Contribution of Genetic Polymorphisms in Human Health
Abstract
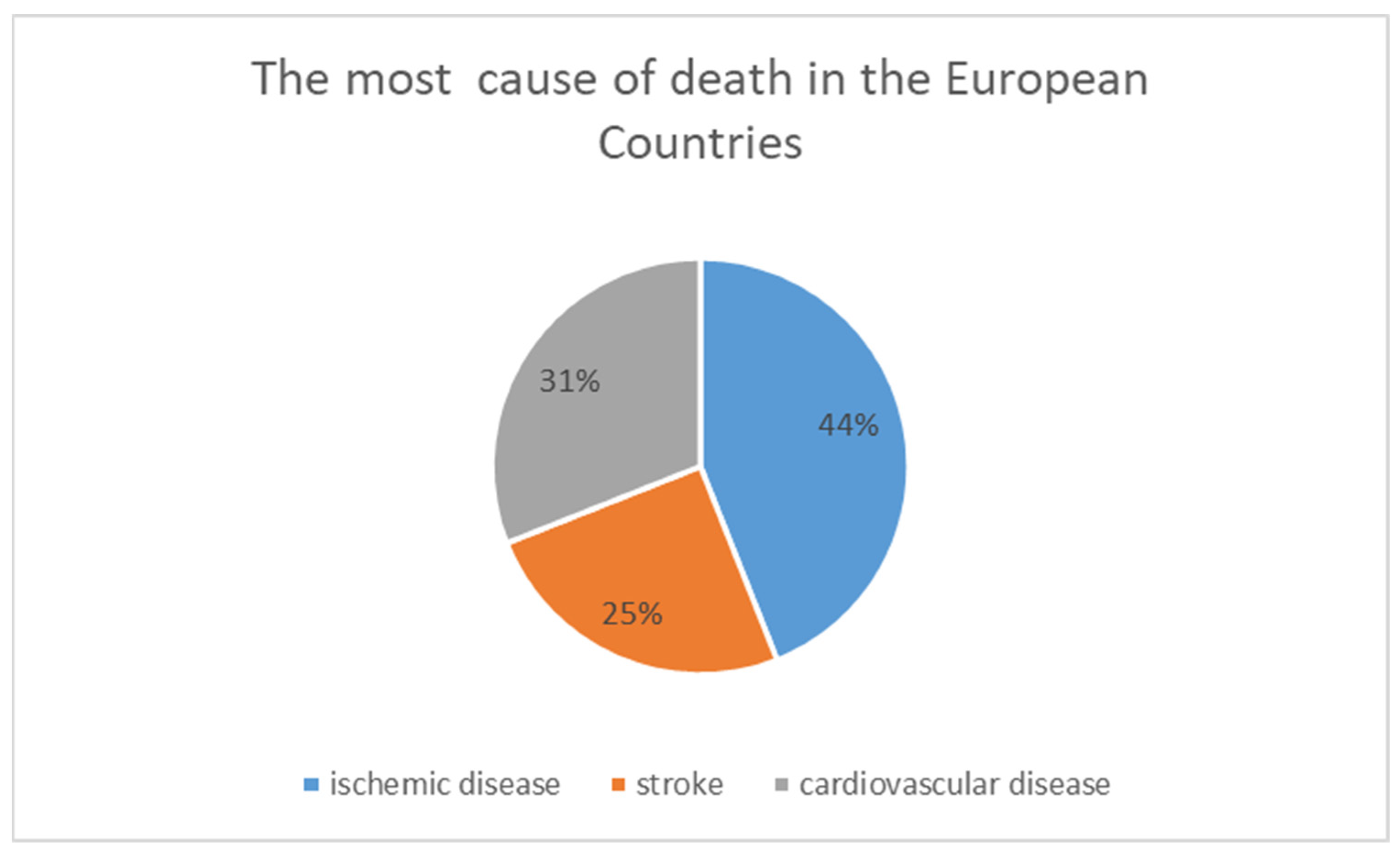
1. Introduction
2. Causative and Susceptibility Genes
3. Genetic Polymorphisms as Biomarkers of Susceptibility
3.1. Genetic Polymorphisms and Differences in the Metabolism
- (1)
- The extensive metabolizer (EM) typical of the normal population. These subjects are either homozygous or heterozygous for the wild-type allele and have a normal metabolism;
- (2)
- The slow metabolizer phenotype (SM) that is associated with the accumulation of specific drug substrates in the body, inherited as a recessive autosomal trait due to the mutation or deletion of both alleles showing a slow metabolism. In some patients, the drug is metabolized very slowly, accumulating the substance in the bloodstream;
- (3)
- The poor metabolizers (PM) carry two defective alleles, showing a complete absence of activity. The higher body concentration of the substance may cause adverse effects due to the substance accumulation;
- (4)
- The rapid metabolizers (RM) clear the drug very quickly, and the therapeutic concentration of the drug in the blood and tissues may not be reached. That means the subject should have a higher dose to produce an effect;
- (5)
- The ultra-extensive metabolizer (UEM) is characterized by enhanced drug metabolism due to gene amplification inherited as an autosomal dominant trait. Individuals with the ultra-extensive phenotype are prone to therapeutic failure because the drug concentrations in the plasma at normal doses are by far too low (faster metabolism) [21]. Here, we distinguish the most common behavioral habits in the following categories.
3.2. Metabolism of Drug
3.3. Metabolism of Smoke
3.4. Metabolism of Ethanol
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dyba, T.; Randi, G.; Bray, F.; Martos, C.; Giusti, F.; Nicholson, N.; Gavin, A.; Flego, M.; Neamtiu, L.; Dimitrova, N.; et al. The European cancer burden in 2020. Incidence and mortality estimates for 40 countries and 25 major cancers. Eur. J. Cancer 2021, 157, 308–347. [Google Scholar] [CrossRef] [PubMed]
- Kotwal, A.A.; Schonberg, M.A. Cancer screening in the elderly: A review of breast, colorectal, lung, and prostate cancer screening. Cancer J. 2017, 23, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Barford, A.; Dorling, D.; Davey Smith, G.; Shaw, M. Life expectancy: Women now on top everywhere. BMJ 2006, 332, 808. [Google Scholar] [CrossRef] [PubMed]
- Chiarella, P.; Capone, P.; Sisto, R. Polymorphic genes and gender difference: Analysis of a European sample. Ital. J. Occup. Environ. Hyg. 2021, 12, 156–166. [Google Scholar] [CrossRef]
- OECD/European Union. Health at a Glance: Europe 2020: State of Health in the EU Cycle; OECD Publishing: Paris, France, 2020; pp. 1–237. [Google Scholar] [CrossRef]
- Townsend, N.; Kazakiewicz, D.; Wright, F.L.; Timmis, A.; Huculeci, R.; Torbica, A.; Gale, C.P.; Achenbach, S.; Weidinger, F.; Vardas, P. Epidemiology of cardiovascular disease in Europe. Nat. Rev. Cardiol. 2022, 19, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, S.; Rosa-Neto, P.; Morais, J.A.; Webster, C. Journey through the diagnosis of dementia. In World Alzheimer Report 2021; Alzheimer’s Disease International: London, England, 2021; pp. 1–313. [Google Scholar]
- Jeorges, J.; Miller, O.; Bintener, C. Dementia in Europe Yearbook 2019: Estimating the Prevalence of Dementia in Europe; Alzheimer Europe: Luxembourg, 2020; pp. 1–109. ISBN 978-99959-995-9-9. [Google Scholar] [CrossRef]
- Regional Epidemiological Service and Epidemiological Data Registers. Available online: https://demenze.regione.veneto.it/PDTA/dati (accessed on 2 September 2022).
- Jackson, S.; Bartek, P.J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Orgogozo, V.; Morizot, B.; Martin, A. The differential view of genotype-phenotype relationships. Front. Genet. 2015, 6, 179. [Google Scholar] [CrossRef] [PubMed]
- Bekris, L.M.; Yum, C.-E.; Bird, T.D.; Tsuang, D. Genetics of Alzheimer disease. Am. J. Neuroradiol. 2010, 23, 213–227. [Google Scholar] [CrossRef]
- Chouinard-Watkins, R.; Plourde, M. Fatty acid metabolism in carriers of Apolipoprotein e Epsilon 4 allele: Is it contributing to higher risk of cognitive decline and coronary heart disease? Nutrients 2014, 6, 4452–4471. [Google Scholar] [CrossRef]
- Tarawneh, R.; Holtzman, D.M. The Clinical Problem of Symptomatic Alzheimer Disease and Mild Cognitive Impairment. Cold Spring Harb Perspect Med. 2012, 2, a006148. [Google Scholar] [CrossRef]
- Giuffrè; Bianchi, A.; Gulotta, G.; Sartori, G. (Eds.) Role of behavioural and genetic factors. In Neuroscience Forensic Manual; Giuffrè: Milan, Italy, 2009; pp. 69–90. ISBN 9788814148590. [Google Scholar]
- Prüss-Üstün, A.; Corvalán, C.; WHO. Preventing Disease through Healthy Environments: Towards an Estimate of the Environmental Burden of Disease; WHO: Geneva, Switzerland, 2006; ISBN 92 4 159382 2.
- Chiarella, P.; Capone, P.; Carbonari, D.; Sisto, R. A predictive model assessing genetic susceptibility risk at workplace. Int. J. Environ. Res. Public Health 2019, 16, 2012. [Google Scholar] [CrossRef]
- Chiarella, P.; Capone, P.; Sisto, R. The Role of Genetic Polymorphisms in the occupational exposure. In The Recent Topics in Genetic Polymorphisms; Caliskan, M., Erol, O., Öz, G.C., Eds.; Intech Open: London, UK, 2019; pp. 1–16. [Google Scholar]
- Topic, E. The role of pharmacogenetics in management of cardiovascular disease. J. Int. Fed. Clin. Chem. Lab. Med. 2003, 14, 78–88. [Google Scholar]
- Rommel, G.T.; Kim, R.B. Introduction to Clinical Pharmacology. In Clinical and Translational Science, 2nd ed.; Robertson, D., Williams, G., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 365–388. [Google Scholar]
- Belle, D.J.; Singh, H. Genetic factors in drug metabolism. Am. Fam. Physician 2008, 77, 1553–1560. [Google Scholar]
- Ingelman-Sundberg, M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functional diversity. Pharm. J. 2005, 5, 6–13. [Google Scholar] [CrossRef] [PubMed]
- World Bank. World Bank Report on the Economics of Tobacco Control; World Bank: Washington, DC, USA, 1999. [Google Scholar]
- Tobacco Advertising Ban Takes Effect 31 July. Available online: https://ec.europa.eu/commission/presscorner/detail/en/IP_05_1013 (accessed on 3 July 2022).
- Murphy, S.E. Nicotine metabolism and smoking: Ethnic differences in the role of P450 2A6. Chem. Res. Toxicol. 2017, 30, 410–419. [Google Scholar] [CrossRef]
- Derby, K.S.; Cuthrell, K.; Caberto, C.; Carmella, S.G.; Franke, A.A.; Hecht, S.S.; Murphy, S.E.; Le Marchand, L. Nicotine metabolism in three ethnic/racial groups with different risks of lung cancer. Cancer Epidemiol. Biomark. Prev. 2008, 17, 3526–3535. [Google Scholar] [CrossRef]
- Haiman, C.A.; Stram, O.D.; Wilkens, L.; Pike, C.M.; Malcolm, C.; Kolonel, L.N.; Henderson, B.E.; Le Marchand, L. Nicotine metabolism in three ethnical/ratial groups with different risk of lung cancer. N. Engl. J. Med. 2006, 26, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Patel, Y.M.; Park, S.L.; Han, Y.; Wilkens, L.R.; Bickeböller, H.; Rosenberger, A.; Caporaso, N.; Landi, M.T.; Brüske, I.; Risch, A.; et al. Novel association of genetic markers affecting CYP2A6 activity and lung cancer risk. Cancer Res. 2016, 76, 5768–5776. [Google Scholar] [CrossRef]
- Rehm, J. The risks associated with alcohol use and alcoholism. Alcohol Res. Health 2011, 34, 135–143. [Google Scholar]
- Edenberg, H.; Foroud, T. The genetics of alcoholism: Identifying specific genes through family studies. Addict. Biol. 2006, 11, 386–396. [Google Scholar] [CrossRef]
- Kitson, K. Regulation of Alcohol and Aldehyde dehydrogenase activity: A metabolic balancing act with important social consequences. Alcohol. Clin. Exp. Res. 1999, 23, 955–957. [Google Scholar] [CrossRef] [PubMed]
- Wall, T.; Luczak, S.E.; Hiller-Sturmhofel, S. Biology, Genetics and Environment: Underlying factor influencing alcohol metabolism. Alcohol Res. Curr. Rev. 2016, 38, 59–68. [Google Scholar]
- Edenberg, H.J. The genetics of alcohol metabolism: Role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health 2007, 30, 5–13. [Google Scholar]
- Rashidi, W.N.W.; Bakar, S.A. Glutathione S-Transferase: An overview on distribution of GSTM1 and GSTT1 polymorphisms in Malaysian and other populations. Mal. J. Med. Health Sci. 2019, 15, 85–95. [Google Scholar]
- Uckun Şahinoğulları, Z. Frequencies of glutathione S-transferase A1 rs3957357 polymorphism in a Turkish population. J. Surg. Med. 2021, 5, 221–225. [Google Scholar] [CrossRef]
- Xiao, J.; Wang, Y.; Wang, Z.; Zhang, Y.; Li, Y.; Xu, C.; Xiao, M.; Wang, H.; Guo, S.; Jin, L.; et al. The relevance analysis of GSTP1 rs1695 and lung cancer in the Chinese Han population. Int. J. Biol. Mark. 2021, 36, 48–54. [Google Scholar] [CrossRef]
- Agudo, A.; Peluso, M.; Sala, N.; Capellà, G.; Munnia, A.; Piro, S.; Marín, F.; Ibáñez, R.; Amiano, P.; Tormo, M.J.; et al. Aromatic DNA adducts and polymorphisms in metabolic genes in healthy adults: Findings from the EPIC-Spain cohort. Carcinogenesis 2009, 30, 968–976. [Google Scholar] [CrossRef]
- Wang, L.N.; Wang, F.; Liu, J.; Ying-Hui, J.; Fang, C.; Ren, X.Q. CYP1A1 Ile462Val polymorphism is associated with cervical cancer risk in Caucasians not Asians: A Meta-Analysis. Front. Physiol. 2017, 8, 1081. [Google Scholar] [CrossRef]
- Haufroid, V.; Jakubowski, M.; Janasikb, B.; Ligocka, D.; Buchet, J.P.; Bergamaschi, E.; Manini, P.; Mutti, A.; Ghittorid, S.; Arande, M.; et al. Interest of genotyping and phenotyping of drug-metabolizing enzymes for the interpretation of biological monitoring of exposure to styrene. Pharmacogenetics 2002, 12, 691–702. [Google Scholar] [CrossRef][Green Version]
- Del Tredici, A.L.; Malhotra, A.; Dedek, M.; Espin, F.; Roach, D.; Zhu, G.D.; Voland, J.; Moreno, T.A. Frequency of CYP2D6 alleles including structural variants in the United States. Front. Pharmacol. 2018, 9, 305. [Google Scholar] [CrossRef]
- Windmill, K.F.; Gaedigk, A.; Hall, P.M.; Samaratunga, H.; Grant, D.M.; McManu, M.E. Localization of N-Acetyltransferases NAT1 and NAT2 in human tissues. Toxicol. Sci. 2000, 54, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Matejcic, M.; Vogelsang, M.; Wang, Y.; Parker, I.M. NAT1 and NAT2 genetic polymorphisms and environmental exposure as risk factors for oesophageal squamous cell carcinoma: A case-control study. BMC Cancer 2015, 15, 150. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, W.F.; Hiltunen, M.; Pirskanen, M.; Mannermaa, A.; Helisalmi, S.; Lehtovirta, M.; Alafuzoff, I.; Soininen, H. MPO and APO Eepsilon4 polymorphisms interact to increase risk for AD in Finnish males. Neurology 2000, 55, 1284–1290. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, W.F.; Rhees, J.; Maciejewski, D.; Paladino, T.; Sieburg, H.; Maki, R.A.; Masliah, E. Myeloperoxidase polymorphism is associated with gender specific risk for Alzheimer’s disease. Exp. Neurol. 1999, 155, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Mateo, I.; Sánchez-Juan, P.; Rodríguez-Rodríguez, E.; Infante, J.; Vázquez-Higuera, J.L.; García-Gorostiaga, I.; Berciano, J.; Combarros, O. Synergistic effect of heme oxygenase-1 and tau genetic variants on Alzheimer’s disease risk. Dement Geriatr. Cogn. Disord. 2008, 26, 339–342. [Google Scholar] [CrossRef]
- Möllsten, A.; Marklund, S.L.; Wessman, M.; Svensson, M.; Forsblom, C.; Parkkonen, M.; Brismar, K.; Groop, P.H.; Dahlquist, G. A functional polymorphism in the manganese superoxide dismutase gene and diabetic nephropathy. Diabetes 2007, 56, 265–269. [Google Scholar] [CrossRef]
- Honkura, Y.; Matsuo, H.; Murakami, S.; Sakiyama, M.; Mizutari, K.; Shiotani, A.; Yamamoto, M.; Morita, I.; Shinomiya, N.; Kawase, T.; et al. NRF2 Is a key target for prevention of noise-induced hearing loss by reducing oxidative damage of cochlea. Sci. Rep. 2016, 6, 19329. [Google Scholar] [CrossRef]
- Okano, Y.; Nezu, U.; Enokida, Y.; Lee, M.T.; Kinoshita, H.; Lezhava, A.; Hayashizaki, Y.; Morita, S.; Taguri, M.; Ichikawa, Y.; et al. SNP (-617C>A) in ARE-like loci of the NRF2 gene: A new biomarker for prognosis of lung adenocarcinoma in Japanese non-smoking women. PLoS ONE 2013, 8, e73794. [Google Scholar] [CrossRef]
- Teena, R.; Dhamodharan, U.; Ali, D.; Rajesh, K.; Ramkumar, K.M. Genetic Polymorphism of the Nrf2 promoter region (rs35652124) is associated with the risk of diabetic foot ulcers. Oxid. Med. Cell Longev. 2020, 2020, 9825028. [Google Scholar] [CrossRef]
- Liu, F.; Luo, L.; Wei, Y.; Wang, W.; Li, B.; Yan, L.; Wen, T. A functional NQO1 609C>T polymorphism and risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 2013, 34, 47–53. [Google Scholar] [CrossRef]
- Hsu, P.C.; Chen, C.C.; Tzeng, H.E.; Hsu, Y.N.; Kuo, C.C.; Lin, M.L.; Chang, W.S.; Wang, Y.C.; Tsai, C.W.; Pei, J.S.; et al. HOGG1 rs1052133 Genotypes and risk of childhood acute lymphoblastic leukemia in a taiwanese population. In Vivo 2019, 33, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, T.; Li, J.; Mo, Z. X-ray repair cross-complementing group 1 (XRCC1) Arg399Gln polymorphism significantly associated with prostate cancer. Int. J. Biol. Mark. 2015, 30, e12–e21. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.R.; Miller, M.S.; Lohman, K.; Lange, E.M.; Case, L.D.; Mohrenweiser, H.W.; Hu, J.J. Polymorphisms of XRCC1 and XRCC3 genes and susceptibility to breast cancer. Cancer Lett. 2003, 190, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Salimzadeh, H.; Lindskog, E.B.; Gustavsson, B.; Wettergren, Y.; Ljungman, D. Association of DNA repair gene variants with colorectal cancer: Risk, toxicity, and survival. BMC Cancer 2020, 20, 409. [Google Scholar] [CrossRef]
- Li, D.; Zhao, H.; Gelernter, J. Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biol. Psychiatry 2011, 70, 504–512. [Google Scholar] [CrossRef]
- Thomasson, H.; Beard, J.D.; Li, T.K. ADH2 gene polymorphisms are determinants of alcohol pharmacokinetics. Alcohol Clin. Exp. Res. 1995, 19, 1494–1499. [Google Scholar] [CrossRef]
- Xue, Y.; Wang, M.; Zhong, D.; Tong, N.; Chu, H.; Sheng, X.; Zhang, Z. ADH1C Ile350Val polymorphism and cancer risk: Evidence from 35 case-control studies. PLoS ONE 2012, 7, e37227. [Google Scholar] [CrossRef]
- Xu, X.-L.; Wang, J.; Zhu, S.-M.; Yang, M.; Fang, Y.; Qian, S.; An, Z.; Mao., W. Impact of alcohol dehydrogenase gene 4 polymorphisms on esophageal squamous-cell carcinoma risk in a Chinese population. J. Clin. Oncol. 2015, 33 (Suppl. S15), e15021. [Google Scholar] [CrossRef]
- Chang, B.; Hao, S.; Zhang, L.; Gao, M.; Sun, Y.; Huang, A.; Teng, G.; Li, B.; Crabb, D.W.; Kusumanchi, P.; et al. Association between aldehyde dehydrogenase 2 Glu504Lys polymorphism and alcoholic liver disease. Am. J. Med. Sci. 2018, 356, 10–14. [Google Scholar] [CrossRef]
- Denny, J.C.; Collins, F.S. Precision medicine in 2030-seven ways to transform healthcare. Cell 2021, 184, 1415–1419. [Google Scholar] [CrossRef]
- Velmovitsky, P.E.; Bevilacqua, T.; Alencar, P.; Cowan, D.; Morita, P.P. Convergence of precision medicine and public health into precision public health: Toward a big data perspective. Front. Public Health 2021, 9, 561873. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Shu, Y.; Cai, Y.D. Genetic differences among ethnic groups. BMC Genom. 2015, 16, 1093. [Google Scholar] [CrossRef] [PubMed]
- Esteban, M.; Castaño, A. Non-invasive matrices in human biomonitoring: A review. Environ. Int. 2009, 35, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Bergdahl, I.A.; Skerfving, S. Biomonitoring of lead exposure-alternatives to blood. J. Toxicol. Environ. Health A 2008, 71, 1235–1243. [Google Scholar] [CrossRef]
- Lum, A.; Le Marchand, L. A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer Epidemiol. Biomark. Prev. 1998, 7, 719–724. [Google Scholar]
- Feigelson, H.S.; Rodriguez, C.; Welch, R.; Hutchinson, A.; Shao, W.; Jacobs, K.; Diver, W.R.; Calle, E.E.; Thun, M.J.; Hunter, D.J.; et al. Successful genome-wide scan in paired blood and buccal samples. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1023–1025. [Google Scholar] [CrossRef]
- Mold, A.; Beccaria, F.; Berridge, V.; Eisenbach-Stangl, I.; Herczyńska, G.; Moskalewicz, J.; Petrilli, E.; Taylor, S. Concepts of addiction in Europe in the 1970s and 1980s: What does a long view tell us about drugs, alcohol, and tobacco? In Concepts of Addictive Substances and Behaviours across Time and Place; Hellman, M., Berridge, V., Duke, K., Mold, A., Eds.; Oxford University Press: Oxford, UK, 2016; pp. 15–32. [Google Scholar] [CrossRef]
- Ghatak, S.; Muthukumaran, R.B.; Nachimuthu, S.K. A simple method of genomic DNA extraction from human samples for PCR-RFLP analysis. J. Biomol. Tech. 2013, 24, 224–231. [Google Scholar] [CrossRef]
- National Human Genome Research Institute. Available online: https://www.genome.gov/ (accessed on 2 September 2022).
- Almazroo, O.A.; Miah, M.K.; Venkataramanan, R. Drug Metabolism in the liver. Clin. Liver Dis. 2017, 21, 1–20. [Google Scholar] [CrossRef]
- Kielholz, P. Alcohol and Depression. Br. J. Addict. Alcohol Other Drugs 1970, 65, 187–193. [Google Scholar] [CrossRef]
- Warner, K.E. Cigarette Smoking in the 1970’s: The Impact of the Antismoking Campaign on Consumption. Science 1981, 211, 729–731. [Google Scholar] [CrossRef]
- Hahn, M.; Roll, S.C. The Influence of pharmacogenetics on the clinical relevance of pharmacokinetic drug–drug interactions: Drug–gene, drug–gene–gene and drug–drug–gene interactions. Pharmaceuticals 2021, 14, 487. [Google Scholar] [CrossRef]
- Krainca, T.; Fuentesa, A. Genetic ancestry in precision medicine is reshaping the race debate. Proc. Natl. Acad. Sci. USA 2022, 119, e2203033119. [Google Scholar] [CrossRef] [PubMed]
- Traversi, D.; Pulliero, A.; Izzotti, A.; Franchitti, E.; Iacoviello, L.; Gianfagna, F.; Gialluisi, A.; Izzi, B.; Agodi, A.; Barchitta, M.; et al. Precision medicine and public health: New challenges for effective and sustainable health. J. Pers. Med. 2021, 11, 135. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.R.; Gaedigk, A. Precision medicine: Does ethnicity information complement genotype-based prescribing decisions? Ther. Adv. Drug Saf. 2018, 9, 45–62. [Google Scholar] [CrossRef] [PubMed]
| Polymorphism Function | Detoxification | European Allele Frequency | Effects on Enzyme Activity |
|---|---|---|---|
| Glutathione S Transferase | |||
| GST-T1 rs17856199 | Detoxification of xenobiotics, carcinogenic substances, therapeutic drugs, environmental toxins, and oxidative stress products. | GST-T1 A = 0.867 C = 0.183 | GST-M1 and GST-T1 homozygous deletions (pos/null) have a decreased ability to detoxify carcinogens, toxicants, and oxidative stress products. The gene deletion has been found in Caucasian and Asian populations compared to Africans [33]. |
| GST-M1 rs366631 | GST-M1 A = 0.488 G = 0.512 | ||
| GST-A1 rs3957357 | GST-A1 A = 0.571 G = 0.429 | The distributions of GSTA1-69C > T promoter haplotypes and diplotypes were significantly different among the human populations. The frequencies of GSTA1-69C > T polymorphism were similar to those of the African American population and the populations with White ancestry, but significantly different from those reported for the populations with Asian ancestry [34]. | |
| GST-P1 rs1695 | GST-P1 A = 0.669 G = 0.331 | Several papers published findings associating GSTP1 Ile105Val genotypes with bronchial, childhood, or atopic asthma. Compared with the AA genotype, the GA + GG genotype decreased lung cancer susceptibility [35]. | |
| Epoxide hydrolase | |||
| EPHX1-Ex_3 rs1051740 | Biotransformation enzymes converting epoxides from the degradation of aromatic compounds to trans-dihydrodiols excreted from the body. EPHX1 was shown to take part in protection against oxidative stress. | EPHX1-Ex3: T = 0.696 C = 0.304 | In vitro polymorphisms in exons 3 (His113Tyr) and 4 (Arg139His) lead to reduced activity (slow allele) and increased activity (fast allele). T to C substitution (slow allele) reduces the enzyme activity. Decreased activity (histidine) A to G substitution (fast allele) increases the enzyme activity. Increased activity (arginine) [36]. |
| EPHX1-Ex_4 rs2234922 | EPHX1-Ex4: A = 0.836 G = 0.164 | ||
| Cytochrome P450 family | |||
| CYP1A1_2A rs4646903 | Catalysis of reactions involved in the drug metabolism and synthesis of cholesterol, steroids, and lipids, some of which are found in cigarette smoke. The enzyme’s endogenous substrate is able to metabolize some polycyclic aromatic hydrocarbons into carcinogenic intermediates. | CYP1A1-2A: A = 0.893 G = 0.107 | The CYP1A1 Ile462Val polymorphism may enhance the susceptibility to cervical cancer in Caucasian females. The gene has been associated with lung cancer risk. Higher inducibility. Increased oxidation [37]. |
| CYP1A1_2C rs1048943 Ile462Val | CYP1A1-2C: T = 0.965 C = 0.035 | ||
| Cytochrome P450 2E1 | |||
| CYP2E1*5B rs3813867 | Cytochrome P450 2E1 (CYP2E1) is one of the major enzymes involved in the metabolism and detoxification of various drugs and xenobiotics. | CYP2E1*5B: G = 0.959 A = 0.041 | The genotype distributions of CYP2E1*5B and *6 were similar to the Caucasian population but were different from East Asian populations. Wang, L.N.; Wang, F.; Liu, J.; Ying-Hui, J.; Fang, C.; Ren, X.Q. CYP1A1 Ile462Val polymorphism is associated with cervical cancer risk in Caucasians not Asians: A Meta-Analysis. Front. Physiol. 2017, 8, 1081. https://doi.org/10.3389/fphys.2017.01081. [38] |
| CYP2E1*6 rs6413432 | CYP2E1*6: T = 0.908 A = 0.091 | CYP2E1*6 polymorphism causes a reduction in CYP2E1 enzyme activity [39]. | |
| Cytochrome P450 family 2 subfamily D6 and Cytochrome P450 family 2 subfamily A6 | |||
| CYP2D6 rs16947 | CYP2D6 is responsible for the oxidative metabolism of 20–25% of drugs. | CYP2D6: G = 0.710 A = 0.290 | Unfunctional alleles represent 26% of the variability mainly in CYP2D6*4. “Frequency of CYP2D6 Alleles Including Structural Variants in the United States” Del Tredici, A.L.; Malhotra, A.; Dedek, M.; Espin, F.; Roach, D.; dan Zhu, Guang.; Voland, J.; Moreno, T.;A. Front. Pharmacol., 2018 Pharmacogenetics and Pharmacogenomics [40] |
| CYP2A6 rs1801272 | Nicotine metabolism is mediated primarily by cytochrome CYP2A6. The genetic variation in this gene has been linked with several smoking behavior phenotypes. | CYP2A6: A = 0.973 T = 0.026 | The frequencies of this allele vary considerably among different ethnic populations, the deletion alleles being most common in Oriental people (up to 20%). Studies of Japanese populations suggest that CYP2A6 poor metabolizer genotypes result in altered nicotine kinetics and may lower cigarette smoking-elicited lung cancer risk, whereas similar studies in Caucasian populations have not revealed any clear associations between variant CYP2A6 genotypes and smoking behavior or lung cancer predisposition [41]. |
| N-Acetyl Transferase | |||
| NAT1 rs4987076 | N-acetyltransferase 1 detoxifies many drugs and chemicals found in the environment eliminated from the body or bioactivated to metabolites causing toxicity/cancer. NAT1 activity is regulated by genetic polymorphisms as well as environmental factors such as substrate-dependent down-regulation and oxidative stress. | NAT-1: G = 0.974 A = 0.025 | NAT-1: a monomorphic form of the enzyme [42]. |
| NAT2 rs1208 | N-acetyltransferase 2 enzyme detoxifies xenobiotics (carcinogens and drugs). Variation at the NAT2 gene has been linked to the human acetylation capacity: slow, intermediate, and fast, which modifies susceptibility to cancer and adverse drug reactions. | NAT-2: G = 0.434 A = 0.565 | Multiple NAT2 alleles (NAT2*5,*6, *7, and *14) have substantially decreased acetylation activity and are common in Caucasian people and populations of African descent. Link between acetylator phenotype and increased risk for bladder and colon cancer [43]. |
| Oxidative Stress | |||
| MPO Myeloperoxidase rs2333227 | Oxidoreductase catalyzing H2O2 to H2O. | MPO: C = 0.910 T = 0.089 | A study of 127 Finnish patients concluded that the rs2333227 allele increased the risk of Alzheimer’s disease [44]. In a further study polymorphism of MPO was statistically associated with AD in a gender-specific manner. Myeloperoxidase polymorphism is associated with a gender-specific risk for Alzheimer’s disease [45]. |
| Heme Oxygenase rs2071746 -413AT | Cytoprotective enzyme activated during cellular stress such as inflammation, ischemia, hypoxia, hyperoxia, and radiation. | HO1: A = 0.440 G = 0.560 | In the Ala16Val, the Ala amino acid seems to be more favorable than the Val amino acid. Higher risk for Alzheimer’s disease (TT) [46]. |
| SOD2 rs4880 | Mitochondrial enzyme with a key role in protecting the cell from oxidative damage. | SOD: A = 0.502 G = 0.498 | Ala16Val: The Val amino acid is less favorable. Association of smoking and homozygosity for the MnSOD Val allele contributing to an increased risk of diabetic nephropathy [47]. |
| NRF2 rs6721961 | Regulator of the cell transcriptional response to oxidative stress induced by exposure to xenobiotics. | NRF2 rs6721961 G = 0.875 (wt) T = 0.125 (mut) | The “G” and “T” alleles resulted in higher and lower expressions of NRF2 mRNA, showing that the G allele is beneficial for protection from pathologies. In contrast, the “T” allele of rs6721961 significantly increases susceptibility to the elevation of the hearing threshold at 4 kHz in the occupational setting [48]. NRF2 (rs6721961) (-617A/A) alleles in the NRF2 gene have been associated with female non-smokers with adenocarcinoma and are regarded as a prognostic biomarker for assessing the overall survival of patients with lung adenocarcinoma [49]. “C” indicates the wild allele, and “T” indicates the mutant allele [50]. |
| NRF2 rs6706649 | NRF2 rs6706649 C = 0.882 T = 0.118 | ||
| NRF2 rs35652124 | NRF2 rs35652124 C = 0.327 T = 0.672 | ||
| NQO1 rs1800566 | Enzyme involved in preventing free-redox radical generation. | NQO1 rs1800566: G = 0.809 A = 0.190 | The variant enzyme, C609T, is ubiquitinated by the proteasome (greater risk for 609TT). Susceptibility risk for hepatocellular carcinoma [51]. |
| NQO1 rs1131341 | (C465T, Arg139Trp) is a polymorphism within NQO1 (NAD(P)H dehydrogenase (quinone 1) | NQO1 rs1131341 G = 0.961 A = 0.038 | The variant enzyme C465T shows reduced enzyme activity. Potential risk of (ALL) Acute Lymphoblastic Leukemia (AML) Acute Myeloid Leukemia [52]. |
| DNA damage Repair | |||
| hOGG1 rs1052133 | The human 8-oxoG DNA glycosylase 1 plays a central role in repairing 8-oxoGs via the base excision repair pathway. Evidence suggests that hOGG1 affects the activity as a genetic marker for the prediction of personal susceptibility to several cancers. | hOGG1 C = 0.778 G = 0.221 | In solid tumors, the genotypes CG and/or GG at hOGG1 rs1052133 have been reported to be associated with an increased risk of various types of cancer. The polymorphic site of hOGG1, i.e., rs1052133 (Ser326Cys), C to G showed that the glycosylase activity of the “G” variant is more sensitive to inactivation by oxidizing agents than that of the “C” wild-type, and cells carrying the “G” allele may accumulate mutations more readily under oxidative stress [53]. |
| XRCC1 rs25487 Arg399Gln | The protein encoded by XRCC1 is involved in the repair of DNA single-strand breaks formed by exposure to ionizing radiation and alkylating agents. | XRCC1 C = 0.642 T = 0.357 | XRCC1 has protective gene polymorphisms due to the enhanced repair activity even though the Arg399Gln polymorphism (rs25487 and rs1799782 Arg194Trp) have been reported to be related to prostate cancer [54]. |
| rs1799782 Arg194Trp | G = 0.937 A = 0.062 | ||
| XRCC3 rs861539 | The gene is involved in the homologous recombination repair pathway (HRR) of double-stranded DNA, deputed to repair chromosomal fragmentation, translocations, and deletions. | XRCC3 G = 0.617 A = 0.382 | XRCC3 promotes the homology-directed repair of DNA damage in mammalian cells. Three amino acid substitution variants of DNA repair genes (XRCC1 Arg194Trp, XRCC1 Arg399Gln, and XRCC3 Thr241Met) showed an association with breast cancer susceptibility. Polymorphisms of XRCC1 and XRCC3 genes and susceptibility to breast cancer [55]. |
| XPD/ERCC2 rs13181 | The ERCC2 contributes to the synthesis of XPD protein. This is an essential subunit of a group of proteins known as the TFIIH complex with two major functions: the activation of gene transcription and the repair of damaged DNA. | XPD/ERCC2 T = 0.626 G = 0.373 | XPD protein is involved in transcription initiation and in the control of the cell cycle and apoptosis. The ERCC2-rs13181 C allele was associated with a significantly increased risk of colorectal cancer risk [56]. |
| Alcohol dehydrogenase | |||
| ADH1B rs1229984 Arg 48 His | Enzymes involved in alcohol metabolism. The functional variant, Arg48His (rs1229984), is located in the ADH1B gene and protects against alcohol dependence. | ADH1B G = 0.951 A = 0.048 | ADH1B has been reported to associate with reduced rates of alcohol and drug dependence. The allele with increased activity, meaning the more rapid oxidation of ethanol to acetaldehyde, is His48, encoded by rs1229984. Carriers with one or two ADH alleles, such as (G/A) or (A/A) have a reduced risk for alcoholism. These findings support that the His allele can greatly lower the risk of AD and alcohol abuse and provide strong evidence for the involvement of the ADH1B gene in the pathogenesis of AD and alcohol-induced diseases in particular in multiple populations such as Asian populations [57]. |
| ADH1B rs2066702 Arg370Cys | ADH1B G = 0.997 A = 0.002 | ADH1B Arg370Cys is monomorphic in European populations, but it has been shown to have a small effect on the rate of alcohol elimination in African Americans [58]. | |
| ADH1C rs698 Ile350Val | The rs698(A) allele is the most common, encoding isoleucine; the rare (T) allele encodes the variant phenylalanine | ADH1C A = 0.609 T = 0.391 | An increasing number of studies have investigated the association between ADH polymorphisms and cancer risk in humans. AA normal enzyme activity AT-TT reduction in the enzyme activity and intolerance to ethanol. Among them, studies of the ADH1C Ile350Val (rs698) variant accounted for more than others. The results indicate that ADH1C Ile350Val polymorphism may contribute to cancer risk among African populations and Asian populations [59]. |
| ADH4 rs3805322 | The alcohol dehydrogenase (ADH) family represents one of the key sets of enzymes responsible for the oxidation of alcohol. This enzyme is an important member of this family, it is a functional candidate for alcohol dependence in human consumption of alcohol. | ADH4 A = 0.998 G = 0.003 | A significantly increased risk of developing esophageal squamous-cell carcinoma was revealed in subjects with the AA genotype for rs3805322 (ADH4) compared with those with the AG or GG genotype [60]. |
| Aldehyde dehydrogenase | |||
| ALDH2 rs671 Glu504Lys | Aldehyde dehydrogenases efficiently oxidize and, in most instances, detoxifies a significant number of chemical aldehydes which otherwise would be harmful to the organism. | ALDH G = 1.000 | Individuals with the ALDH2 504Lys variant were less associated with alcoholic liver disease compared to those with ALDH2 504Glu by genotypic and allelic analyses [61]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiarella, P.; Capone, P.; Sisto, R. Contribution of Genetic Polymorphisms in Human Health. Int. J. Environ. Res. Public Health 2023, 20, 912. https://doi.org/10.3390/ijerph20020912
Chiarella P, Capone P, Sisto R. Contribution of Genetic Polymorphisms in Human Health. International Journal of Environmental Research and Public Health. 2023; 20(2):912. https://doi.org/10.3390/ijerph20020912
Chicago/Turabian StyleChiarella, Pieranna, Pasquale Capone, and Renata Sisto. 2023. "Contribution of Genetic Polymorphisms in Human Health" International Journal of Environmental Research and Public Health 20, no. 2: 912. https://doi.org/10.3390/ijerph20020912
APA StyleChiarella, P., Capone, P., & Sisto, R. (2023). Contribution of Genetic Polymorphisms in Human Health. International Journal of Environmental Research and Public Health, 20(2), 912. https://doi.org/10.3390/ijerph20020912








