Characterisation of Salmonella Enteritidis ST11 and ST1925 Associated with Human Intestinal and Extra-Intestinal Infections in Singapore
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Salmonella Isolates
2.2. Genomic DNA Extraction for Whole Genome Sequencing (WGS) and Analysis
2.3. Phenotypic Antimicrobial Susceptibility Testing
2.4. Statistical Calculation and Analysis
3. Results
3.1. Phenotypic Antimicrobial Resistance (Broth Micro Dilution)
3.2. Genotypic Antimicrobial Resistance, Virulence, SPI and Plasmid Profiles
3.3. Phylogenetic Analysis
4. Discussion
4.1. Sequence Type in S. Enteritidis Isolates
4.2. Antimicrobial Resistance Phenotype in S. Enteritidis Isolates
4.3. Phylogenetic Relation between Some Human, Food and Farm/Slaughterhouse Isolates
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Afshari, A.; Baratpour, A.; Khanzade, S.; Jamshidi, A. Salmonella Enteritidis and Salmonella Typhimurium identification in poultry carcasses. Iran. J. Microbiol. 2018, 10, 45–50. [Google Scholar] [PubMed]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef] [PubMed]
- Guard-Petter, J. The chicken, the egg and Salmonella Enteritidis. Environ. Microbiol. 2001, 3, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Feasey, N.A.; Dougan, G.; Kingsley, R.A.; Heyderman, R.S.; Gordon, M.A. Invasive non-typhoidal salmonella disease: An emerging and neglected tropical disease in Africa. Lancet 2012, 379, 2489–2499. [Google Scholar] [CrossRef]
- Oneko, M.; Kariuki, S.; Muturi-Kioi, V.; Otieno, K.; Otieno, V.O.; Williamson, J.M.; Folster, J.; Parsons, M.B.; Slutsker, L.; Mahon, B.E.; et al. Emergence of Community-Acquired, Multidrug-Resistant Invasive Nontyphoidal Salmonella Disease in Rural Western Kenya, 2009–2013. Clin. Infect. Dis. 2015, 61, S310–S316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanaway, J.D.; Parisi, A.; Sarkar, K.; Blacker, B.F.; Reiner, R.C.; Hay, S.I.; Nixon, M.R.; Dolecek, C.; James, S.L.; Mokdad, A.H.; et al. The global burden of non-typhoidal salmonella invasive disease: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019, 19, 1312–1324. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, S.D.; Flores, F.S.; dos Santos, L.R.; Brandelli, A. Antimicrobial resistance in Salmonella Enteritidis strains isolated from broiler carcasses, food, human and poultry-related samples. Int. J. Food Microbiol. 2005, 97, 297–305. [Google Scholar] [CrossRef]
- Rodríguez, I.; Rodicio, M.R.; Guerra, B.; Hopkins, K.L. Potential International Spread of Multidrug-Resistant InvasiveSalmonella entericaSerovar Enteritidis. Emerg. Infect. Dis. 2012, 18, 1173–1176. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available online: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 16 March 2018).
- Yan, S.S.; Pendrak, M.L.; Abela-Ridder, B.; Punderson, J.W.; Fedorko, D.P.; Foley, S.L. An overview of Salmonella typingPublic health perspectives. Clin. Appl. Immunol. Rev. 2004, 4, 189–204. [Google Scholar] [CrossRef]
- Minor, L. The Genus Salmonella. In The Prokaryotes an Evolving Electronic Resource for the Microbiological Community, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Eds.; Springer: New York, NY, USA, 2013; p. 314. [Google Scholar]
- Vestby, L.; Moretro, T.; Langsrud, S.; Heir, E.; Nesse, L. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal-and feed factories. BMC Vet. Res. 2009, 5, 20. [Google Scholar] [CrossRef] [Green Version]
- Davies, R.H.; Wray, C. Seasonal Variations in the Isolation of Salmonella typhimurium, Salmonella Enteritidis, Bacillus cereus and Clostridium perfringens from Environmental Samples. J. Veter- Med. Ser. B 1996, 43, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Cozorici, D.; Măciucă, R.-A.; Stancu, C.; Tihăuan, B.-M.; Uță, R.B.; Codrea, C.I.; Matache, R.; Pop, C.-E.; Wolff, R.; Fendrihan, S. Microbial Contamination and Survival Rate on Different Types of Banknotes. Int. J. Environ. Res. Public Health 2022, 19, 4310. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.; Chiu, T.; Pang, J.; Hsuan-Yuan, C.; Chang, G.; Tsen, H. Characterisation of antimicrobial resistatnce patterns and class 1 integrons among Escherichia coli and Salmonella enterica serovar Choleraesuis strains isolated from humans and swine in Taiwan. Int. J. Antimicrob. Agents 2006, 27, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zhang, Y.; Zhang, J.; Xue, C.; Yan, J.; Li, X.; Zheng, X.; Dong, R.; Bai, J.; Su, Y.; et al. Analysis of antibiotic-induced drug resistance of Salmonella Enteritidis and its biofilm formation mechanism. Bioengineered 2021, 12, 10254–10263. [Google Scholar] [CrossRef]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Aung, K.T.; Khor, W.C.; Octavia, S.; Ye, A.; Leo, J.; Chan, P.P.; Lim, G.; Wong, W.K.; Tan, B.Z.Y.; Schlundt, J.; et al. Distribution of Salmonella Serovars in Humans, Foods, Farm Animals and Environment, Companion and Wildlife Animals in Singapore. Int. J. Environ. Res. Public Health 2020, 17, 5774. [Google Scholar] [CrossRef]
- Ehuwa, O.; Jaiswal, A.; Jaiswal, S. Salmonella, Food Safety and Food Handling Practices. Foods 2021, 10, 907. [Google Scholar] [CrossRef]
- Ministry of Health Singapore. Communicable Diseases Surveillance in Singapore 2016. Available online: https://www.moh.gov.sg/docs/librariesprovider5/resources-statistics/reports/full-version.pdf (accessed on 7 December 2019).
- Lin, Y.; Fong, R.; Leo, J.; Kwan, W.; Ye, A.; Chan, P.; Chong, N.; Lim, G.; Octavia, S.; Lin, M.; et al. Distribution of Salmonella spp. along the food chain in Singapore, 2008–2016. ENB Q. 2019, 45, 44–54. [Google Scholar]
- Aung, K.; Chau, M.; Mak, K.; Lim, N.; Oh, J.; Kang, J.; Lim, Y.; Goh, T.; Yap, H.; Gutierrez, R.; et al. Microbiological assessment of chicken meat sold at chicken rice stalls in Singapore. Southeast Asian J. Trop. Med. Public Health 2018, 49, 1043–1052. [Google Scholar]
- Octavia, S.; Ang, M.; La, M.; Zulaina, S.; Saat, Z.; Tien, W.; Han, H.; Ooi, P.; Cui, L.; Lin, R. Retrospective genome-wide comparisons of Salmonella enterica serovar Enteritidis from suspected outbreaks in Singapore. Infect. Genet. Evol. 2018, 61, 229–233. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilm, A.; Aw, P.; Bertrand, D.; Yeo, G.; Ong, S.; Wong, C.; Khor, C.; Petric, R.; Hibberd, M.; Nagarajan, N. LoFreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012, 40, 11189–11201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.; Dehal, P.; Arkin, A. FastTree 2—approximately Maximum-Likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.; Zhu, H.; Guan, Y.; Lam, T.-Y. ggtree: An R package for visualisation and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Inouye, M.; Dashnow, H.; Raven, L.-A.; Schultz, M.; Pope, B.; Tomita, T.; Zobel, J.; Holt, K. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014, 6, 90–106. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421–430. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Gupta, S.; Padmanabhan, B.; Diene, S.; Lopez-Rojas, R.; Kempf, M.; Landraud, L. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Yin, Y.; Jones, M.; Zhang, Z.; Deatherage Kaiser, B.; Dinsmore, B.; Fitzgerald, C.; Fields, P.; Deng, X. Salmonella serotype determination utilizing high-throughput genome sequencing data. J. Clin. Microbiol. 2015, 53, 1685–1692. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Integrated Surveillance of Antimicrobial Resistance in Food-Borne Bacteria: Application of a One Health Approach; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Food and Agriculture Organization of the United Nations. Monitoring and surveillance of antimicrobial resistance in bacteria from healthy food animals intended for consumption. In Regional Antimicrobial Resistance Monitoring and Surveillance Guidelines; Food and Agriculture Organization of the United Nations: Bangkok, Thailand, 2019; Volume 1. [Google Scholar]
- Britto, C.; John, J.; Verghese, V.; Valsan, P.; Pollard, A. A systematic review of antimicrobial resistance of typhoidal Salmonella in India. Indian J. Med. Res. 2019, 149, 151–163. [Google Scholar] [CrossRef]
- Han, J.; Lynne, A.; David, D.; Tang, H.; Xu, J.; Nayak, R.; Kaldhone, P.; Logue, C.; Foley, S. DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype Heidelberg isolates. PLoS ONE 2012, 7, e51160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khajanchi, B.; Hasan, N.; Choi, S.; Han, J.; Zhao, S.; Colwell, R.; Cerniglia, C.; Foley, S. Comparative genomic analysis and characterisation of incompatibility group FIB plasmid encoded virulence factors of Salmonella enterica isolated from food sources. BMC Genom. 2017, 18, 570–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sethuvel, D.; Anandan, S.; Ragupathi, N.; Gajendiran, R.; Kuroda, M.; Shibayama, K.; Veeraraghavan, B. IncFII plasmid carrying antimicrobial resistance genes in Shigella flexneri: Vehicle for dissemination. J. Glob. Antimicrob. Resist. 2019, 16, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Mayer, L. Use of plasmid profiles in epidemiologic surveillance of disease outbreaks and in tracing the transmission of antibiotic resistance. Clin. Microbiol. Rev. 1988, 1, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Dziewit, L.; Pyzik, A.; Szuplewska, M.; Matlakowska, R.; Mielnicki, S.; Wibberg, D.; Schluter, A.; Puhler, A.; Bartosik, D. Diversity and role of plasmids in adaptation of bacteria inhabiting the Lubin copper mine in Polan, an environment rich in heavy metals. Front. Microbiol. 2015, 6, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsen, J.; Skov, M.; Threlfall, E.; Brown, D. Clonal lines of Salmonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field gel electrophoresis and RFLP typing. J. Med. Microbiol. 1994, 40, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Mezal, E.; Sabol, A.; Khan, M.; Ali, N.; Stefanova, R.; Khan, A. Isolation and molecular characterization of Salmonella enterica serovva Enteritidis from poultry house and clinical samples during 2010. Food Microbiol. 2014, 38, 67–74. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and foodborne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar]
- Centers for Disease Control and Prevention. National Enteric Disease Surveillance: Salmonella Annual Reports; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2016. Available online: https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf (accessed on 20 March 2020).
- Zakaria, Z.; Hassan, L.; Sharif, Z.; Ahmad, N.; Ali, R.; Husin, S.; Hazis, N.; Sohaimi, N.; Bakar, S.; Garba, B. Analysis of Salmonella enterica serovar Enteritidis isolates from chickens and chicken meat products in Malaysia using PFGE, and MLST. BMC Vet. Res. 2020, 16, 393. [Google Scholar] [CrossRef]
- Zakaria, Z.; Hassan, L.; Sharif, Z.; Ahmad, N.; Ali, R.; Husin, S.; Sohaimi, N.; Bakar, S.; Garba, B. Virulence gene profile, antimicrobial resistance and multilocus sequence typing of Salmonella enterica subsp. enterica serovar Enteritidis from chickens and chicken products. Animals 2022, 12, 97. [Google Scholar] [CrossRef]
- Toro, M.; Retamal, P.; Ayers, S.; Barreto, M.; Allard, M.; Brown, E.; Gonzalez-Escalona, N. Whole-genome sequencing analysis of Salmonella enterica serovar Enteritidis isolates in Chile provides insights into possible transmission between gulls, poultry and humans. Appl. Environ. Microbiol. 2016, 82, 6223–6232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, R.; Hiley, L.; Rathnayake, I.; Jennison, A. Comparative genomics identifies distinct lineages of S. Enteritidis from Queensland, Australia. PLoS ONE 2018, 13, e0191042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fandino, L.; Verjan-Garcia, N. A common Salmonella Enteritidis sequence type from poultry and human gastroenteritis in Ibagué, Colombia. Biomédica 2019, 39, 50–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patchanee, P.; Boonkhot, P.; Kittiwan, N.; Tadee, P.; Chotinun, S. Dissemination of Salmonella enterica sequence types among ASEAN economic community countries. Southeast Asian J. Trop. Med. Public Health 2015, 46, 707–719. [Google Scholar] [PubMed]
- Kariuki, S.; Mbae, C.; Onsare, R.; Kavai, S.; Wairimu, C.; Ngetich, R.; Ali, M.; Clemens, J.; Dougan, G. Multidrug-resistant nontyphoidal Salmonella hotspots as targets for vaccine use in management of infections in endemic settings. Clin. Infect. Dis. 2019, 68, S10–S15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aung, K.; Chen, H.; Chau, M.; Yap, G.; Lim, X.; Humaidi, M.; Chua, C.; Yeo, G.; Yap, H.; Oh, J.; et al. Salmonella in retail food and wild birds in Singapore-prevalence, antimicrobial resistance, and sequence types. Int. J. Environ. Res. Public Health 2019, 16, 4235–4247. [Google Scholar] [CrossRef] [Green Version]
- Gu, D.; Wang, Z.; Tian, Y.; Kang, X.; Meng, C.; Chen, X.; Pan, Z.; Jiao, X. Prevalence of Salmonella isolates and their distribution based on whole-genome sequence in a chicken slaughterhouse in Jiangsu, China. Front. Vet. Sci. 2020, 7, 29–39. [Google Scholar] [CrossRef]
- Lan, N.; Phuong, T.; Huu, H.; Thuy, L.; Mather, A.; Park, S.; Marks, F.; Thwaites, G.; Chau, N.; Thompson, C.; et al. Invasive non-typhoidal Salmonella infections in Asia: Clinical observations, disease outcome and dominant serovars from an infectious disease hospital in Vietnam. PLoS Negl. Trop. Dis. 2016, 10, e0004857. [Google Scholar] [CrossRef] [Green Version]
- European Centre for Disease Prevention and Control; European Food Safety Authority. Multi-country outbreak of Salmonella Enteritidis sequence type (ST)11 infections linked to eggs and egg products–8 February 2022. Jt. ECDC-EFSA Rapid Outbreak 2022, 19, 7180E. [Google Scholar]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X.; Fang, W.; Yue, M. Antibiotic resistance in Salmonella Typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 2019, 10, 985–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granados-Chinchilla, F.; Rodriguez, C. Tetracyclines in food and feeding stuffs: From regulation to analytical methods, bacterial resistance, and environmental and health implications. J. Anal. Methods Chem. 2017, 2017, 1315497. [Google Scholar] [CrossRef] [PubMed]
- Asperilla, M.; Smego, R.J.; Scott, L. Quinolone antibiotics in the treatment of Salmonella infections. Rev. Infect. Dis. 1990, 12, 873–889. [Google Scholar] [CrossRef] [PubMed]
- Sirinavin, S.; Thavornnunth, J.; Sakchainanont, B.; Bangtrakulnonth, A.; Chongthawonsatid, S.; Junumporn, S. Norfloxacin and azithromycin for treatment of nontyphoidal Salmonella carriers. Clin. Infect. Dis. 2003, 37, 685–691. [Google Scholar] [CrossRef] [Green Version]
- Kaur, J. Increasing antimicrobial resistance and narrowing therapeutics in typhoidal salmonellae. J. Clin. Diagn. Res. 2013, 7, 576–579. [Google Scholar] [CrossRef]
- Singapore Food Agency. AVA Annual Report. Available online: https://www.sfa.gov.sg/files/annualreport/AR2018-19/ (accessed on 20 March 2020).
- Centers for Disease Control and Prevention. Salmonella Enteritidis Infections Linked to Raw, Frozen, Stuffed Chicken Entrees (Final Update). Available online: https://www.cdc.gov/salmonella/frozen-chicken-entrees-part2-07-15/index.html (accessed on 24 March 2020).
- Thung, T.; Mahyudin, N.; Basri, D.; Wan Mohamed Radzi, C.; Nakaguchi, Y.; Nishibuchi, M.; Radu, S. Prevalence and antibiotic resistance of Salmonella Enteritidis and Salmonella Typhimurium in raw chicken meat at retail markets in Malaysia. Poulry Sci. 2016, 95, 1888–1893. [Google Scholar] [CrossRef]
- Campioni, F.; Cao, G.; Kastanis, G.; Janies, D.; Bergamini, A.; dos Prazeres Rodrigues, D.; Stones, R.; Brown, E.; Allard, M.; Falcao, J. Changing of the genomic pattern of Salmonella Enteritidis strains isolated in Brazil over a 48 year-period revealed by whole genome SNP analyses. Sci. Rep. 2018, 8, 10478–10485. [Google Scholar] [CrossRef]
- Taylor, A.; Lappi, V.; Wolfgang, W.; Lapierre, P.; Palumbo, M.; Medus, C.; Boxrud, D. Characterisation of foodborne outbreaks of Salmonella enterica serovar Enteritidis with whole-genome sequencing single nucleotide polymorphism-based analysis for surveillance and outbreak detection. J. Clin. Microbiol. 2015, 53, 3334–3340. [Google Scholar] [CrossRef] [Green Version]
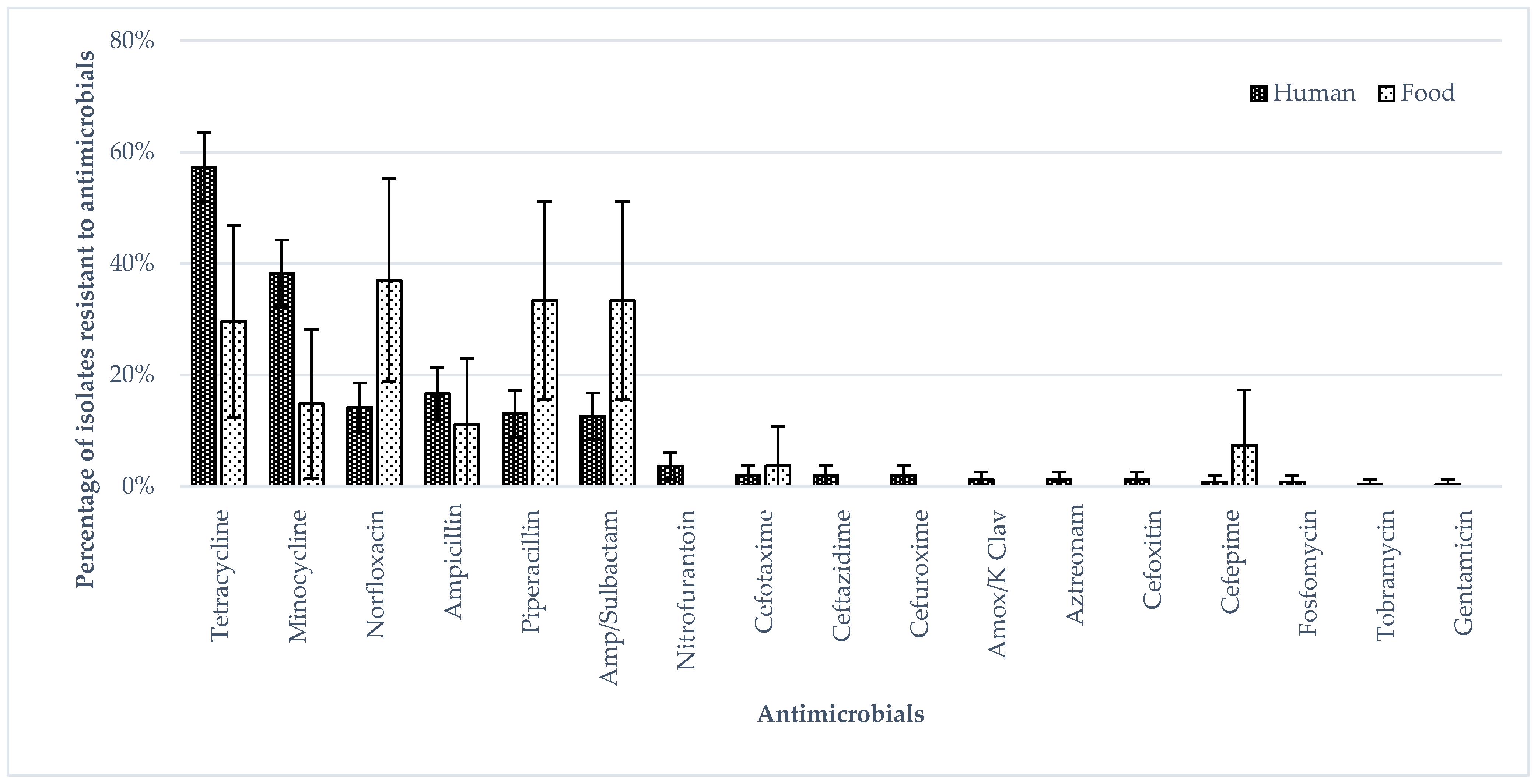

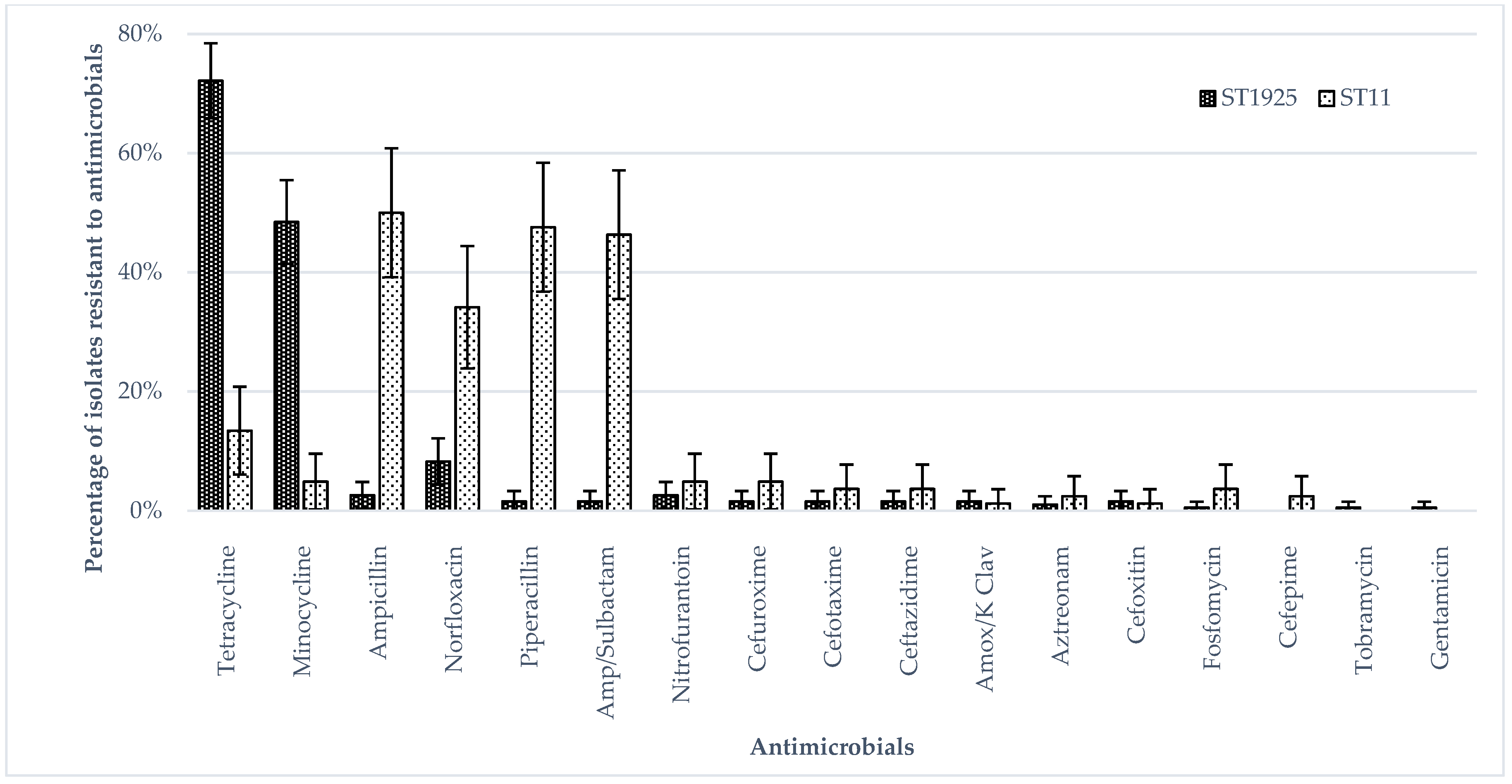

| Types of Samples | Isolates (n) | ST1925 (n) | ST11 (n) |
| Human | 246 | 184 | 62 |
| Human (intestinal) | 171 | 135 | 36 |
| Human (extra-intestinal) | 75 | 49 | 26 |
| Food | 27 | 8 | 19 |
| Chicken meat | 23 | 6 | 17 |
| Cooked or ready-to-eat food | 2 | 1 | 1 |
| Duck meat | 2 | 1 | 1 |
| Farm and slaughter house environment | 3 | 2 | 1 |
| Environmental swab (drag swab/farm) | 2 | 2 | 0 |
| Water (ice block/slaughter house) | 1 | 0 | 1 |
| Human (n = 246) | Food (n = 27) | Animal (n = 3) | ||||||
|---|---|---|---|---|---|---|---|---|
| Intestinal (n = 171) | Extra-Intestinal (n = 75) | Chicken (n = 23) | Non-Chicken (n = 4) | Environmental Swab (n = 2) | Water (n = 1) | Total | ||
| Aminoglycosides | Aac6-Iaa_AGly | 171 | 75 | 22 | 4 | 2 | 1 | 276 |
| Aac3-Iva_AGly | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| AadA_AGly | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Aph4-Ia_AGly | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Tetracycline | tetA_tet | 108 * | 33 | 5 | 3 | 2 | 1 | 153 |
| Beta-lactam | TEM-1D_bla | 12 | 16 * | 7 | 1 | 0 | 0 | 36 |
| CMY_bla | 2 | 0 | 0 | 0 | 1 | 0 | 3 | |
| CTX-M-1_bla | 1 | 1 | 0 | 0 | 0 | 0 | 2 | |
| Polymyxin | mcr1_colistin | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Fluoroquinolone | qnr-S_flq | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Folate pathway inhibitor | SulII_sul | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| SulIII_Sul | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aung, K.T.; Khor, W.C.; Ong, K.H.; Tan, W.L.; Wong, Z.N.; Oh, J.Q.; Wong, W.K.; Tan, B.Z.Y.; Maiwald, M.; Tee, N.W.S.; et al. Characterisation of Salmonella Enteritidis ST11 and ST1925 Associated with Human Intestinal and Extra-Intestinal Infections in Singapore. Int. J. Environ. Res. Public Health 2022, 19, 5671. https://doi.org/10.3390/ijerph19095671
Aung KT, Khor WC, Ong KH, Tan WL, Wong ZN, Oh JQ, Wong WK, Tan BZY, Maiwald M, Tee NWS, et al. Characterisation of Salmonella Enteritidis ST11 and ST1925 Associated with Human Intestinal and Extra-Intestinal Infections in Singapore. International Journal of Environmental Research and Public Health. 2022; 19(9):5671. https://doi.org/10.3390/ijerph19095671
Chicago/Turabian StyleAung, Kyaw Thu, Wei Ching Khor, Kar Hui Ong, Wei Ling Tan, Zhi Ning Wong, Jia Quan Oh, Wai Kwan Wong, Brian Zi Yan Tan, Matthias Maiwald, Nancy Wen Sim Tee, and et al. 2022. "Characterisation of Salmonella Enteritidis ST11 and ST1925 Associated with Human Intestinal and Extra-Intestinal Infections in Singapore" International Journal of Environmental Research and Public Health 19, no. 9: 5671. https://doi.org/10.3390/ijerph19095671
APA StyleAung, K. T., Khor, W. C., Ong, K. H., Tan, W. L., Wong, Z. N., Oh, J. Q., Wong, W. K., Tan, B. Z. Y., Maiwald, M., Tee, N. W. S., Barkham, T., Koh, T. H., Dalsgaard, A., Chen, S. L., Schlundt, J., & Ng, L. C. (2022). Characterisation of Salmonella Enteritidis ST11 and ST1925 Associated with Human Intestinal and Extra-Intestinal Infections in Singapore. International Journal of Environmental Research and Public Health, 19(9), 5671. https://doi.org/10.3390/ijerph19095671







