032. [Google Scholar] [CrossRef]
- LeCun, Y.; Kavukcuoglu, K.; Farabet, C. Convolutional networks and applications in vision. In Proceedings of the ISCAS 2010–2010 IEEE International Symposium on Circuits and Systems: Nano-Bio Circuit Fabrics and Systems, Paris, France, 30 May–2 June 2010; pp. 253–256. [Google Scholar] [CrossRef]
- Kujan, O.; Glenny, A.-M.; Oliver, R.; Thakker, N.; Sloan, P. Screening programmes for the early detection and prevention of oral cancer. Cochrane Database Syst. Rev. 2006, 54, CD004150. [Google Scholar] [CrossRef]
- Almangush, A.; Alabi, R.O.; Mäkitie, A.A.; Leivo, I. Machine learning in head and neck cancer: Importance of a web-based prognostic tool for improved decision making. Oral Oncol. 2022, 124, 105452. [Google Scholar] [CrossRef]
- Bouaoud, J.; Bossi, P.; Elkabets, M.; Schmitz, S.; van Kempen, L.C.; Martinez, P.; Jagadeeshan, S.; Breuskin, I.; Puppels, G.J.; Hoffmann, C.; et al. Unmet needs and perspectives in oral cancer prevention. Cancers 2022, 14, 1815. [Google Scholar] [CrossRef]
- Rethman, M.P.; Carpenter, W.; Cohen, E.E.; Epstein, J.; Evans, C.A.; Flaitz, C.M.; Graham, F.J.; Hujoel, P.P.; Kalmar, J.R.; Koch, W.M.; et al. Evidence-based clinical recommendations regarding screening for oral squamous cell carcinomas. J. Am. Dent. Assoc. 2010, 141, 509–520. [Google Scholar] [CrossRef]
- Warin, K.; Limprasert, W.; Suebnukarn, S.; Jinaporntham, S.; Jantana, P. Performance of deep convolutional neural network for classification and detection of oral potentially malignant disorders in photographic images. Int. J. Oral Maxillofac. Surg. 2022, 51, 699–704. [Google Scholar] [CrossRef]
- Uthoff, R.D.; Song, B.; Sunny, S.; Patrick, S.; Suresh, A.; Kolur, T.; Keerthi, G.; Spires, O.; Anbarani, A.; Wilder-Smith, P.; et al. Point-of-care, smartphone-based, dual-modality, dual-view, oral cancer screening device with neural network classification for low-resource communities. PLoS ONE 2018, 13, e0207493. [Google Scholar] [CrossRef]
- Chi, A.C.; Neville, B.W.; Damm, D.D.; Allen, C.M. Oral and Maxillofacial Pathology-E-Book; Elsevier Health Sciences: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Speight, P.M.; Khurram, S.A.; Kujan, O. Oral potentially malignant disorders: Risk of progression to malignancy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 125, 612–627. [Google Scholar] [CrossRef]
- Miranda-Filho, A.; Bray, F. Global patterns and trends in cancers of the lip, tongue and mouth. Oral Oncol. 2020, 102, 104551. [Google Scholar] [CrossRef]
- Woo, S.-B. Oral epithelial dysplasia and premalignancy. Head Neck Pathol. 2019, 13, 423–439. [Google Scholar] [CrossRef]
- Fu, Q.; Chen, Y.; Li, Z.; Jing, Q.; Hu, C.; Liu, H.; Bao, J.; Hong, Y.; Shi, T.; Li, K.; et al. A deep learning algorithm for detection of oral cavity squamous cell carcinoma from photographic images: A retrospective study. EClinicalMedicine 2020, 27, 100558. [Google Scholar] [CrossRef]
- Xie, L.; Shang, Z. Burden of oral cancer in asia from 1990 to 2019: Estimates from the global burden of disease 2019 study. PLoS ONE 2022, 17, e0265950. [Google Scholar] [CrossRef]
- Ilhan, B.; Guneri, P.; Wilder-Smith, P. The contribution of artificial intelligence to reducing the diagnostic delay in oral cancer. Oral Oncol. 2021, 116, 105254. [Google Scholar] [CrossRef]
- Cleveland, J.L.; Robison, V.A. Clinical oral examinations may not be predictive of dysplasia or oral squamous cell carcinoma. J. Evid. Based Dent. Pract. 2013, 13, 151–154. [Google Scholar] [CrossRef]
- Guo, J.; Wang, H.; Xue, X.; Li, M.; Ma, Z. Real-time classification on oral ulcer images with residual network and image enhancement. IET Image Process. 2022, 16, 641–646. [Google Scholar] [CrossRef]
- Welikala, R.A.; Remagnino, P.; Lim, J.H.; Chan, C.S.; Rajendran, S.; Kallarakkal, T.G.; Zain, R.B.; Jayasinghe, R.D.; Rimal, J.; Kerr, A.R.; et al. Automated detection and classification of oral lesions using deep learning for early detection of oral cancer. IEEE Access 2020, 8, 132677–132693. [Google Scholar] [CrossRef]
- Tanriver, G.; Tekkesin, M.S.; Ergen, O. Automated detection and classification of oral lesions using deep learning to detect oral potentially malignant disorders. Cancers 2021, 13, 2766. [Google Scholar] [CrossRef]
- Gomes, R.F.T.; Schuch, L.F.; Martins, M.D.; Honório, E.F.; de Figueiredo, R.M.; Schmith, J.; Machado, G.N.; Carrard, V.C. Use of Deep Neural Networks in the Detection and Automated Classification of Lesions Using Clinical Images in Ophthalmology, Dermatology, and Oral Medicine—A Systematic Review. J. Digit. Imaging 2023. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Jurczyszyn, K.; Gedrange, T.; Kozakiewicz, M. Theoretical background to automated diagnosing of oral leukoplakia: A preliminary report. J. Healthc. Eng. 2020, 2020, 8831161. [Google Scholar] [CrossRef]
- Gupta, R.K.; Kaur, M.; Manhas, J. Tissue level based deep learning framework for early detection of dysplasia in oral squamous epithelium. J. Multimed. Inf. Syst. 2019, 6, 81–86. [Google Scholar] [CrossRef]
- Jurczyszyn, K.; Kazubowska, K.; Kubasiewicz-Ross, P.; Ziółkowski, P.; Dominiak, M. Application of fractal dimension analysis and photodynamic diagnosis in the case of differentiation between lichen planus and leukoplakia: A preliminary study. Adv. Clin. Exp. Med. 2018, 27, 1729–1736. [Google Scholar] [CrossRef]
- Thomas, B.; Kumar, V.; Saini, S. Texture analysis based segmentation and classification of oral cancer lesions in color images using ann. In Proceedings of the 2013 IEEE International Conference on Signal Processing, Computing and Control (ISPCC), Solan, India, 26–28 September 2013; pp. 1–5. [Google Scholar] [CrossRef]
- Mortazavi, H.; Safi, Y.; Baharvand, M.; Jafari, S.; Anbari, F.; Rahmani, S. Oral White Lesions: An Updated Clinical Diagnostic Decision Tree. Dent. J. 2019, 7, 15. [Google Scholar] [CrossRef]
- Mortazavi, H.; Safi, Y.; Baharvand, M.; Rahmani, S.; Jafari, S. Peripheral Exophytic Oral Lesions: A Clinical Decision Tree. Int. J. Dent. 2017, 2017, 9193831. [Google Scholar] [CrossRef]
- Mortazavi, H.; Safi, Y.; Baharvand, M.; Rahmani, S. Diagnostic features of common oral ulcerative lesions: An updated decision tree. Int. J. Dent. 2016, 2016, 7278925. [Google Scholar] [CrossRef]
- Brasil, Ministério da Saúde. Diretrizes e normas regulamentadoras de pesquisa envolvendo seres humanos. Conselho Nacional de Saúde. Brasília 2012, 150, 59–62. [Google Scholar]
- LEI Nº 10.973, DE 2 DE DEZEMBRO DE 2004—Publicação OriginalDiário Oficial da União-Seção 1-3/12/2004, Página 2. Disponível em. Available online: https://www2.camara.leg.br/legin/fed/lei/2004/lei-10973-2-dezembro-2004-534975-publicacaooriginal-21531-pl.html (accessed on 19 February 2023).
- Ministério da Defesa—Lei Geral de Proteção de Dados Pessoais (LGPD), Lei nº 13.709, de 14 de Agosto de 2018. Disponível em. Available online: https://www.gov.br/defesa/pt-br/acesso-a-informacao/lei-geral-de-protecao-de-dados-pessoais-lgpd (accessed on 19 February 2023).
- Heaton, J.; Goodfellow, I.; Bengio, Y.; Courville, A. Deep learning. Genet. Program. Evolvable Mach. 2018, 19, 305–307. [Google Scholar] [CrossRef]
- Wong, K.K.; Fortino, G.; Abbott, D. Deep learning-based cardiovascular image diagnosis: A promising challenge. Future Gener. Comput. Syst. 2020, 110, 802–811. [Google Scholar] [CrossRef]
- Ovalle-Magallanes, E.; Avina-Cervantes, J.G.; Cruz-Aceves, I.; Ruiz-Pinales, J. Transfer learning for stenosis detection in X-ray Coronary Angiography. Mathematics 2020, 8, 1510. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Mishkin, D.; Matas, J. All you need is a good init. arXiv 2015, arXiv:1511.06422. [Google Scholar] [CrossRef]
- Zeng, X.; Jin, X.; Zhong, L.; Zhou, G.; Zhong, M.; Wang, W.; Fan, Y.; Liu, Q.; Qi, X.; Guan, X.; et al. Difficult and complicated oral ulceration: An expert consensus guideline for diagnosis. Int. J. Oral Sci. 2022, 14, 28. [Google Scholar] [CrossRef]
- Van der Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef]
- Uthoff, R.; Song, B.; Sunny, S.; Patrick, S.; Suresh, A.; Kolur, T.; Gurushanth, K.; Wooten, K.; Gupta, V.; Platek, M.E.; et al. Small form factor, flexible, dual-modality handheld probe for smartphone-based, point-of-care oral and oropharyngeal cancer screening. J. Biomed. Opt. 2019, 24, 106003. [Google Scholar] [CrossRef]
- Shamim, M.Z.M.; Syed, S.; Shiblee, M.; Usman, M.; Ali, S.J.; Hussein, H.S.; Farrag, M. Automated detection of oral pre-cancerous tongue lesions using deep learning for early diagnosis of oral cavity cancer. Comput. J. 2022, 65, 91–104. [Google Scholar] [CrossRef]

| Architecture | Size | Parameters | Reference |
|---|---|---|---|
| VGG16 | 533 MB | 138 × 106 | [43] |
| ResNet-50 | 102 MB | 25 × 106 | [44] |
| InceptionV3 | 96 MB | 24 × 106 | [29] |
| Xception | 91 MB | 23 × 106 | [45] |
| Class | Number of Images | Augmentation Factor | Resulting Dataset |
|---|---|---|---|
| Plaque | 1618 | 1× | 1618 |
| Erosion | 159 | 10× | 1590 |
| Vesicle/Bullous | 308 | 5× | 1540 |
| Ulcer | 1506 | 1× | 1506 |
| Papule/Nodule | 1417 | 1× | 1417 |
| Macule/Spot | 61 | 30× | 1830 |
| Class | Sensitivity | Specificity | Accuracy | Precision | F1-Score |
|---|---|---|---|---|---|
| Ulcer | 71.71 | 95.19 | 91.30 | 74.73 | 73.19 |
| Papule/Nodule | 79.79 | 94.58 | 92.14 | 74.52 | 77.07 |
| Macule/Spot | 94.00 | 100.00 | 98.99 | 100.00 | 96.90 |
| Plaque | 87.00 | 93.17 | 92.14 | 71.90 | 78.73 |
| Erosion | 79.00 | 99.79 | 96.32 | 98.75 | 87.77 |
| Vesicle/Bullous | 100.00 | 99.59 | 99.66 | 98.03 | 99.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, R.F.T.; Schmith, J.; Figueiredo, R.M.d.; Freitas, S.A.; Machado, G.N.; Romanini, J.; Carrard, V.C. Use of Artificial Intelligence in the Classification of Elementary Oral Lesions from Clinical Images. Int. J. Environ. Res. Public Health 2023, 20, 3894. https://doi.org/10.3390/ijerph20053894
Gomes RFT, Schmith J, Figueiredo RMd, Freitas SA, Machado GN, Romanini J, Carrard VC. Use of Artificial Intelligence in the Classification of Elementary Oral Lesions from Clinical Images. International Journal of Environmental Research and Public Health. 2023; 20(5):3894. https://doi.org/10.3390/ijerph20053894
Chicago/Turabian StyleGomes, Rita Fabiane Teixeira, Jean Schmith, Rodrigo Marques de Figueiredo, Samuel Armbrust Freitas, Giovanna Nunes Machado, Juliana Romanini, and Vinicius Coelho Carrard. 2023. "Use of Artificial Intelligence in the Classification of Elementary Oral Lesions from Clinical Images" International Journal of Environmental Research and Public Health 20, no. 5: 3894. https://doi.org/10.3390/ijerph20053894
APA StyleGomes, R. F. T., Schmith, J., Figueiredo, R. M. d., Freitas, S. A., Machado, G. N., Romanini, J., & Carrard, V. C. (2023). Use of Artificial Intelligence in the Classification of Elementary Oral Lesions from Clinical Images. International Journal of Environmental Research and Public Health, 20(5), 3894. https://doi.org/10.3390/ijerph20053894






