Estimating Soil Salinity with Different Levels of Vegetation Cover by Using Hyperspectral and Non-Negative Matrix Factorization Algorithm
Abstract
1. Introduction
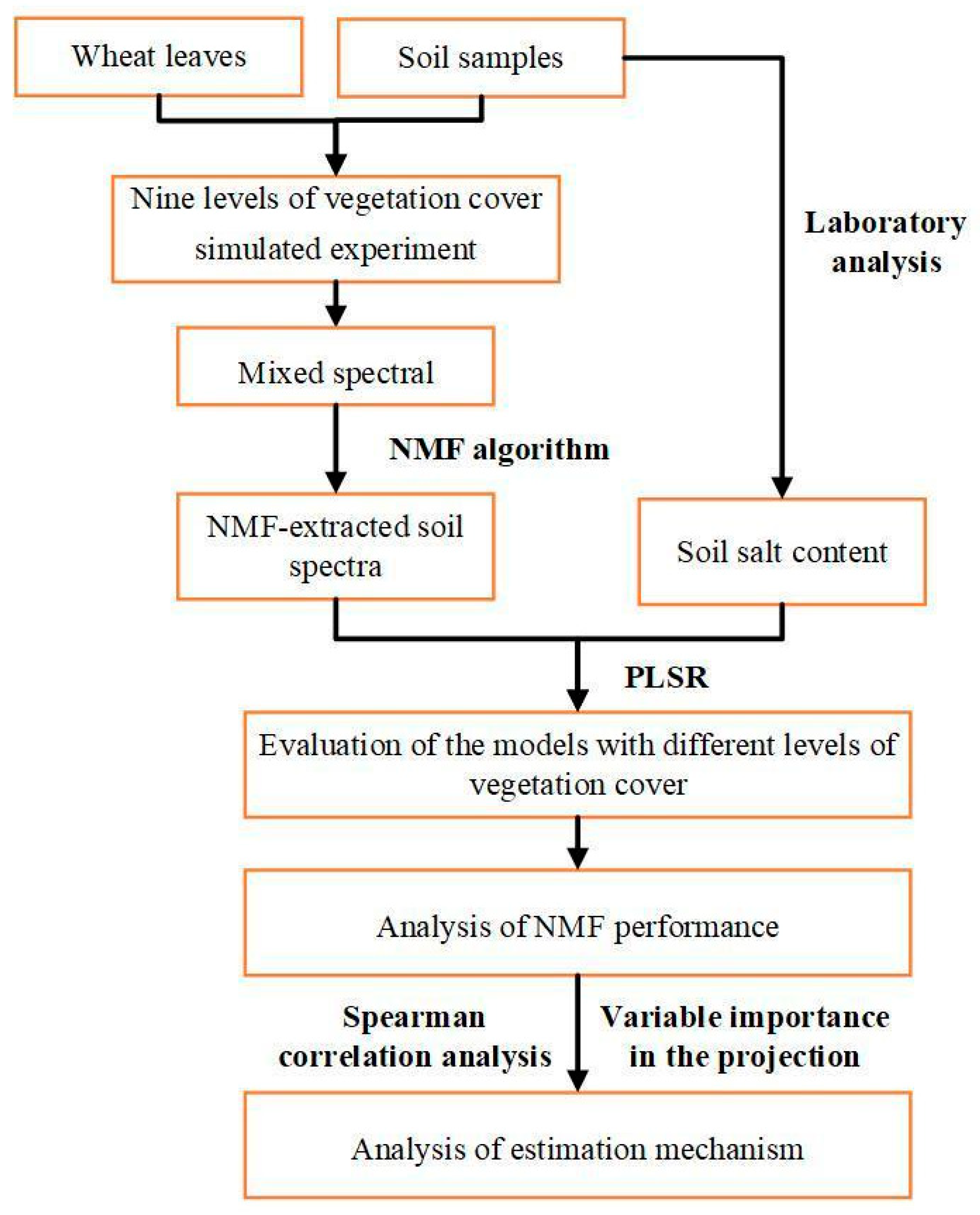
2. Materials and Methods
2.1. Data Sources
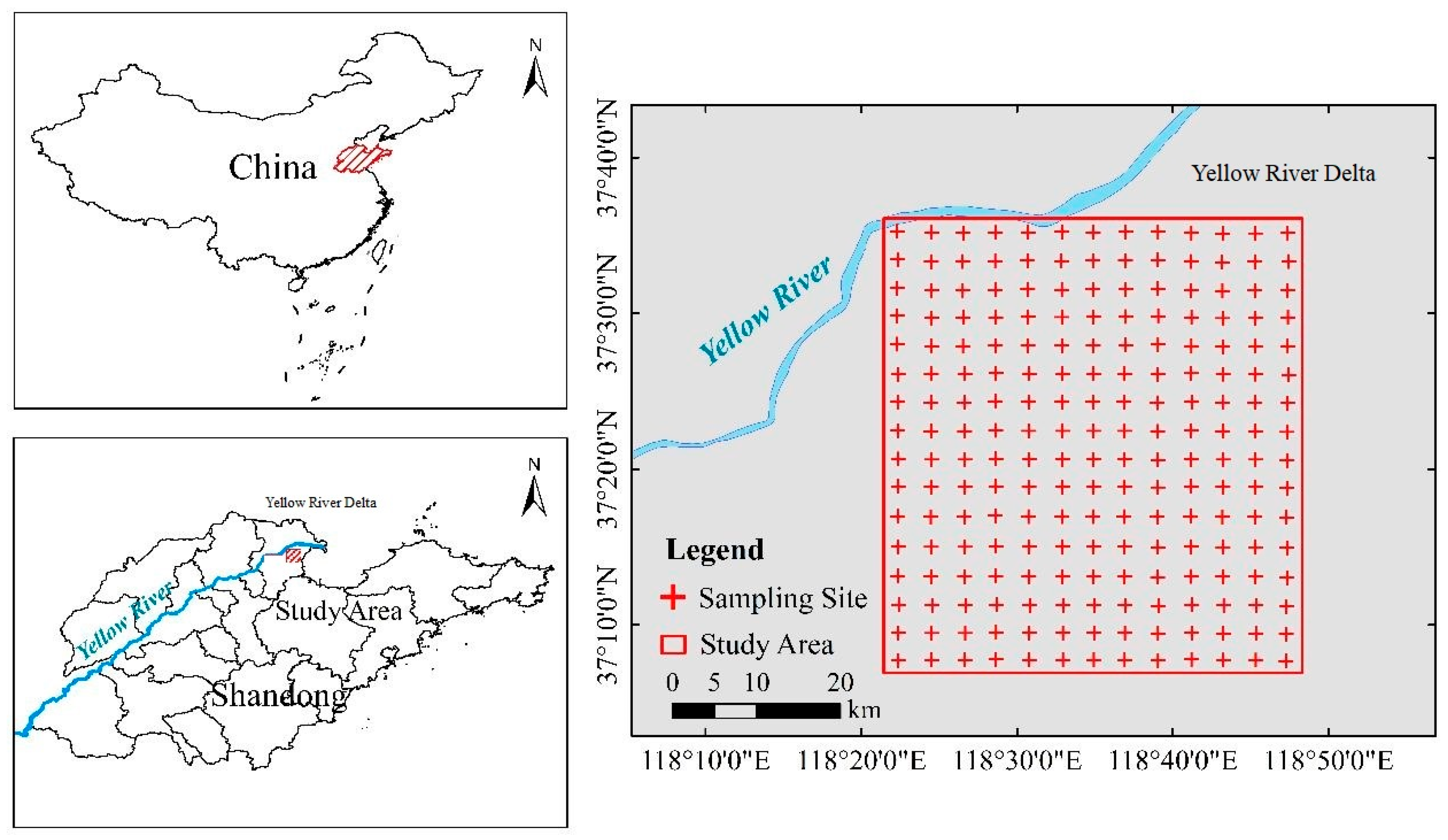
2.1.1. Study Site and Soil Sampling
2.1.2. Laboratory Analysis
2.2. Experiments
- (1)
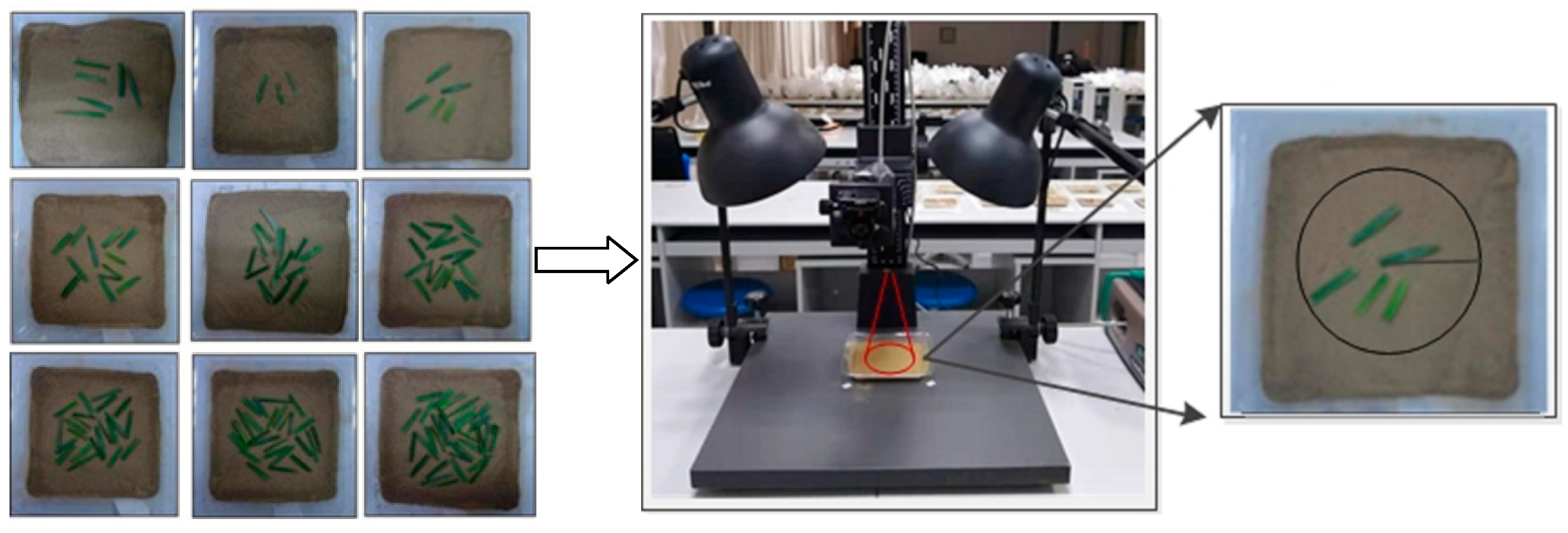
- Soil sample preparation: As shown in Figure 2, the soil was placed into the plastic tray (7.5 cm radius). It was filled with approximately 150 g of soil at a depth of approximately 3 cm.
- (2)
- Vegetation cover simulation: Wheat leaves were cut into 3 cm sections (1 cm height) and placed on the soil surface in the scene. Without damaging the soil sample, 3 prepared leaves were added at a time. Digital photographs of each scene were taken using a digital camera for the vegetation coverage calculation [28]. The FVC was calculated using the method proposed by Zhang et al. (2013) [29]. For each soil sample, nine levels of mean FVC were designed as follows: 5.61%, 9.21%, 16.34%, 25.76%, 37.81%, 48.28%, 56.42%, 63.55%, and 76.42%.
- (3)
- Spectrum measurement: The spectrometer was placed vertically approximately 15 cm above the soil sample with a 25° field of view. The reflectance of the mixed scenes was measured within a radius of approximately 3.3 cm (Figure 2). Spectra were measured with an ASD FieldSpec HandHeld 3 portable spectroradiometer (Analytical Spectra Devices, Inc., Boulder, CO, USA), which covered the wavelengths of 350–2500 nm with a sampling interval of 1.4 nm (350–1000 nm) and 2 nm (1000–2500 nm) [30]. Soil samples were illuminated using two 50 W halogen lamps with an incident angle of 45°, which were positioned 50 cm from the sample [31,32]. The spectrometer probe was positioned vertically, approximately 30 cm above the sample. The spectrometer was preheated for 20–30 min and calibrated with a white panel before each measurement [27]. Ten spectral measurements were repeated for each soil sample [31]. As a result, a total of 9 × 208 × 10 = 18,720 mixed spectra were obtained. Spectral preprocessing was performed using ASD software to remove physical variability from light scattering and highlight the spectral features of interest [33]. The spectra were subjected to Savitzky–Golay smoothing with a moving window width of nine and transformed into the first derivative. Because of the presence of high-frequency noise at the edges of the spectrum, after the preprocessing stage, all the spectra were reduced to 500–2350 nm.
2.3. Methodology
2.3.1. Non-Negative Matrix Factorization
2.3.2. Model Calibration and Validation
2.3.3. Estimation Mechanism Analysis
3. Results and Discussion
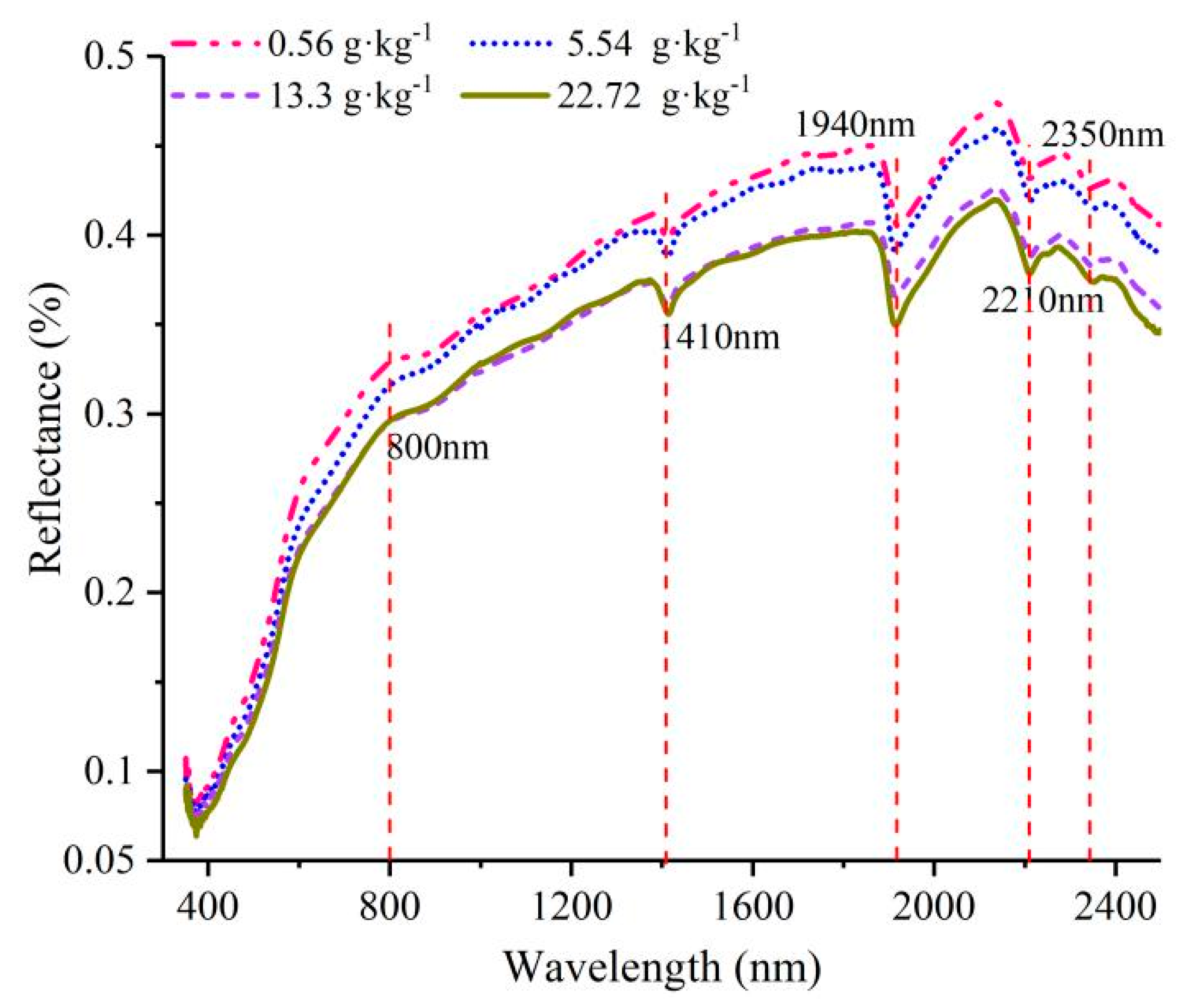
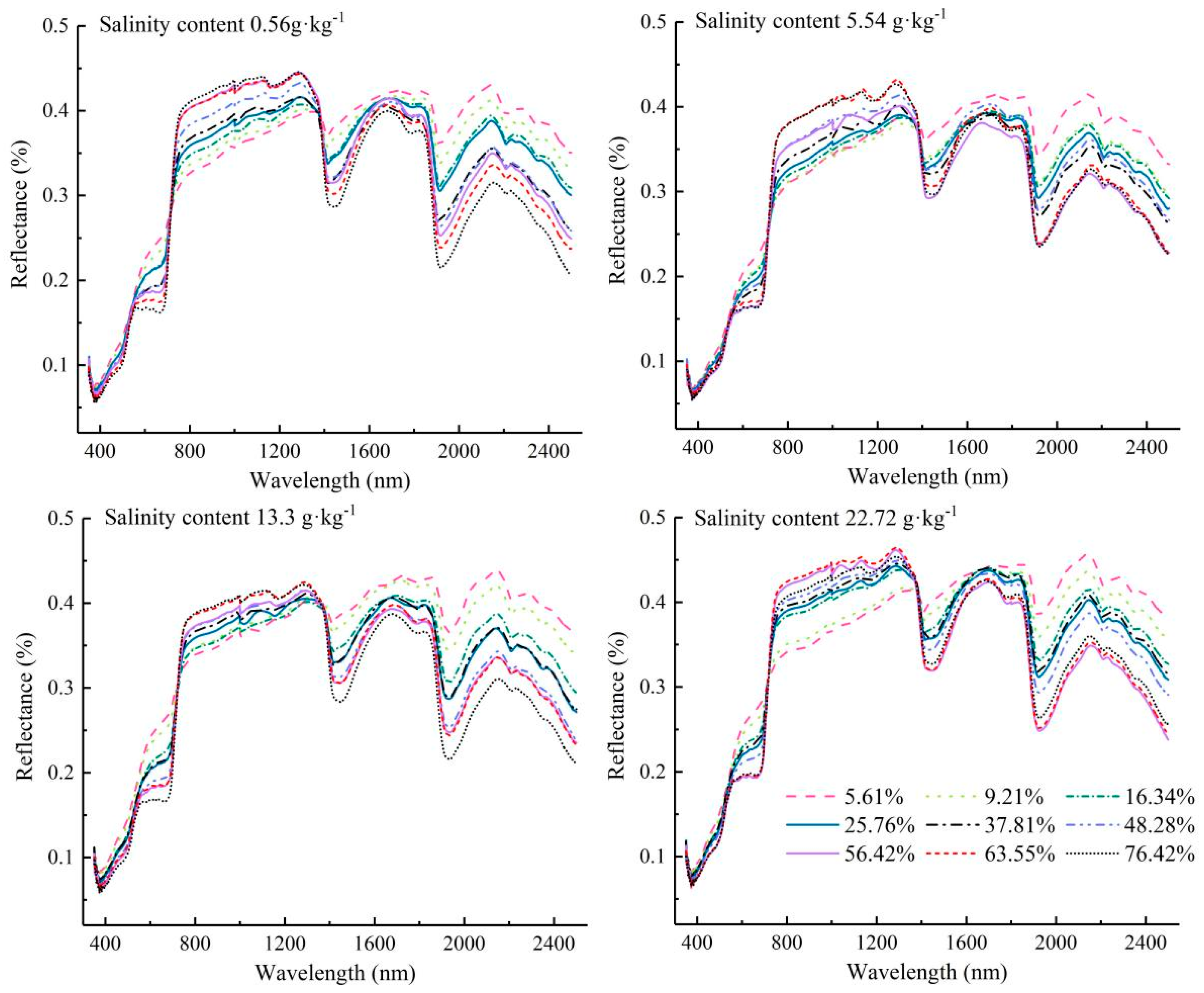
3.1. Analysis of Spectral Characteristic Analysis
3.2. Estimation SSC with Different Levels of Vegetation
3.3. Performance of NMF
3.3.1. Performance of Soil Spectra Extraction
3.3.2. Performance of SSC Estimation
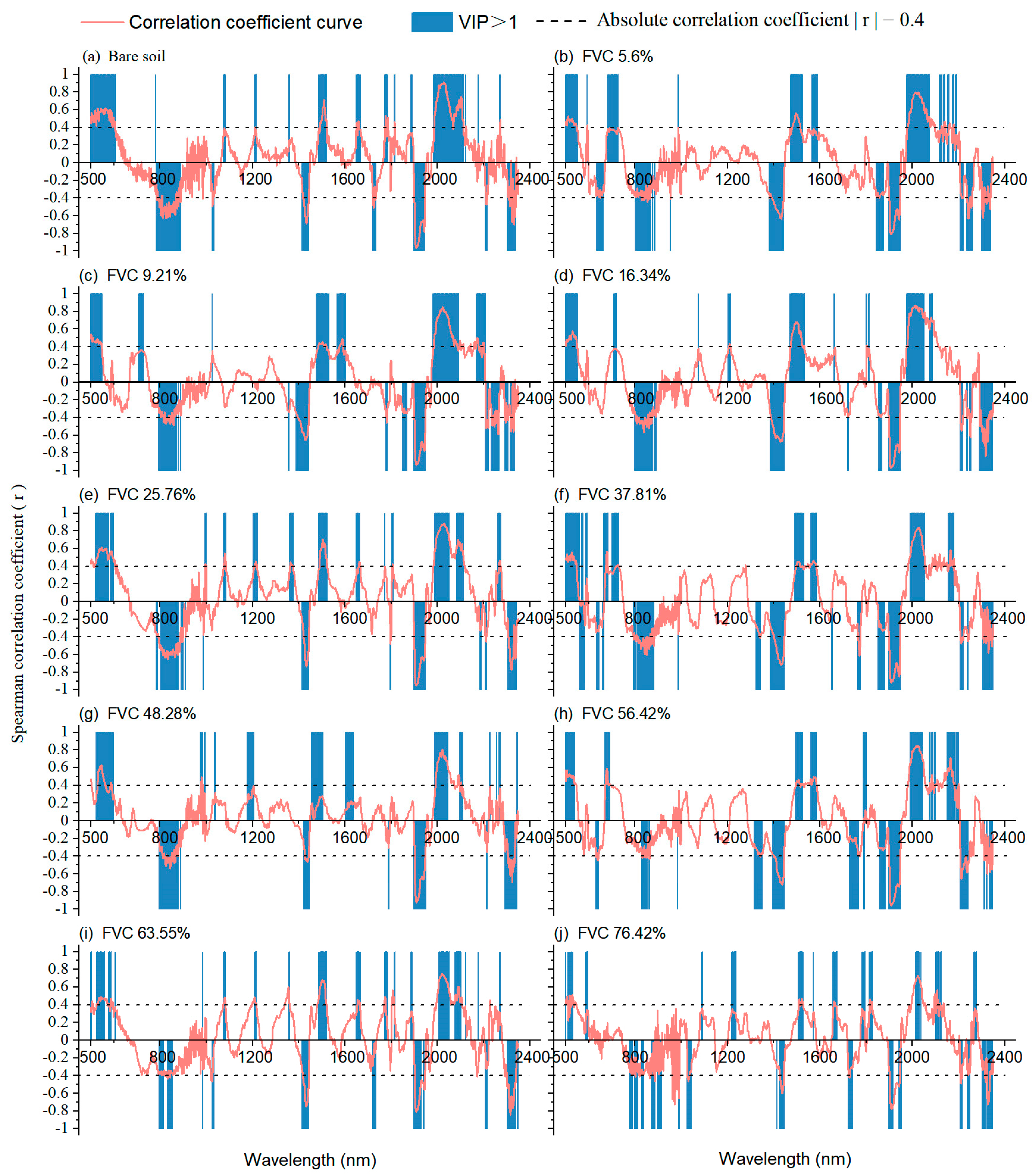
3.4. Estimation Mechanism Analysis
3.4.1. Explanation of Influence Mechanism
3.4.2. Explanation of Optimization Mechanism
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, J.; Peng, J.; Zhou, Y.; Xu, D.Y.; Shi, Z. Quantitative estimation of soil salinity using UVA-borne hyperspectral and satellite multispectral images. Remote Sens. 2019, 11, 736. [Google Scholar] [CrossRef]
- Wei, Y.; Ding, J.L.; Yang, S.T.; Wang, F.; Wang, C. Soil salinity prediction based on scale-dependent relationships with environmental variables by discrete wavelet transform in the Tarim Basin. Catena 2021, 196, 104939. [Google Scholar] [CrossRef]
- Chi, Y.; Zheng, W.; Shi, H.; Sun, J.; Fu, Z. Spatial heterogeneity of estuarine wetland ecosystem health influenced by complex natural and anthropogenic factors. Sci. Total Environ. 2018, 634, 1445–1462. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhao, Y.H.; Cao, W.; Xing, M.X.; Xu, X.Y.; Wang, Z.Y.; Sun, J. Metagenomic analysis reveals antibiotic resistance genes and virulence factors in the saline-alkali soils from the Yellow River Delta, China. Environ. Res. 2022, 214, 113823. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Zang, W.Q.; Luo, W.; Wen, Y.; Yang, F.; Han, B.M.; Fan, Y.W.; Chen, X.; Qi, Z.; Wang, Z.; et al. Detection model of soil salinization information in the Yellow River Delta based on feature space models with typical surface parameters derived from landsat8 OLI image. Geomatics. Nat. Hazards Risk 2020, 11, 288–300. [Google Scholar] [CrossRef]
- Cao, X.Y.; Chen, W.Q.; Ge, X.Y.; Chen, X.Y.; Wang, J.Z.; Ding, J.L. Multidimensional soil salinity data mining and evaluation from different satellites. Sci. Total Environ. 2022, 846, 157416. [Google Scholar] [CrossRef]
- Yu, H.; Kong, B.; Wang, Q.; Liu, X.; Liu, X. Hyperspectral remote sensing applications in soil: A review. Hyperspectral Remote Sens. 2020, 269–291. [Google Scholar] [CrossRef]
- Wang, J.Z.; Ding, J.L.; Yu, D.L.; Ma, X.K.; Zhang, Z.P.; Ge, X.Y.; Teng, D.X.; Li, X.H.; Liang, J.; Lizaga, I.; et al. Capability of Sentinel-2 MSI data for monitoring and mapping of soil salinity in dry and wet seasons in the Ebinur Lake region, Xinjiang, China. Geoderma 2019, 353, 172–187. [Google Scholar] [CrossRef]
- Yue, J.B.; Tian, Q.J.; Tang, S.F.; Xu, K.J.; Zhou, C.Q. A dynamic soil endmember spectrum selection approach for soil and crop residue linear spectral unmixing analysis. Int. J. Appl. Earth Obs. Geoinf. 2019, 78, 306–317. [Google Scholar] [CrossRef]
- Tilley, D.R.; Ahmed, M.; Son, J.H.; Badrinarayanan, H. Hyperspectral reflectance response of freshwater macrophytes to salinity in a brackish subtropical marsh. J. Environ. Qual. 2007, 36, 780–789. [Google Scholar] [CrossRef]
- Fernández-Buces, N.; Siebe, C.; Cram, S.; Palacio, J.L. Mapping soil salinity using a combined spectral response index for bare soil and vegetation: A case study in the former lake Texcoco, Mexico. J. Arid Environ. 2006, 65, 644–667. [Google Scholar] [CrossRef]
- Douaoui, A.E.K.; Nicolas, H.; Walter, C. Detecting salinity hazards within a semiarid context by means of combining soil and remote-sensing data. Geoderma 2017, 134, 217–230. [Google Scholar] [CrossRef]
- Zhang, M.; Ustin, S.L.; Rejmankova, E.; Sanderson, E.W. Monitoring pacific coast salt marshes using remote sensing. Ecol. Appl. 1997, 7, 1039–1053. [Google Scholar] [CrossRef]
- Bartholomeus, H.; Kooistra, L.; Stevens, A.; Leeuwen, M.V.; Wesemael, B.V.; Ben-Dor, E.; Tychon, B. Soil organic carbon mapping of partially vegetated agricultural fields with imaging spectroscopy. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 81–88. [Google Scholar] [CrossRef]
- Ouerghemmi, W.; Gomez, C.; Naceur, S.; Lagacherie, P. Applying blind source separation on hyperspectral data for clay content estimation over partially vegetated surfaces. Geoderma 2011, 163, 227–237. [Google Scholar] [CrossRef]
- Ouerghemmi, W.; Gomez, C.; Naceur, S.; Lagacherie, P. Semi-blind source separation for the estimation of the clay content over semi-vegetated areas using VNIR/SWIR hyperspectral airborne data. Remote Sens. Environ. 2016, 181, 251–263. [Google Scholar] [CrossRef]
- Liu, Y.; Pan, X.Z.; Shi, R.J.; Li, Y.L.; Wang, C.K.; Li, Z.T. Predicting soil salt content over partially vegetated surfaces using non-negative matrix factorization. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 5305–5316. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, Y.; Wu, S.W.; Wang, C.K.; Xu, A.A.; Pan, X.Z. Hyper-spectral estimation of wheat biomass after alleviating of soil effects on spectra by non-negative matrix factorization. Eur. J. Agron. 2017, 84, 58–66. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, F.F.; Wang, C.K.; Wu, S.W.; Liu, J.; Xu, A.A.; Pan, K.; Pan, X.Z. Estimating the soil salinity over partially vegetated surfaces from multispectral remote sensing image using non-negative matrix factorization. Geoderma 2019, 354, 113887. [Google Scholar] [CrossRef]
- Zhang, X.L.; Zhang, Z.H.; Wang, W.; Fang, W.T.; Liu, X.J.; Ju, H.R. Vegetation successions of coastal wetlands in southern Laizhou Bay, Bohai Sea, northern China, influenced by the changes in relative surface elevation and soil salinity. J. Environ. Manag. 2021, 293, 112964. [Google Scholar] [CrossRef]
- Liu, Y.L.; Chen, Y.Y. Feasibility of estimating Cu contamination in floodplain soils using VNIR spectroscopy—A case study in the Le’an River floodplain, China. Soil Sediment Contam 2012, 21, 951–969. [Google Scholar] [CrossRef]
- Jiang, Q.H.; Chen, Y.Y.; Guo, L.; Fei, T.; Qi, K. Estimating Soil Organic Carbon of Cropland Soil at Different Levels of Soil Moisture Using VIS-NIR Spectroscopy. Remote Sens. 2016, 8, 755. [Google Scholar] [CrossRef]
- Cheng, H.; Shen, R.; Chen, Y.; Wan, Q.; Shi, T.; Wang, J.; Wan, Y.; Hong, Y.; Li, X. Estimating heavy metal concentrations in suburban soils with reflectance spectroscopy. Geoderma 2019, 336, 59–67. [Google Scholar] [CrossRef]
- Chi, Y.; Shi, H.H.; Zheng, W.; Sun, J.K.; Fu, Z.Y. Spatiotemporal characteristics and ecological effects of the human interference index of the Yellow River Delta in the last 30 years. Ecol. Indic. 2018, 39, 1219–1220. [Google Scholar] [CrossRef]
- Jiang, C.; Chen, S.; Pan, S.; Fan, Y.; Ji, H. Geomorphic evolution of the Yellow River Delta: Quantification of basin-scale natural and anthropogenic impacts. Catena 2018, 163, 361–377. [Google Scholar] [CrossRef]
- Zhu, W.; Yang, J.; Yao, R.; Xie, W.; Wang, X.; Liu, Y. Soil water-salt control and yield improvement under the effect of compound control in saline soil of the Yellow River Delta, China. Agric. Water Manag. 2022, 263, 107455. [Google Scholar] [CrossRef]
- Chen, T.; Chang, Q.R.; Liu, J.; Clevers, J.G.P.W.; Kooistra, L. Identification of soil heavy metal sources and improvement in spatial mapping based on soil spectral information: A case study in northwest China. Sci. Total Environ. 2016, 565, 155–164. [Google Scholar] [CrossRef]
- Liu, S.L.; Hou, X.Y.; Yang, M.; Cheng, F.Y.; Coxixo, A.; Wu, X.; Zhang, Y.Q. Factors driving the relationships between vegetation and soil properties in the Yellow River Delta, China. Catena 2018, 165, 279–285. [Google Scholar] [CrossRef]
- Wu, C.S.; Liu, G.H.; Huang, C.; Liu, Q.S. Soil quality assessment in Yellow River Delta: Establishing a minimum data set and fuzzy logic model. Geoderma 2019, 334, 82–89. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Hunt, E.R. Mitigating the effects of soil and residue water contents on remotely sensed estimates of crop residue cover. Remote Sensing. Environ. 2008, 112, 1647–1657. [Google Scholar] [CrossRef]
- Zhang, X.F.; Liao, C.H.; Li, J.; Sun, Q. Fractional vegetation cover estimation in arid and semi-arid environments using HJ-1 satellite hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 506–512. [Google Scholar] [CrossRef]
- Shi, T.Z.; Chen, Y.; Liu, Y.Y.; Wu, G.F. Visible and near-infrared reflectance spectroscopy-an alternative for monitoring soil contamination by heavy metals. J. Hazard. Mater. 2014, 265, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.S.; Yu, L.; Chen, Y.Y.; Liu, Y.F.; Liu, Y.L.; Liu, Y.; Cheng, H. Prediction of soil organic matter by VIS–NIR spectroscopy using normalized soil moisture index as a proxy of soil moisture. Remote Sens. 2017, 10, 28. [Google Scholar] [CrossRef]
- Hong, Y.S.; Chen, Y.; Yu, L.; Liu, Y.F.; Liu, Y.L.; Zhang, Y.; Liu, Y.; Cheng, H. Combining fractional order derivative and spectral variable selection for organic matter estimation of homogeneous soil samples by VIS–NIR spectroscopy. Remote Sens. 2018, 10, 479. [Google Scholar] [CrossRef]
- Dotto, A.C.; Dalmolin, R.S.D.; Caten, T. A systematic study on the application of scatter-corrective and spectral-derivative preprocessing for multivariate prediction of soil organic carbon by Vis-NIR spectra. Geoderma 2018, 314, 262–274. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- Guillamet, D.; Vitrià, J.; Schiele, B. Introducing a weighted non-negative matrix factorization for image classification. Pattern Recogn. Lett. 2003, 24, 2447–2454. [Google Scholar] [CrossRef]
- Heinz, D.C.; Chein, I.C. Fully constrained least squares linear spectral mixture analysis method for material quantification in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2002, 39, 529–545. [Google Scholar] [CrossRef]
- Farifteh, A.J.; Van der Meer, F.; Atzberger, C.; Carranza, E.J.M. Quantitative analysis of salt-affected soil reflectance spectra: A comparison of two adaptive methods (PLSR and ANN). Remote Sens. Environ. 2007, 110, 59–78. [Google Scholar] [CrossRef]
- Liu, Y.L.; Jiang, Q.H.; Fei, T.; Wang, J.J.; Shi, T.Z.; Guo, K.; Li, X.R.; Chen, Y.Y. Transferability of a visible and near-infrared model for soil organic matter estimation in riparian landscapes. Remote Sens. 2014, 6, 4305–4322. [Google Scholar] [CrossRef]
- Liu, H.; Shi, T.; Chen, Y.; Wang, J.; Fei, T.; Wu, G. Improving spectral estimation of soil organic carbon content through semi-supervised regression. Remote Sens. 2017, 9, 29. [Google Scholar] [CrossRef]
- Chang, C.W.; Laird, D.A.; Mausbach, M.J.; Hurburgh, C.R. Near-Infrared Reflectance Spectroscopy-Principal Components Regression Analyses of Soil Properties. Soil Sci. Soc. Am. J. 2001, 65, 480–490. [Google Scholar] [CrossRef]
- Terhoeven-Urselmans, T.; Vagen, T.G.; Spaargaren, O.; Shepherd, K.D. Prediction of soil fertility properties from a globally distributed soil mid-infrared spectral library. Soil Sci. Soc. Am. J. 2010, 74, 1792–1799. [Google Scholar] [CrossRef]
- Chong, I.G.; Jun, G.H. Performance of some variable selection methods when multicollinearity is present. Chemom. Intell. Lab. Syst. 2005, 78, 103–112. [Google Scholar] [CrossRef]
- Gomez, C.; Lagacherie, P.; Coulouma, G. Continuum removal versus PLSR method for clay and calcium carbonate content estimation from laboratory and airborne hyperspectral measurements. Geoderma 2008, 148, 141–148. [Google Scholar] [CrossRef]
- Clark, R.N.; King, T.; Klejwa, M.; Swayze, G.A.; Vergo, N. High spectral resolution reflectance spectroscopy of minerals. J. Geophys. Res. Solid Earth 1990, 95, 12653–12680. [Google Scholar] [CrossRef]
- Whiting, M.L.; Lin, L.; Ustin, S.L. Predicting water content using Gaussian model on soil spectra. Remote Sens. Environ. 2004, 89, 535–552. [Google Scholar] [CrossRef]
- Weng, Y.L.; Peng, G.; Zhu, Z.L. A spectral index for estimating soil salinity in the Yellow River Delta region of China using EO-1 Hyperion Data. Pedosphere 2010, 20, 378–388. [Google Scholar] [CrossRef]
- Berry, M.W.; Browne, M.; Langville, A.N.; Pauca, V.P.; Plemmons, R.J. Algorithms and applications for approximate nonnegative matrix factorization. Comput. Stat. Data An. 2007, 52, 155–173. [Google Scholar] [CrossRef]
- Buda, A.; Jarynowski, A. Life time of correlations and its applications. Wydaw. Niezależne 2010, 1, 5–21. [Google Scholar]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Academic Press: Cambridge, MA, USA, 1988; p. 567. [Google Scholar]
- Quan, W.; Li, P.H.; Xi, C. Modeling salinity effects on soil reflectance under various moisture conditions and its inverse application: A laboratory experiment. Geoderma 2012, 170, 103–111. [Google Scholar]
- Miao, L.D.; Qi, H.R. Endmember Extraction from Highly Mixed Data Using Minimum Volume Constrained Nonnegative Matrix Factorization. IEEE Trans. Geosci. Remote Sens. 2017, 45, 765–777. [Google Scholar] [CrossRef]
- Yue, Y.; Shan, G.; Sun, W.D. Minimum distance constrained non-negative matrix factorization for the endmember extraction of hyperspectral images. Proc. SPIE—Int. Soc. Opt. Eng. 2007, 6790, 309–317. [Google Scholar]


| FVC (%) | Calibration Set | Validation Set | |||
|---|---|---|---|---|---|
| R2c | RMSEc | R2cv | RMSEcv | RPD | |
| Bare | 0.97 | 2.75 | 0.95 | 2.55 | 2.93 |
| 5.61 | 0.88 | 3.98 | 0.82 | 3.72 | 2.02 |
| 9.21 | 0.82 | 4.91 | 0.77 | 4.06 | 1.84 |
| 16.34 | 0.78 | 5.46 | 0.72 | 4.88 | 1.53 |
| 25.76 | 0.76 | 5.83 | 0.68 | 5.18 | 1.43 |
| 37.81 | 0.71 | 6.01 | 0.65 | 5.67 | 1.32 |
| 48.28 | 0.66 | 5.52 | 0.62 | 5.32 | 1.41 |
| 56.42 | 0.61 | 6.11 | 0.59 | 5.94 | 1.26 |
| 63.55 | 0.57 | 6.51 | 0.54 | 6.35 | 1.18 |
| 76.42 | 0.36 | 8.29 | 0.34 | 7.93 | 0.94 |
| FVC (%) | Calibration Set | Validation Set | |||
|---|---|---|---|---|---|
| R2c | RMSEc | R2cv | RMSEcv | RPD | |
| 5.61 | 0.93 | 3.57 | 0.88 | 3.89 | 1.93 |
| 9.21 | 0.92 | 3.88 | 0.88 | 3.79 | 1.98 |
| 16.34 | 0.91 | 3.74 | 0.85 | 3.86 | 1.94 |
| 25.76 | 0.92 | 3.41 | 0.79 | 3.27 | 2.30 |
| 37.81 | 0.90 | 3.42 | 0.83 | 3.98 | 1.89 |
| 48.28 | 0.88 | 3.73 | 0.86 | 3.68 | 2.03 |
| 56.42 | 0.83 | 3.92 | 0.74 | 3.65 | 2.05 |
| 63.55 | 0.77 | 4.67 | 0.69 | 4.15 | 1.80 |
| 76.42 | 0.37 | 5.67 | 0.36 | 5.45 | 1.37 |
| Calibration Set | Validation Set | ||||
|---|---|---|---|---|---|
| R2c | RMSEc | R2cv | RMSEcv | RPD | |
| Mixed spectra | 0.74 | 7.54 | 0.69 | 7.24 | 1.68 |
| NMF-extracted soil spectra | 0.90 | 4.20 | 0.84 | 5.99 | 2.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Yang, H.; Lv, J.; Wu, Q.; Zhang, B. Estimating Soil Salinity with Different Levels of Vegetation Cover by Using Hyperspectral and Non-Negative Matrix Factorization Algorithm. Int. J. Environ. Res. Public Health 2023, 20, 2853. https://doi.org/10.3390/ijerph20042853
Cao J, Yang H, Lv J, Wu Q, Zhang B. Estimating Soil Salinity with Different Levels of Vegetation Cover by Using Hyperspectral and Non-Negative Matrix Factorization Algorithm. International Journal of Environmental Research and Public Health. 2023; 20(4):2853. https://doi.org/10.3390/ijerph20042853
Chicago/Turabian StyleCao, Jianfei, Han Yang, Jianshu Lv, Quanyuan Wu, and Baolei Zhang. 2023. "Estimating Soil Salinity with Different Levels of Vegetation Cover by Using Hyperspectral and Non-Negative Matrix Factorization Algorithm" International Journal of Environmental Research and Public Health 20, no. 4: 2853. https://doi.org/10.3390/ijerph20042853
APA StyleCao, J., Yang, H., Lv, J., Wu, Q., & Zhang, B. (2023). Estimating Soil Salinity with Different Levels of Vegetation Cover by Using Hyperspectral and Non-Negative Matrix Factorization Algorithm. International Journal of Environmental Research and Public Health, 20(4), 2853. https://doi.org/10.3390/ijerph20042853








