Microbial Degradation of Azo Dyes: Approaches and Prospects for a Hazard-Free Conversion by Microorganisms
Abstract
:1. Introduction
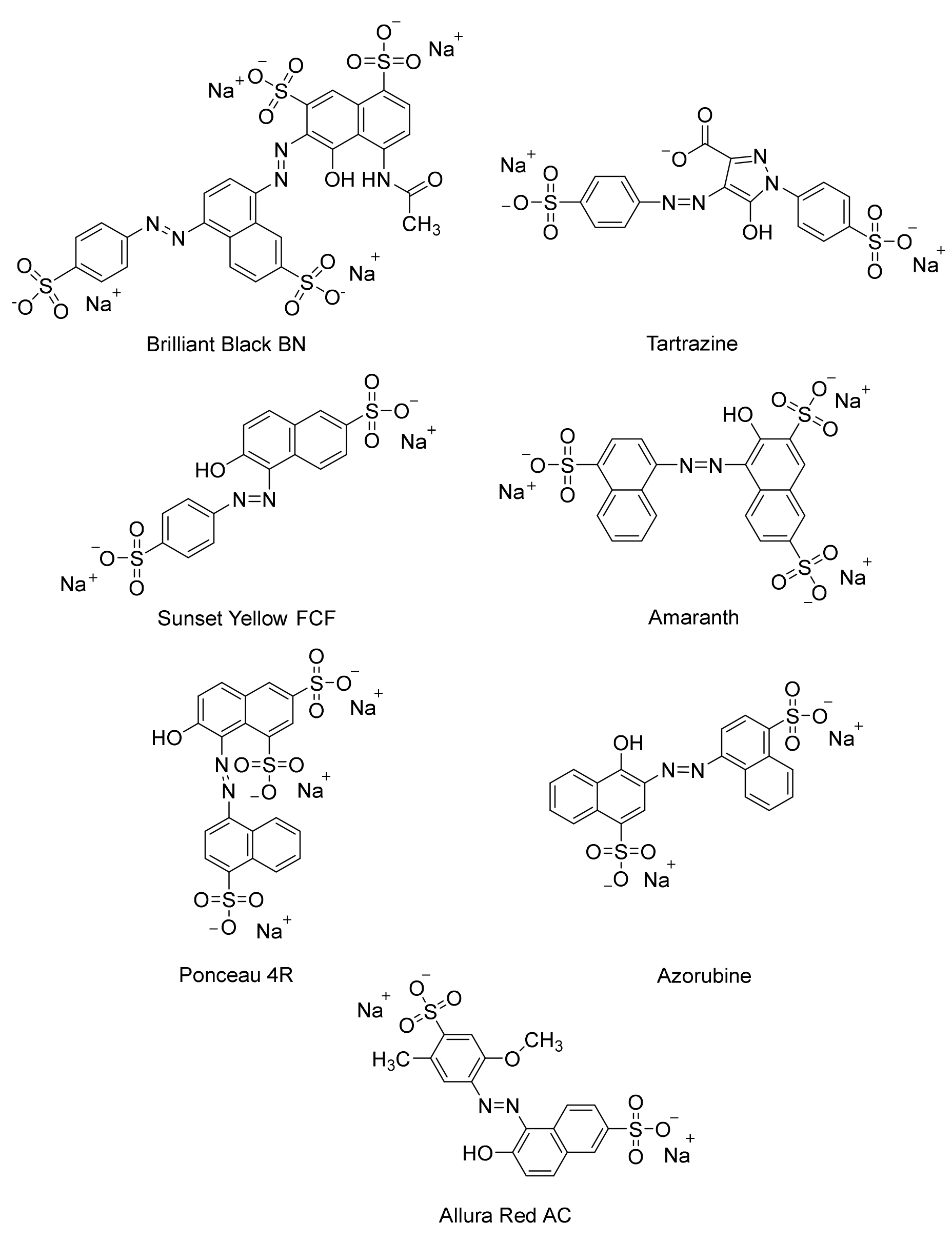
2. Impacts of Azo Dyes on Human Health and the Environment
3. Impact of Azo Dye Metabolites
4. Physical and Chemical Treatment of Dyes
5. Biological Treatment of Dyes
5.1. Biological Treatment of Dyes Using Filamentous Fungi
5.2. Biological Treatment of Dyes Using Yeasts
5.3. Biological Treatment of Dyes Using Algae
5.4. Biological Treatment of Dyes Using Bacteria
6. Enzymatic Degradation of Azo Dyes
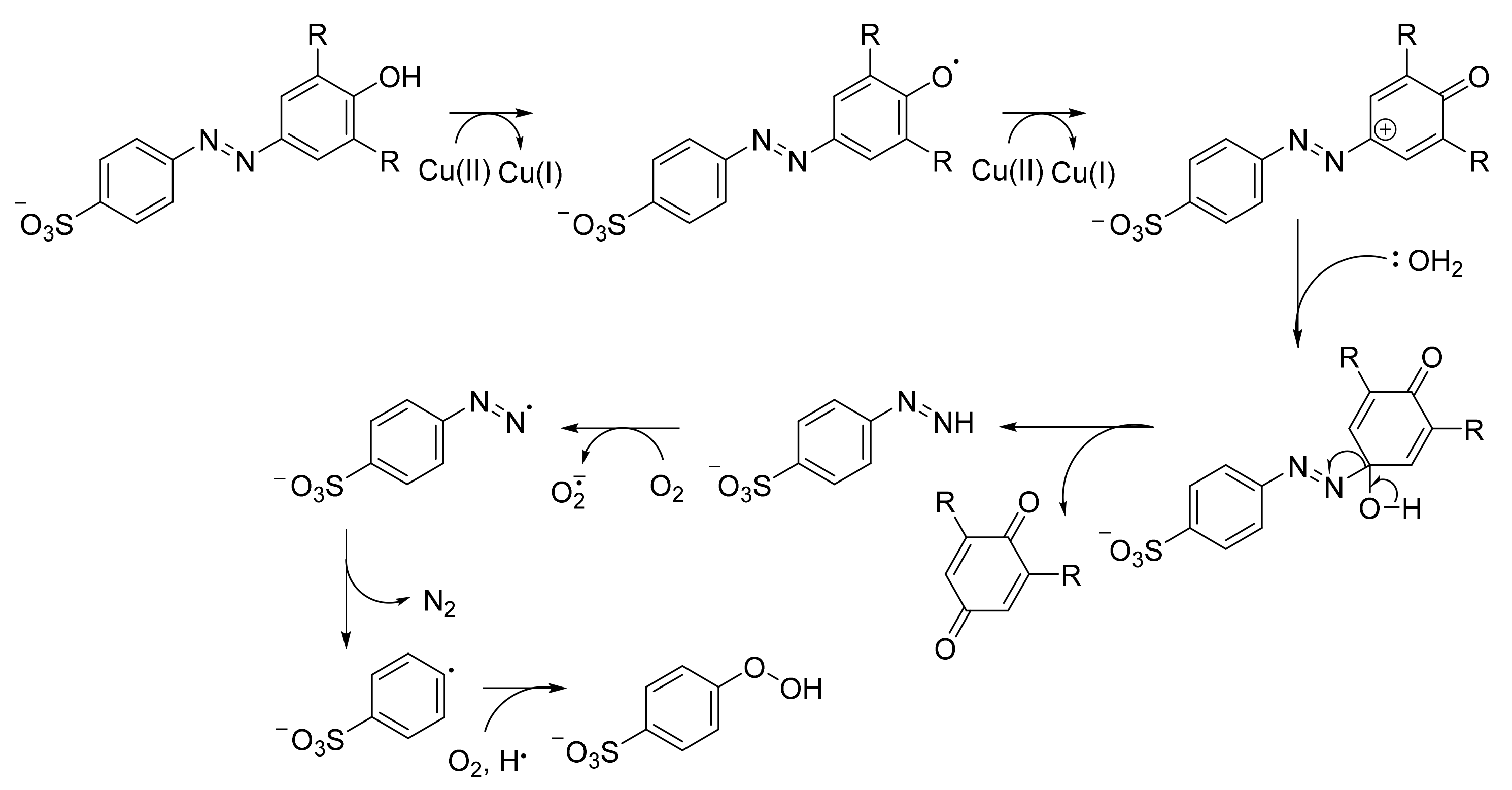
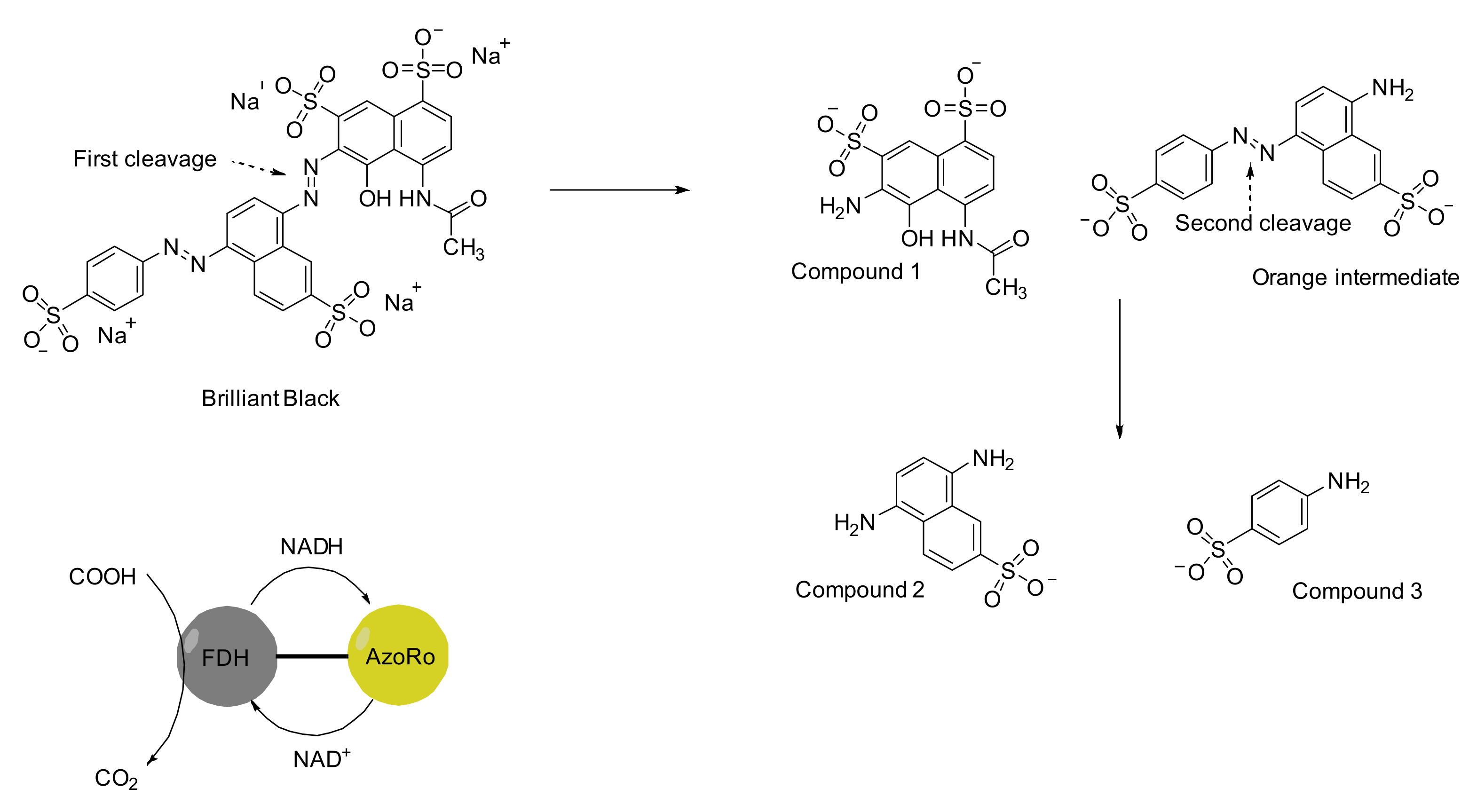
7. An Insight to Azoreductases
8. Mediators and Varying Energy Sources for a More Efficient Dye Degradation
9. Prospects on Azo Dye Degradation
9.1. Immobilization
9.2. Bioreactors
9.3. Microbial Fuel Cells
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Saxena, S.; Raja, A.S.M. Natural dyes: Sources, chemistry, application and sustainability issues. In Roadmap to Sustainable Textiles and Clothing; Springer: Singapore, 2014; pp. 37–80. [Google Scholar]
- Datta, S.; Uddin, M.A.; Afreen, K.S.; Akter, S.; Bandyopadhyay, A. Assessment of antimicrobial effectiveness of natural dyed fabrics. Bangladesh J. Sci. Ind. Res. 2013, 48, 179–184. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.K. An evaluation of UV protection imparted by cotton fabrics dyed with natural colorants. BMC Dermatol. 2004, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.N.; Pan, N.C.; Roy, A.K.; Saxena, S.; Khan, A. Development of natural dyed jute fabric with improved colour yield and UV protection characteristics. J. Text. Inst. 2013, 104, 808–818. [Google Scholar]
- Prabhu, K.H.; Teli, M.D. Eco-dyeing using Tamarindus indica L. seed coat tannin as a natural mordant for textiles with antibacterial activity. J. Saudi Chem. Soc. 2014, 18, 864–872. [Google Scholar] [CrossRef] [Green Version]
- Cooksey, C. Tyrian purple: The first four thousand years. Sci. Prog. 2013, 96, 171–186. [Google Scholar] [CrossRef]
- Tyrian Purple, Genuine. Available online: https://www.kremer-pigmente.com/en/shop/pigments/36010-tyrian-purple-genuine.html (accessed on 9 February 2022).
- Popli, S.; Patel, U. Destruction of azo dyes by anaerobic–aerobic sequential biological treatment: A review. Int. J. Environ. Sci. Technol. 2015, 12, 405–420. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.K.; Singh, R.L. Bio-removal of azo dyes: A review. Int. J. Appl. Sci. Biotechnol. 2017, 5, 108–126. [Google Scholar] [CrossRef]
- Zollinger, H. Azo dyes and pigments. Colour Chem.-Synth. Prop. Appl. Org. Dyes Pigment. 1987, 1st edition, 92–100. [Google Scholar]
- Padamavathy, S. Aerobic decolorization of reactive azo dyes in presence of various cosubstrates. Chem. Biochem. Eng. Q. 2003, 17, 147–152. [Google Scholar]
- Sandhya, S.; Padmavathy, S.; Swaminathan, K.; Subrahmanyam, Y.V.; Kaul, S.N. Microaerophilic–aerobic sequential batch reactor for treatment of azo dyes containing simulated wastewater. Process Biochem. 2005, 40, 885–890. [Google Scholar] [CrossRef]
- Sarkar, S.; Banerjee, A.; Halder, U.; Biswas, R.; Bandopadhyay, R. Degradation of synthetic azo dyes of textile industry: A sustainable approach using microbial enzymes. Water Conserv. Sci. Eng. 2017, 2, 121–131. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, F.H.; Bustos-Obregon, E.; Salvadori, D.M.F. Disperse Red 1 (textile dye) induces cytotoxic and genotoxic effects in mouse germ cells. Reprod. Toxicol. 2015, 53, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Ayed, L.; Bakir, K.; Mansour, H.B.; Hammami, S.; Cheref, A.; Bakhrouf, A. In vitro mutagenicity, NMR metabolite characterization of azo and triphenylmethanes dyes by adherents bacteria and the role of the “cna” adhesion gene in activated sludge. Microb. Pathog. 2017, 103, 29–39. [Google Scholar] [CrossRef] [PubMed]
- De Vasconcelos, D.; Maria, G.; Mulinari, J.; Ulson de Souza, A.A.; De Oliveira, D.; De Andrade, C.J. Biodegradation of azo dye-containing wastewater by activated sludge: A critical review. World J. Microbiol. Biotechnol. 2021, 37, 101. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.M.; Nigam, P.; Singh, D.; Marchant, R. Microbial decolorization of textile-dyecontaining effluents: A review. Bioresour. Technol. 1996, 58, 217–227. [Google Scholar] [CrossRef]
- Forgacs, E.; Cserháti, T.; Oros, G. Removal of synthetic dyes from wastewaters: A review. Environ. Int. 2004, 30, 953–971. [Google Scholar] [CrossRef]
- Dos Santos, A.B.; Cervantes, F.J.; Van Lier, J.B. Review paper on current technologies for decolourisation of textile wastewaters: Perspectives for anaerobic biotechnology. Bioresour. Technol. 2007, 98, 2369–2385. [Google Scholar] [CrossRef]
- Stolz, A. Basic and applied aspects in the microbial degradation of azo dyes. Appl. Microbiol. Biotechnol. 2001, 56, 69–80. [Google Scholar] [CrossRef]
- Pandey, A.; Singh, P.; Iyengar, L. Bacterial decolorization and degradation of azo dyes. Int. Biodeterior. Biodegrad. 2007, 59, 73–84. [Google Scholar] [CrossRef]
- Saratale, R.G.; Saratale, G.D.; Chang, J.S.; Govindwar, S.P. Bacterial decolorization and degradation of azo dyes: A review. J. Taiwan Inst. Chem. Eng. 2011, 42, 138–157. [Google Scholar] [CrossRef]
- Mota, I.G.C.; Neves, R.A.M.D.; Nascimento, S.S.D.C.; Maciel, B.L.L.; Morais, A.H.D.A.; Passos, T.S. Artificial dyes: Health risks and the need for revision of international regulations. Food Rev. Int. 2021, 27, 1–16. [Google Scholar] [CrossRef]
- Chung, K.T. The significance of azoreduction in the mutagenesis and carcinogenesis of azo dyes. Mutat. Res. Rev. Genet. Toxicol. 1983, 114, 269–281. [Google Scholar] [CrossRef]
- Chung, K.T.; Stevens, S.E.; Cerniglia, C.E. The reduction of azo dyes by the intestinal microflora. Crit. Rev. Microbiol. 1992, 18, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Puvaneswari, N.; Muthukrishnan, J.; Gunasekaran, P. Toxicity assessment and microbial degradation of azo dyes. Indian J. Exp. Biol. 2006, 44, 618–626. [Google Scholar]
- Chung, K.T.; Cerniglia, C.E. Mutagenicity of azo dyes: Structure-activity relationships. Mutat. Res. Rev. Genet. Toxicol. 1992, 277, 201–220. [Google Scholar] [CrossRef]
- Chen, H. Recent advances in azo dye degrading enzyme research. Curr. Protein Pept. Sci. 2006, 7, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Carmen, Z.; Daniela, S. Textile Organic Dyes-Characteristics, Polluting Effects and Separation/Elimination Procedures from Industrial Effluents—A Critical Overview; IntechOpen: Rijeka, Croatia, 2012; Volume 3, pp. 55–86. [Google Scholar]
- Khan, S.; Malik, A. Environmental and health effects of textile industry wastewater. In Environmental Deterioration and Human Health; Springer: Dordrecht, The Netherlands, 2014; pp. 55–71. [Google Scholar]
- Chung, K.T.; Fulk, G.E.; Andrews, A.W. Mutagenicity testing of some commonly used dyes. Appl. Environ. Microbiol. 1981, 42, 641–648. [Google Scholar] [CrossRef] [Green Version]
- Mishra, V.; Mishra, M.; Chaudhari, B.P.; Khanna, R.; Das, M. Argemone oil and butter yellow induced toxicity in hepatic and extra hepatic tissues. Bioenergetics 2014, 3, 111. [Google Scholar]
- Tsuda, S.; Murakami, M.; Matsusaka, N.; Kano, K.; Taniguchi, K.; Sasaki, Y.F. DNA damage induced by red food dyes orally administered to pregnant and male mice. Toxicol. Sci. 2001, 61, 92–99. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, Y.F.; Kawaguchi, S.; Kamaya, A.; Ohshita, M.; Kabasawa, K.; Iwama, K.; Taniguchi, K.; Tsuda, S. The comet assay with 8 mouse organs: Results with 39 currently used food additives. Mutat. Res. /Genet. Toxicol. Environ. Mutagenesis 2002, 519, 103–119. [Google Scholar] [CrossRef]
- Shimada, C.; Kano, K.; Sasaki, Y.F.; Sato, I.; Tsuda, S. Differential colon DNA damage induced by azo food additives between rats and mice. J. Toxicol. Sci. 2010, 35, 547–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macioszek, V.K.; Kononowicz, A.K. The evaluation of the genotoxicity of two commonly used food colors: Quinoline Yellow (E 104) and Brilliant Black BN (E 151). Cell. Mol. Biol. Lett. 2004, 9, 107–122. [Google Scholar] [PubMed]
- Ali, M.Y.; Hassan, G.M.; Hassan, A.M.S.; Mohamed, Z.A.; Ramadan, M.F. In vivo genotoxicity assessment of sunset yellow and sodium benzoate in female rats. Drug Chem. Toxicol. 2020, 43, 504–513. [Google Scholar] [CrossRef]
- El-Borm, H.T.; Badawy, G.M.; Hassab El-Nabi, S.; El-Sherif, W.A.; Atallah, M.N. Toxicity of sunset yellow FCF and tartrazine dyes on DNA and cell cycle of liver and kidneys of the chick embryo: The alleviative effects of curcumin. Egypt. J. Zool. 2020, 74, 43–55. [Google Scholar] [CrossRef]
- Chung, K.T. Azo dyes and human health: A review. J. Environ. Sci. Health Part C 2016, 34, 233–261. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, T.B.; Lizier, T.M.; Das Dores Assis, M.; Zanoni, M.V.B.; De Oliveira, D.P. CYP-450 isoenzymes catalyze the generation of hazardous aromatic amines after reaction with the azo dye Sudan III. Food Chem. Toxicol. 2013, 57, 217–226. [Google Scholar] [CrossRef] [Green Version]
- Gadallah, M.A.A. Phytotoxic effects of industrial and sewage waste waters on growth, chlorophyll content, transpiration rate and relative water content of potted sunflower plants. Water Air Soil Pollut. 1996, 89, 33–47. [Google Scholar] [CrossRef]
- Jagruti, B. Evaluation of azo dye toxicity using some haematological and histopathological alterations in fish Catla catla. Int. J. Biol. Biomol. Agric. Food Biotechnol. Eng. 2015, 9, 415–418. [Google Scholar]
- Parrott, J.L.; Bartlett, A.J.; Balakrishnan, V.K. Chronic toxicity of azo and anthracenedione dyes to embryo-larval fathead minnow. Environ. Pollut. 2016, 210, 40–47. [Google Scholar] [CrossRef]
- Katsumi, I.; Yuichi, I.; Osamu, N.; Keisuke, N.; Nobuyuki, I. Carcinogenicity and toxicity tests on p-phenylenediamine in F344 rats. Toxicol. Lett. 1983, 16, 259–269. [Google Scholar] [CrossRef]
- Burnett, C.; Loehr, R.; Corbett, J. Dominant lethal mutagenicity study on hair dyes. J. Toxicol. Environ. Health Part A Curr. Issues 1977, 2, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.H.; Solodar, W.E. Structure—Activity relationship studies on the mutagenicity of some azo dyes in the Salmonella/microsome assay. Mutagenesis 1988, 3, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Ishidate, M., Jr.; Nohmi, T. Sensitive method for the detection of mutagenic nitroarenes and aromatic amines: New derivatives of Salmonella typhimurium tester strains possessing elevated O-acetyltransferase levels. Mutat. Res. Environ. Mutagenesis Relat. Subj. 1990, 234, 337–348. [Google Scholar] [CrossRef]
- Sontag, J.M. Carcinogenicity of substituted-benzenediamines (phenylenediamines) in rats and mice. J. Natl. Cancer Inst. 1981, 66, 591–602. [Google Scholar] [PubMed]
- Chen, S.C.; Chen, C.H.; Chern, C.L.; Hsu, L.S.; Huang, Y.C.; Chung, K.T.; Chye, S.M. P-phenylenediamine induces p53-mediated apoptosis in Mardin–Darby canine kidney cells. Toxicol. Vitr. 2006, 20, 801–807. [Google Scholar] [CrossRef]
- Rollison, D.E.; Helzlsouer, K.J.; Pinney, S.M. Personal hair dye use and cancer: A systematic literature review and evaluation of exposure assessment in studies published since 1992. J. Toxicol. Environ. Health Part B 2006, 9, 413–439. [Google Scholar] [CrossRef]
- Vineis, P.; Pirastu, R. Aromatic amines and cancer. Cancer Causes Control. 1997, 8, 346–355. [Google Scholar] [CrossRef]
- Vandevivere, P.C.; Bianchi, R.; Verstraete, W. Treatment and reuse of wastewater from the textile wet-processing industry: Review of emerging technologies. J. Chem. Technol. Biotechnol. Int. Res. Process Environ. Clean Technol. 1998, 72, 289–302. [Google Scholar] [CrossRef]
- Amin, N.K. Removal of reactive dye from aqueous solutions by adsorption onto activated carbons prepared from sugarcane bagasse pith. Desalination 2008, 223, 152–161. [Google Scholar] [CrossRef]
- Faria, P.C.C.; Órfão, J.J.M.; Figueiredo, J.L.; Pereira, M.F.R. Adsorption of aromatic compounds from the biodegradation of azo dyes on activated carbon. Appl. Surf. Sci. 2008, 254, 3497–3503. [Google Scholar] [CrossRef]
- Velmurugan, P.; Rathinakumar, V.; Dhinakaran, G. Dye removal from aqueous solution using low cost adsorbent. Int. J. Environ. Sci. 2011, 1, 1492–1503. [Google Scholar]
- Das, S.; Singh, S.; Garg, S. Agri-residual waste, wheat bran as a biosorbent for mitigation of dye pollution in industrial wastewaters. J. Basic Microbiology 2022, 62, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Quevedo, L.C.; González-Caicedo, C.; Torres-Luna, J.A.; Carriazo, J.G. Removal of a textile azo-dye (Basic Red 46) in water by efficient adsorption on a natural clay. Water Air Soil Pollut. 2021, 232, 4. [Google Scholar] [CrossRef]
- Wang, F.; Li, L.; Iqbal, J.; Yang, Z.; Du, Y. Preparation of magnetic chitosan corn straw biochar and its application in adsorption of amaranth dye in aqueous solution. Int. J. Biol. Macromolecules 2022, 199, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Golob, V.; Vinder, A.; Simonič, M. Efficiency of the coagulation/flocculation method for the treatment of dyebath effluents. Dye. Pigment. 2005, 67, 93–97. [Google Scholar] [CrossRef]
- Sonal, S.; Mishra, B.K. Role of coagulation/flocculation technology for the treatment of dye wastewater: Trend and future aspects. In Water Pollution and Management Practices; Springer: Singapore, 2021; pp. 303–331. [Google Scholar]
- Azami, M.; Bahram, M.; Nouri, S.; Naseri, A. Central composite design for the optimization of removal of the azo dye, methyl orange, from wastewater using fenton reaction. J. Serb. Chem. Soc. 2012, 77, 235–246. [Google Scholar] [CrossRef]
- Tizaoui, C.; Grima, N. Kinetics of the ozone oxidation of Reactive Orange 16 azo-dye in aqueous solution. Chem. Eng. J. 2011, 173, 463–473. [Google Scholar] [CrossRef]
- Garg, S.K.; Tripathi, M. Microbial strategies for discoloration and detoxification of azo dyes from textile effluents. Res. J. Microbiol. 2017, 12, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Vidali, M. Bioremediation. An overview. Pure Appl. Chem. 2001, 73, 1163–1172. [Google Scholar] [CrossRef]
- Dangi, A.K.; Sharma, B.; Hill, R.T.; Shukla, P. Bioremediation through microbes: Systems biology and metabolic engineering approach. Crit. Rev. Biotechnol. 2019, 39, 79–98. [Google Scholar] [CrossRef]
- Mrozik, A.; Piotrowska-Seget, Z.; Labuzek, S. Bacterial degradation and bioremediation of polycyclic aromatic hydrocarbons. Pol. J. Environ. Stud. 2003, 12, 15–25. [Google Scholar]
- Chaudhary, P.; Sahay, H.; Sharma, R.; Pandey, A.K.; Singh, S.B.; Saxena, A.K.; Nain, L. Identification and analysis of polyaromatic hydrocarbons (PAHs)—Biodegrading bacterial strains from refinery soil of India. Environ. Monit. Assess. 2015, 187, 391. [Google Scholar] [CrossRef]
- Nanca, C.L.; Neri, K.D.; Ngo, A.C.R.; Bennett, R.M.; Dedeles, G.R. Degradation of polycyclic aromatic hydrocarbons by moderately halophilic bacteria from luzon salt beds. J. Health Pollut. 2018, 8, 180915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, R.M.; Cordero, P.R.F.; Bautista, G.S.; Dedeles, G.R. Reduction of hexavalent chromium using fungi and bacteria isolated from contaminated soil and water samples. Chem. Ecol. 2013, 29, 320–328. [Google Scholar] [CrossRef]
- Kang, C.H.; Kwon, Y.J.; So, J.S. Bioremediation of heavy metals by using bacterial mixtures. Ecol. Eng. 2016, 89, 64–69. [Google Scholar] [CrossRef]
- Paul, D.; Pandey, G.; Meier, C.; Roelof van der Meer, J.; Jain, R.K. Bacterial community structure of a pesticide-contaminated site and assessment of changes induced in community structure during bioremediation. FEMS Microbiol. Ecol. 2006, 57, 116–127. [Google Scholar] [CrossRef] [Green Version]
- Góngora-Echeverría, V.R.; García-Escalante, R.; Rojas-Herrera, R.; Giácoman-Vallejos, G.; Ponce-Caballero, C. Pesticide bioremediation in liquid media using a microbial consortium and bacteria-pure strains isolated from a biomixture used in agricultural areas. Ecotoxicol. Environ. Saf. 2020, 200, 110734. [Google Scholar] [CrossRef] [PubMed]
- Barsing, P.; Tiwari, A.; Joshi, T.; Garg, S. Application of a novel bacterial consortium for mineralization of sulphonated aromatic amines. Bioresour. Technol. 2011, 102, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, M.; Liyanage, N.; Herath, C.; Rathnayake, S.; Fernando, E.Y. Mineralization of persistent azo dye pollutants by a microaerophilic tropical lake sediment mixed bacterial consortium. Environ. Adv. 2021, 3, 100038. [Google Scholar] [CrossRef]
- Khan, R.; Bhawana, P.; Fulekar, M.H. Microbial decolorization and degradation of synthetic dyes: A review. Rev. Environ. Sci. Bio/Technol. 2013, 12, 75–97. [Google Scholar] [CrossRef]
- Yesilada, O.; Birhanli, E.; Geckil, H. Bioremediation and decolorization of textile dyes by white rot fungi and laccase enzymes. In Mycoremediation and Environmental Sustainability; Springer: Cham, Switzerland, 2018; pp. 121–153. [Google Scholar]
- Akansha, K.; Chakraborty, D.; Sachan, S.G. Decolorization and degradation of methyl orange by Bacillus stratosphericus SCA1007. Biocatal. Agric. Biotechnol. 2019, 18, 101044. [Google Scholar] [CrossRef]
- Barathi, S.; Aruljothi, K.N.; Karthik, C.; Padikasan, I.A. Optimization for enhanced ecofriendly decolorization and detoxification of Reactive Blue160 textile dye by Bacillus subtilis. Biotechnol. Rep. 2020, 28, e00522. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Luo, H.; Cheng, W.; Jiang, K.; Lu, L.; Ling, L. Decolorization characteristics and mechanism of methyl orange dye by using Stenotrophomonas acidaminiphila EFS1. Int. J. Environ. Sci. Technol. 2022, 1–10. [Google Scholar] [CrossRef]
- Dhir, B. Degradation of dyes using filamentous fungi. In Dye Biodegradation, Mechanisms and Techniques; Springer: Singapore, 2022; pp. 51–66. [Google Scholar]
- Solís, M.; Solís, A.; Pérez, H.I.; Manjarrez, N.; Flores, M. Microbial decolouration of azo dyes: A review. Process Biochem. 2012, 47, 1723–1748. [Google Scholar] [CrossRef]
- Sen, S.K.; Raut, S.; Bandyopadhyay, P.; Raut, S. Fungal decolouration and degradation of azo dyes: A review. Fungal Biol. Rev. 2016, 30, 112–133. [Google Scholar] [CrossRef]
- Kyzas, G.Z.; Fu, J.; Matis, K.A. The change from past to future for adsorbent materials in treatment of dyeing wastewaters. Materials 2013, 6, 5131–5158. [Google Scholar] [CrossRef]
- Fomina, M.; Gadd, G.M. Biosorption: Current perspectives on concept, definition and application. Bioresour. Technol. 2014, 160, 3–14. [Google Scholar] [CrossRef]
- Du, L.N.; Wang, B.; Li, G.; Wang, S.; Crowley, D.E.; Zhao, Y.H. Biosorption of the metal-complex dye Acid Black 172 by live and heat-treated biomass of Pseudomonas sp. strain DY1: Kinetics and sorption mechanisms. J. Hazard. Mater. 2012, 205, 47–54. [Google Scholar] [CrossRef]
- Hernández-Zamora, M.; Cristiani-Urbina, E.; Martínez-Jerónimo, F.; Perales-Vela, H.V.; Ponce-Noyola, T.; Montes-Horcasitas, M.D.C.; Cañizares-Villanueva, R.O. Bioremoval of the azo dye Congo Red by the microalga Chlorella vulgaris. Environ. Sci. Pollut. Res. 2015, 22, 10811–10823. [Google Scholar] [CrossRef]
- Singh, S.; Pakshirajan, K. Enzyme activities and decolourization of single and mixed azo dyes by the white-rot fungus Phanerochaete chrysosporium. Int. Biodeterior. Biodegrad. 2010, 64, 146–150. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Das, A. Mycoremediation of Congo red dye by filamentous fungi. Braz. J. Microbiol. 2011, 42, 1526–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, A.A.; Lucas, M.S.; Sampaio, A.; Peres, J.A.; Bezerra, R.M. Decolorization of azo dyes by yeasts. In Biodegradation of Azo Dyes; Springer: Berlin/Heidelberg, Germany, 2010; pp. 183–193. [Google Scholar]
- Ngo, A.C.R.; Devanadera, M.K.P.; Dedeles, G.R. Decolorization of selected synthetic textile dyes by yeasts from leaves and fruit peels. J. Health Pollut. 2016, 6, 42–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shetty, K.; Krishnakumar, G. Algal and cyanobacterial biomass as potential dye biodecolorizing material: A review. Biotechnol. Lett. 2020, 42, 2467–2488. [Google Scholar] [CrossRef] [PubMed]
- El-Sheekh, M.M.; El-Shanshoury, A.R.; Abou-El-Souod, G.W.; Gharieb, D.Y.; El Shafay, S.M. Decolorization of dyestuffs by some species of green algae and cyanobacteria and its consortium. Int. J. Environ. Sci. Technol. 2021, 18, 3895–3906. [Google Scholar] [CrossRef]
- Ghodake, G.; Jadhav, U.; Tamboli, D.; Kagalkar, A.; Govindwar, S. Decolorization of textile dyes and degradation of mono-azo dye amaranth by Acinetobacter calcoaceticus NCIM 2890. Indian J. Microbiol. 2011, 51, 501–508. [Google Scholar] [CrossRef] [Green Version]
- Shah, M.P.; Patel, K.A.; Nair, S.S.; Darji, A.M. Microbial decolourization of methyl orange dye by Pseudomonas spp. OA Biotechnology 2013, 2, 10. [Google Scholar] [CrossRef]
- Haque, M.M.; Haque, M.A.; Mosharaf, M.K.; Marcus, P.K. Decolorization, degradation and detoxification of carcinogenic sulfonated azo dye methyl orange by newly developed biofilm consortia. Saudi J. Biol. Sci. 2021, 28, 793–804. [Google Scholar] [CrossRef]
- Fu, Y.; Viraraghavan, T. Removal of Congo Red from an aqueous solution by fungus Aspergillus niger. Adv. Environ. Res. 2002, 7, 239–247. [Google Scholar] [CrossRef]
- Claus, H.; Faber, G.; König, H.J.A.M. Redox-mediated decolorization of synthetic dyes by fungal laccases. Appl. Microbiol. Biotechnol. 2002, 59, 672–678. [Google Scholar] [CrossRef]
- Fu, Y.; Viraraghavan, T. Removal of a dye from an aqueous solution by the fungus Aspergillus niger. Water Qual. Res. J. 2000, 35, 95–112. [Google Scholar] [CrossRef]
- Gallagher, K.A.; Healy, M.G.; Allen, S.J. Biosorption of synthetic dye and metal ions from aqueous effluents using fungal biomass. In Studies in Environmental Science; Elsevier: Amsterdam, The Netherlands, 1997; Volume 66, pp. 27–50. [Google Scholar]
- Brahimi-Horn, M.C.; Lim, K.K.; Liang, S.L.; Mou, D.G. Binding of textile azo dyes by Myrothecium verrucaria. J. Ind. Microbiol. Biotechnol. 1992, 10, 31–36. [Google Scholar]
- Jeon, S.J.; Lim, S.J. Purification and characterization of the laccase involved in dye decolorization by the white-rot fungus Marasmius scorodonius. J. Microbiol. Biotechnol. 2017, 27, 1120–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Telke, A.A.; Kadam, A.A.; Jagtap, S.S.; Jadhav, J.P.; Govindwar, S.P. Biochemical characterization and potential for textile dye degradation of blue laccase from Aspergillus ochraceus NCIM-1146. Biotechnol. Bioprocess Eng. 2010, 15, 696–703. [Google Scholar] [CrossRef]
- Ollikka, P.; Alhonmäki, K.; Leppänen, V.M.; Glumoff, T.; Raijola, T.; Suominen, I. Decolorization of azo, triphenyl methane, heterocyclic, and polymeric dyes by lignin peroxidase isoenzymes from Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1993, 59, 4010–4016. [Google Scholar] [CrossRef] [Green Version]
- Shaheen, R.; Asgher, M.; Hussain, F.; Bhatti, H.N. Immobilized lignin peroxidase from Ganoderma lucidum IBL-05 with improved dye decolorization and cytotoxicity reduction properties. Int. J. Biol. Macromol. 2017, 103, 57–64. [Google Scholar] [CrossRef]
- Bouacem, K.; Rekik, H.; Jaouadi, N.Z.; Zenati, B.; Kourdali, S.; El Hattab, M.; Badis, A.; Annane, R.; Bejar, S.; Hacene, H.; et al. Purification and characterization of two novel peroxidases from the dye-decolorizing fungus Bjerkandera adusta strain CX-9. Int. J. Biol. Macromol. 2018, 106, 636–646. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Zhang, X.; Geng, A. Purification and characterization of a novel manganese peroxidase from white-rot fungus Cerrena unicolor BBP6 and its application in dye decolorization and denim bleaching. Process Biochem. 2018, 66, 222–229. [Google Scholar] [CrossRef]
- Mou, D.G.; Lim, K.K.; Shen, H.P. Microbial agents for decolorization of dye wastewater. Biotechnol. Adv. 1991, 9, 613–622. [Google Scholar] [CrossRef]
- Polman, K.; Breckenridge, C.R. Biomass-mediated binding and recovery of textile dyes from waste effluents. Text. Chem. Colorist 1996, 28, 31–35. [Google Scholar]
- Przystaś, W.; Zabłocka-Godlewska, E.; Grabińska-Sota, E. Efficiency of decolorization of different dyes using fungal biomass immobilized on different solid supports. Braz. J. Microbiol. 2018, 49, 285–295. [Google Scholar] [CrossRef]
- Yu, Z.; Wen, X. Screening and identification of yeasts for decolorizing synthetic dyes in industrial wastewater. Int. Biodeterior. Biodegrad. 2005, 56, 109–114. [Google Scholar] [CrossRef]
- Chivukula, M.; Renganathan, V. Phenolic azo dye oxidation by laccase from Pyricularia oryzae. Appl. Environ. Microbiol. 1995, 61, 4374–4377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jadhav, J.P.; Parshetti, G.K.; Kalme, S.D.; Govindwar, S.P. Decolourization of azo dye methyl red by Saccharomyces cerevisiae MTCC 463. Chemosphere 2007, 68, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Mahalakshmi, S.; Lakshmi, D.; Menaga, U. Biodegradation of different concentration of dye (Congo red dye) by using green and blue green algae. Int. J. Environ. Res. 2015, 9, 735–744. [Google Scholar]
- Abd Ellatif, S.; El-Sheekh, M.M.; Senousy, H.H. Role of microalgal ligninolytic enzymes in industrial dye decolorization. Int. J. Phytoremediation 2021, 23, 41–52. [Google Scholar] [CrossRef]
- Acuner, E.; Dilek, F.B. Treatment of tectilon yellow 2G by Chlorella vulgaris. Process Biochem. 2004, 39, 623–631. [Google Scholar] [CrossRef]
- Qi, J.; Schlömann, M.; Tischler, D. Biochemical characterization of an azoreductase from Rhodococcus opacus 1CP possessing methyl red degradation ability. J. Mol. Catal. B Enzym. 2016, 130, 9–17. [Google Scholar] [CrossRef]
- Deller, S.; Sollner, S.; Trenker-El-Toukhy, R.; Jelesarov, I.; Gübitz, G.M.; Macheroux, P. Characterization of a thermostable NADPH: FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis. Biochemistry 2006, 45, 7083–7091. [Google Scholar] [CrossRef]
- Ngo, A.C.R.; Qi, J.; Juric, C.; Bento, I.; Tischler, D. Identification of molecular basis that underlie enzymatic specificity of AzoRo from Rhodococcus opacus 1CP: A potential NADH: Quinone oxidoreductase. Arch. Biochem. Biophys. 2022, 717, 109123. [Google Scholar] [CrossRef]
- Rafii, F.; Cerniglia, C.E. Reduction of azo dyes and nitroaromatic compounds by bacterial enzymes from the human intestinal tract. Environ. Health Perspect. 1995, 103 (Suppl. S5), 17–19. [Google Scholar]
- Ryan, A. Azoreductases in drug metabolism. Br. J. Pharmacol. 2017, 174, 2161–2173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, H. Remarkable diversification of bacterial azoreductases: Primary sequences, structures, substrates, physiological roles, and biotechnological applications. Appl. Microbiol. Biotechnol. 2019, 103, 3965–3978. [Google Scholar] [CrossRef] [PubMed]
- Misal, S.A.; Gawai, K.R. Azoreductase: A key player of xenobiotic metabolism. Bioresour. Bioprocess. 2018, 5, 17. [Google Scholar] [CrossRef]
- Shanmugam, B.K.; Mahadevan, S. Metabolism and biotransformation of azo dye by bacterial consortium studied in a bioreaction calorimeter. Bioresour. Technol. 2015, 196, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, B.K.; Easwaran, S.N.; Lakra, R.; Deepa, P.R.; Mahadevan, S. Metabolic pathway and role of individual species in the bacterial consortium for biodegradation of azo dye: A biocalorimetric investigation. Chemosphere 2017, 188, 81–89. [Google Scholar] [CrossRef]
- Anjaneya, O.; Souche, S.Y.; Santoshkumar, M.; Karegoudar, T.B. Decolorization of sulfonated azo dye Metanil Yellow by newly isolated bacterial strains: Bacillus sp. strain AK1 and Lysinibacillus sp. strain AK2. J. Hazard. Mater. 2011, 190, 351–358. [Google Scholar] [CrossRef]
- Ayed, L.; Mahdhi, A.; Cheref, A.; Bakhrouf, A. Decolorization and degradation of azo dye Methyl Red by an isolated Sphingomonas paucimobilis: Biotoxicity and metabolites characterization. Desalination 2011, 274, 272–277. [Google Scholar] [CrossRef]
- Chen, K.C.; Huang, W.T.; Wu, J.Y.; Houng, J.Y. Microbial decolorization of azo dyes by Proteus mirabilis. J. Ind. Microbiol. Biotechnol. 1999, 23, 686–690. [Google Scholar] [CrossRef]
- Chen, K.C.; Wu, J.Y.; Liou, D.J.; Hwang, S.C.J. Decolorization of the textile dyes by newly isolated bacterial strains. J. Biotechnol. 2003, 101, 57–68. [Google Scholar] [CrossRef]
- Franciscon, E.; Grossman, M.J.; Paschoal, J.A.R.; Reyes, F.G.R.; Durrant, L.R. Decolorization and biodegradation of reactive sulfonated azo dyes by a newly isolated Brevibacterium sp. strain VN-15. SpringerPlus 2012, 1, 37. [Google Scholar] [CrossRef] [Green Version]
- Guembri, M.; Neifar, M.; Saidi, M.; Ferjani, R.; Chouchane, H.; Mosbah, A.; Cherif, A.; Saidi, N.; Ouzari, H.I. Decolorization of textile azo dye Novacron Red using bacterial monoculture and consortium: Response surface methodology optimization. Water Environ. Res. 2021, 93, 1346–1360. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Shimada, Y.; Suto, T. Potential use of bacteria collected from human hands for textile dye decolorization. Water Resour. Ind. 2018, 20, 46–53. [Google Scholar] [CrossRef]
- Kalyani, D.C.; Patil, P.S.; Jadhav, J.P.; Govindwar, S.P. Biodegradation of reactive textile dye Red BLI by an isolated bacterium Pseudomonas sp. SUK1. Bioresour. Technol. 2008, 99, 4635–4641. [Google Scholar] [CrossRef] [PubMed]
- Kalyani, D.C.; Telke, A.A.; Dhanve, R.S.; Jadhav, J.P. Ecofriendly biodegradation and detoxification of Reactive Red 2 textile dye by newly isolated Pseudomonas sp. SUK1. J. Hazard. Mater. 2009, 163, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Arshad, M.; Crowley, D.E. Accelerated decolorization of structurally different azo dyes by newly isolated bacterial strains. Appl. Microbiol. Biotechnol. 2008, 78, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, S.; Ngo, A.C.R.; Schultes, F.P.J.; Tischler, D. Draft genome sequence of Kocuria indica DP-K7, a methyl red degrading actinobacterium. 3 Biotech 2020, 10, 175. [Google Scholar] [CrossRef]
- Kumaran, S.; Ngo, A.C.R.; Schultes, F.; Saravanan, V.S.; Tischler, D. In vitro and in silico analysis of Brilliant Black degradation by Actinobacteria and a Paraburkholderia sp. Genomics 2022, 114, 110266. [Google Scholar] [CrossRef]
- Maniyam, M.N.; Ibrahim, A.L.; Cass, A.E. Decolourization and biodegradation of azo dye methyl red by Rhodococcus strain UCC 0016. Environ. Technol. 2020, 41, 71–85. [Google Scholar] [CrossRef]
- Park, E.H.; Jang, M.S.; Cha, I.H.; Choi, Y.L.; Cho, Y.S.; Kim, C.H.; Lee, Y.C. Decolorization of a sulfonated azo dye, Congo Red, by Staphylococcus sp. EY-3. J. Microbiol. Biotechnol. 2005, 15, 221–225. [Google Scholar]
- Parshetti, G.K.; Telke, A.A.; Kalyani, D.C.; Govindwar, S.P. Decolorization and detoxification of sulfonated azo dye methyl orange by Kocuria rosea MTCC 1532. J. Hazard. Mater. 2010, 176, 503–509. [Google Scholar] [CrossRef]
- Wang, H.; Su, J.Q.; Zheng, X.W.; Tian, Y.; Xiong, X.J.; Zheng, T.L. Bacterial decolorization and degradation of the reactive dye Reactive Red 180 by Citrobacter sp. CK3. Int. Biodeterior. Biodegrad. 2009, 63, 395–399. [Google Scholar] [CrossRef]
- Wang, Z.W.; Liang, J.S.; Liang, Y. Decolorization of Reactive Black 5 by a newly isolated bacterium Bacillus sp. YZU1. Int. Biodeterior. Biodegrad. 2013, 76, 41–48. [Google Scholar] [CrossRef]
- Biju, L.M.; Pooshana, V.; Kumar, P.S.; Gayathri, K.V.; Ansar, S.; Govindaraju, S. Treatment of textile wastewater containing mixed toxic azo dye and chromium (VI) BY haloalkaliphilic bacterial consortium. Chemosphere 2022, 287, 132280. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Wenrong, H.; Yuezhong, L. Investigation of isolation and immobilization of a microbial consortium for decoloring of azo dye 4BS. Water Res. 2004, 38, 3596–3604. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Jain, K.; Soni, A.; Madamwar, D. Microaerophilic degradation of sulphonated azo dye–Reactive Red 195 by bacterial consortium AR1 through co-metabolism. Int. Biodeterior. Biodegrad. 2014, 94, 167–175. [Google Scholar] [CrossRef]
- Khehra, M.S.; Saini, H.S.; Sharma, D.K.; Chadha, B.S.; Chimni, S.S. Comparative studies on potential of consortium and constituent pure bacterial isolates to decolorize azo dyes. Water Res. 2005, 39, 5135–5141. [Google Scholar] [CrossRef]
- Kurade, M.B.; Waghmode, T.R.; Jadhav, M.U.; Jeon, B.H.; Govindwar, S.P. Bacterial–yeast consortium as an effective biocatalyst for biodegradation of sulphonated azo dye Reactive Red 198. RSC Adv. 2015, 5, 23046–23056. [Google Scholar] [CrossRef]
- Lade, H.S.; Waghmode, T.R.; Kadam, A.A.; Govindwar, S.P. Enhanced biodegradation and detoxification of disperse azo dye Rubine GFL and textile industry effluent by defined fungal-bacterial consortium. Int. Biodeterior. Biodegrad. 2012, 72, 94–107. [Google Scholar] [CrossRef]
- Masarbo, R.S.; Niranjana, S.R.; Monisha, T.R.; Nayak, A.S.; Karegoudar, T.B. Efficient decolorization and detoxification of sulphonated azo dye Ponceau 4R by using single and mixed bacterial consortia. Biocatal. Biotransformation 2019, 37, 367–376. [Google Scholar] [CrossRef]
- Moosvi, S.; Kher, X.; Madamwar, D. Isolation, characterization and decolorization of textile dyes by a mixed bacterial consortium JW-2. Dye. Pigment. 2007, 74, 723–729. [Google Scholar] [CrossRef]
- Yogesh, P.; Chitra, M.; Akshaya, G. Assessment of biological decolorization and degradation of sulfonated di-azo dye Acid Maroon V by isolated bacterial consortium EDPA. Int. Biodeterior. Biodegrad. 2012, 75, 187–193. [Google Scholar]
- Patil, P.S.; Phugare, S.S.; Jadhav, S.B.; Jadhav, J.P. Communal action of microbial cultures for Red HE3B degradation. J. Hazard. Mater. 2010, 181, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Phugare, S.S.; Kalyani, D.C.; Surwase, S.N.; Jadhav, J.P. Ecofriendly degradation, decolorization and detoxification of textile effluent by a developed bacterial consortium. Ecotoxicol. Environ. Saf. 2011, 74, 1288–1296. [Google Scholar] [CrossRef]
- Saratale, R.G.; Saratale, G.D.; Kalyani, D.C.; Chang, J.S.; Govindwar, S.P. Enhanced decolorization and biodegradation of textile azo dye Scarlet R by using developed microbial consortium-GR. Bioresour. Technol. 2009, 100, 2493–2500. [Google Scholar] [CrossRef]
- Thiruppathi, K.; Rangasamy, K.; Ramasamy, M.; Muthu, D. Evaluation of textile dye degrading potential of ligninolytic bacterial consortia. Environ. Chall. 2021, 4, 100078. [Google Scholar] [CrossRef]
- Lang, W.; Sirisansaneeyakul, S.; Martins, L.O.; Ngiwsara, L.; Sakairi, N.; Pathom-Aree, W.; Okuyama, M.; Mori, H.; Kimura, A. Biodecolorization of a food azo dye by the deep sea Dermacoccus abyssi MT1. 1T strain from the Mariana Trench. J. Environ. Manag. 2014, 132, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Harazono, K.; Watanabe, Y.; Nakamura, K. Decolorization of azo dye by the white-rot basidiomycete Phanerochaete sordida and by its manganese peroxidase. J. Biosci. Bioeng. 2003, 95, 455–459. [Google Scholar] [CrossRef]
- Urek, R.O.; Pazarlioglu, N.K. Enhanced production of manganese peroxidase by Phanerochaete chrysosporium. Braz. Arch. Biol. Technol. 2007, 50, 913–920. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.S.; Al-Tohamy, R.; Sun, J. Performance of Meyerozyma caribbica as a novel manganese peroxidase-producing yeast inhabiting wood-feeding termite gut symbionts for azo dye decolorization and detoxification. Sci. Total Environ. 2022, 806, 150665. [Google Scholar] [CrossRef]
- Ghodake, G.S.; Kalme, S.D.; Jadhav, J.P.; Govindwar, S.P. Purification and partial characterization of lignin peroxidase from Acinetobacter calcoaceticus NCIM 2890 and its application in decolorization of textile dyes. Appl. Biochem. Biotechnol. 2009, 152, 6–14. [Google Scholar] [CrossRef]
- Hao, J.; Song, F.; Huang, F.; Yang, C.; Zhang, Z.; Zheng, Y.; Tian, X. Production of laccase by a newly isolated deuteromycete fungus Pestalotiopsis sp. and its decolorization of azo dye. J. Ind. Microbiol. Biotechnol. 2007, 34, 233. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Mendes, S.; Brissos, V.; Martins, L.O. New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: Towards biotechnological applications. Appl. Microbiol. Biotechnol. 2014, 98, 2053–2065. [Google Scholar] [CrossRef] [PubMed]
- Ngo, A.C.R.; Conrad, C.; Gómez Baraibar, Á.; Matura, A.; Van Pee, K.H.; Tischler, D. Characterization of two hydrogen peroxide resistant peroxidases from Rhodococcus opacus 1CP. Appl. Sci. 2021, 11, 7941. [Google Scholar] [CrossRef]
- Hu, T.L. Degradation of azo dye RP2B by Pseudomonas luteola. Water Sci. Technol. 1998, 38, 299–306. [Google Scholar] [CrossRef]
- Suzuki, Y.; Yoda, T.; Ruhul, A.; Sugiura, W. Molecular cloning and characterization of the gene coding for azoreductase from Bacillus sp. OY1-2 isolated from soil. J. Biol. Chem. 2001, 276, 9059–9065. [Google Scholar] [CrossRef] [Green Version]
- Moutaouakkil, A.; Zeroual, Y.; Dzayri, F.Z.; Talbi, M.; Lee, K.; Blaghen, M. Purification and partial characterization of azoreductase from Enterobacter agglomerans. Arch. Biochem. Biophys. 2003, 413, 139–146. [Google Scholar] [CrossRef]
- Leelakriangsak, M.; Borisut, S. Characterization of the decolorizing activity of azo dyes by Bacillus subtilis azoreductase AzoR1. Songklanakarin J. Sci. Technol. 2012, 34, 509–516. [Google Scholar]
- Gold, M.H.; Youngs, H.L.; Gelpke, M.D.S. Manganese peroxidase. In Metal Ions in Biological Systems; Sigel, A., Sigel, H., Eds.; Marcel Dekker Inc.: New York, NY, USA, 2000; pp. 559–586. [Google Scholar]
- Hofrichter, M. lignin conversion by manganese peroxidase (MnP). Enzym. Microb. Technol. 2002, 30, 454–466. [Google Scholar] [CrossRef]
- Husain, Q. Peroxidase mediated decolorization and remediation of wastewater containing industrial dyes: A review. Rev. Environ. Sci. Bio/Technol. 2010, 9, 117–140. [Google Scholar] [CrossRef]
- Heinfling, A.; Martinez, M.J.; Martínez, A.T.; Bergbauer, M.; Szewzyk, U. Transformation of industrial dyes by manganese peroxidases from Bjerkandera adusta and Pleurotus eryngii in a manganese-independent reaction. Appl. Environ. Microbiol. 1998, 64, 2788–2793. [Google Scholar] [CrossRef] [Green Version]
- Moreira, M.T.; Palma, C.; Mielgo, I.; Feijoo, G.; Lema, J.M. In vitro degradation of a polymeric dye (Poly R-478) by manganese peroxidase. Biotechnol. Bioeng. 2001, 75, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Madamwar, D. Decolourization of synthetic dyes by a newly isolated strain of Serratia marcescens. World J. Microbiol. Biotechnol. 2003, 19, 615–618. [Google Scholar] [CrossRef]
- Novotný, Č.; Svobodová, K.; Erbanová, P.; Cajthaml, T.; Kasinath, A.; Lang, E.; Šašek, V. Ligninolytic fungi in bioremediation: Extracellular enzyme production and degradation rate. Soil Biol. Biochem. 2004, 36, 1545–1551. [Google Scholar] [CrossRef]
- Dawkar, V.V.; Jadhav, U.U.; Ghodake, G.S.; Govindwar, S.P. Effect of inducers on the decolorization and biodegradation of textile azo dye Navy blue 2GL by Bacillus sp. VUS. Biodegradation 2009, 20, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Kalme, S.D.; Parshetti, G.K.; Jadhav, S.U.; Govindwar, S.P. Biodegradation of benzidine based dye Direct Blue-6 by Pseudomonas desmolyticum NCIM 2112. Bioresour. Technol. 2007, 98, 1405–1410. [Google Scholar] [CrossRef] [PubMed]
- Sugano, Y.; Muramatsu, R.; Ichiyanagi, A.; Sato, T.; Shoda, M. DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family: ASP171 replaces the distal histidine of classical peroxidases. J. Biol. Chem. 2007, 282, 36652–36658. [Google Scholar] [CrossRef] [Green Version]
- Sugano, Y.; Yoshida, T. DyP-type peroxidases: Recent advances and perspectives. Int. J. Mol. Sci. 2021, 22, 5556. [Google Scholar] [CrossRef]
- Hofrichter, M.; Ullrich, R.; Pecyna, M.J.; Liers, C.; Lundell, T. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 2010, 87, 871–897. [Google Scholar] [CrossRef]
- Krahe, N.K.; Berger, R.G.; Ersoy, F. A DyP-type peroxidase of Pleurotus sapidus with alkene cleaving activity. Molecules 2020, 25, 1536. [Google Scholar] [CrossRef] [Green Version]
- Colpa, D.I.; Fraaije, M.W.; Van Bloois, E. DyP-type peroxidases: A promising and versatile class of enzymes. J. Ind. Microbiol. Biotechnol. 2014, 41, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Eltis, L.D. The multihued palette of dye-decolorizing peroxidases. Arch. Biochem. Biophys. 2015, 574, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Madhavi, V.; Lele, S.S. Laccase: Properties and applications. BioResources 2009, 4, 1694–1717. [Google Scholar]
- Wong, Y.; Yu, J. Laccase-catalyzed decolorization of synthetic dyes. Water Res. 1999, 33, 3512–3520. [Google Scholar] [CrossRef]
- Hou, H.; Zhou, J.; Wang, J.; Du, C.; Yan, B. Enhancement of laccase production by Pleurotus ostreatus and its use for the decolorization of anthraquinone dye. Process Biochem. 2004, 39, 1415–1419. [Google Scholar] [CrossRef]
- Li, J.F.; Hong, Y.Z.; Xiao, Y.Z.; Xu, Y.H.; Fang, W. High production of laccase B from Trametes sp. in Pichia pastoris. World J. Microbiol. Biotechnol. 2007, 23, 741–745. [Google Scholar] [CrossRef]
- Lu, L.; Zhao, M.; Liang, S.C.; Zhao, L.Y.; Li, D.B.; Zhang, B.B. Production and synthetic dyes decolourization capacity of a recombinant laccase from Pichia pastoris. J. Appl. Microbiol. 2009, 107, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Chi, Y.; Yi, H.; Shao, S. Decolorization of Alizarin Red and other synthetic dyes by a recombinant laccase from Pichia pastoris. Biotechnol. Lett. 2014, 36, 39–45. [Google Scholar] [CrossRef]
- Chen, H.; Wang, R.F.; Cerniglia, C.E. Molecular cloning, overexpression, purification, and characterization of an aerobic FMN-dependent azoreductase from Enterococcus faecalis. Protein Expr. Purif. 2004, 34, 302–310. [Google Scholar] [CrossRef] [Green Version]
- Bürger, S.; Stolz, A. Characterisation of the flavin-free oxygen-tolerant azoreductase from Xenophilus azovorans KF46F in comparison to flavin-containing azoreductases. Appl. Microbiol. Biotechnol. 2010, 87, 2067–2076. [Google Scholar] [CrossRef]
- Mendes, S.; Pereira, L.; Batista, C.; Martins, L.O. Molecular determinants of azo reduction activity in the strain Pseudomonas putida MET94. Appl. Microbiol. Biotechnol. 2011, 92, 393–405. [Google Scholar] [CrossRef]
- Ooi, T.; Ogata, D.; Matsumoto, K.I.; Nakamura, G.; Yu, J.; Yao, M.; Kitamura, M.; Taguchi, S. Flavin-binding of azoreductase: Direct evidences for dual-binding property of apo-azoreductase with FMN and FAD. J. Mol. Catal. B Enzym. 2012, 74, 204–208. [Google Scholar] [CrossRef]
- Matsumoto, K.I.; Mukai, Y.; Ogata, D.; Shozui, F.; Nduko, J.M.; Taguchi, S.; Ooi, T. Characterization of thermostable FMN-dependent NADH azoreductase from the moderate thermophile Geobacillus stearothermophilus. Appl. Microbiol. Biotechnol. 2010, 86, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Bin, Y.; Jiti, Z.; Jing, W.; Cuihong, D.; Hongman, H.; Zhiyong, S.; Yongming, B. Expression and characteristics of the gene encoding azoreductase from Rhodobacter sphaeroides AS1. 1737. FEMS Microbiol. Lett. 2004, 236, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, W.; Yoda, T.; Matsuba, T.; Tanaka, Y.; Suzuki, Y. Expression and characterization of the genes encoding azoreductases from Bacillus subtilis and Geobacillus stearothermophilus. Biosci. Biotechnol. Biochem. 2006, 70, 1655–1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macwana, S.R.; Punj, S.; Cooper, J.; Schwenk, E.; John, G.H. Identification and isolation of an azoreductase from Enterococcus faecium. Curr. Issues Mol. Biol. 2009, 12, 43–48. [Google Scholar] [PubMed]
- Mugerfeld, I.; Law, B.A.; Wickham, G.S.; Thompson, D.K. A putative azoreductase gene is involved in the Shewanella oneidensis response to heavy metal stress. Appl. Microbiol. Biotechnol. 2009, 82, 1131–1141. [Google Scholar] [CrossRef]
- Qi, J.; Paul, C.E.; Hollmann, F.; Tischler, D. Changing the electron donor improves azoreductase dye degrading activity at neutral pH. Enzym. Microb. Technol. 2017, 100, 17–19. [Google Scholar] [CrossRef] [Green Version]
- Eslami, M.; Amoozegar, M.A.; Asad, S. Isolation, cloning and characterization of an azoreductase from the halophilic bacterium Halomonas elongata. Int. J. Biol. Macromol. 2016, 85, 111–116. [Google Scholar] [CrossRef]
- Blümel, S.; Busse, H.J.; Stolz, A.; Kämpfer, P. Xenophilus azovorans gen. nov., sp. nov., a soil bacterium that is able to degrade azo dyes of the Orange II type. Int. J. Syst. Evol. Microbiol. 2001, 51, 1831–1837. [Google Scholar] [CrossRef]
- Hua, J.Q.; Yu, L. Cloning and characterization of a Flavin-free oxygen-insensitive azoreductase from Klebsiella oxytoca GS-4-08. Biotechnol. Lett. 2019, 41, 371–378. [Google Scholar] [CrossRef]
- Kim, M.H.; Kim, Y.; Park, H.J.; Lee, J.S.; Kwak, S.N.; Jung, W.H.; Lee, S.G.; Kim, D.; Lee, Y.C.; Oh, T.K. Structural insight into bioremediation of triphenylmethane dyes by Citrobacter sp. triphenylmethane reductase. J. Biol. Chem. 2008, 283, 31981–31990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Feng, J.; Kweon, O.; Xu, H.; Cerniglia, C.E. Identification and molecular characterization of a novel flavin-free NADPH preferred azoreductase encoded by azoB in Pigmentiphaga kullae K24. BMC Biochem. 2010, 11, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngo, A.C.R.; Josef Schultes, F.P.; Maier, A.; Hadewig, S.N.H.; Tischler, D. Improving biocatalytic properties of an azoreductase via the N-terminal fusion of formate dehydrogenase. ChemBioChem 2022, 23, e202100643. [Google Scholar] [CrossRef] [PubMed]
- BluȠmel, S.; Contzen, M.; Lutz, M.; Stolz, A.; Knackmuss, H.J. Isolation of a bacterial strain with the ability to utilize the sulfonated azo compound 4-carboxy-4′-sulfoazobenzene as the sole source of carbon and energy. Appl. Environ. Microbiol. 1998, 64, 2315–2317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.; Ning, S.; Xia, H.; Sun, J. Aerobic decolorization and mineralization of azo dyes by a microbial community in the absence of an external carbon source. Int. Biodeterior. Biodegrad. 2013, 85, 210–216. [Google Scholar] [CrossRef]
- Manogaran, M.; Yasid, N.A.; Othman, A.R.; Gunasekaran, B.; Halmi, M.I.E.; Shukor, M.Y.A. Biodecolourisation of Reactive Red 120 as a sole carbon source by a bacterial consortium—Toxicity assessment and statistical optimisation. Int. J. Environ. Res. Public Health 2021, 18, 2424. [Google Scholar] [CrossRef]
- Saranraj, P.; Sivasakthi, S.; Jayaprakash, A. Studies on the effect of carbon and nitrogen sources for the decolourization of reactive textile dyes by bacterial isolates. World Appl. Sci. J. 2018, 36, 767–773. [Google Scholar]
- Sun, J.; Li, W.; Li, Y.; Hu, Y.; Zhang, Y. Redox mediator enhanced simultaneous decolorization of azo dye and bioelectricity generation in air-cathode microbial fuel cell. Bioresour. Technol. 2013, 142, 407–414. [Google Scholar] [CrossRef]
- Guo, J.; Liu, H.; Qu, J.; Lian, J.; Zhao, L.; Jefferson, W.; Yang, J. The structure activity relationship of non-dissolved redox mediators during azo dye bio-decolorization processes. Bioresour. Technol. 2012, 112, 350–354. [Google Scholar] [CrossRef]
- Walker, R.; Ryan, A.J. Some molecular parameters influencing rate of reduction of azo compounds by intestinal microflora. Xenobiotica 1971, 1, 483–486. [Google Scholar] [CrossRef]
- Datta, S.; Christena, L.R.; Rajaram, Y.R.S. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech 2013, 3, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.C.; Wu, J.Y.; Huang, C.C.; Liang, Y.M.; Hwang, S.C.J. Decolorization of azo dye using PVA-immobilized microorganisms. J. Biotechnol. 2003, 101, 241–252. [Google Scholar] [CrossRef]
- Pandey, K.; Saha, P.; Rao, K.B. A study on the utility of immobilized cells of indigenous bacteria for biodegradation of reactive azo dyes. Prep. Biochem. Biotechnol. 2020, 50, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, M.; Mishra, S.; Sreekrishnan, T.R. Mediator-assisted decolorization and detoxification of textile dyes/dye mixture by Cyathus bulleri laccase. Appl. Biochem. Biotechnol. 2008, 151, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Nadaroglu, H.; Mosber, G.; Gungor, A.A.; Adıguzel, G.; Adiguzel, A. Biodegradation of some azo dyes from wastewater with laccase from Weissella viridescens LB37 immobilized on magnetic chitosan nanoparticles. J. Water Process Eng. 2019, 31, 100866. [Google Scholar] [CrossRef]
- Ayed, L.; Achour, S.; Bakhrouf, A. Application of the mixture design to decolourise effluent textile wastewater using continuous stirred bed reactor. Water Sa 2011, 37, 21–26. [Google Scholar] [CrossRef] [Green Version]
- Al-Ansari, M.M.; Li, Z.; Masood, A.; Rajaselvam, J. Decolourization of azo dye using a batch bioreactor by an indigenous bacterium Enterobacter aerogenes ES014 from the waste water dye effluent and toxicity analysis. Environ. Res. 2022, 205, 112189. [Google Scholar] [CrossRef]
- Iqbal, A.; Ali, N.; Shang, Z.H.; Malik, N.H.; Rehman, M.M.U.; Sajjad, W.; Rehman, M.L.U.; Khan, S. Decolorization and toxicity evaluation of simulated textile effluent via natural microbial consortia in attached growth reactors. Environ. Technol. Innov. 2022, 26, 102284. [Google Scholar] [CrossRef]
- Sodaneath, H.; Lee, J.I.; Yang, S.O.; Jung, H.; Ryu, H.W.; Cho, K.S. Decolorization of textile dyes in an air-lift bioreactor inoculated with Bjerkandera adusta OBR105. J. Environ. Sci. Health Part A 2017, 52, 1099–1111. [Google Scholar] [CrossRef]
- Teerapatsakul, C.; Parra, R.; Keshavarz, T.; Chitradon, L. Repeated batch for dye degradation in an airlift bioreactor by laccase entrapped in copper alginate. Int. Biodeterior. Biodegrad. 2017, 120, 52–57. [Google Scholar] [CrossRef]
- Pakshirajan, K.; Singh, S. Decolorization of synthetic wastewater containing azo dyes in a batch-operated rotating biological contactor reactor with the immobilized fungus Phanerochaete chrysosporium. Ind. Eng. Chem. Res. 2010, 49, 7484–7487. [Google Scholar] [CrossRef]
- Liu, L.; Li, F.B.; Feng, C.H.; Li, X.Z. Microbial fuel cell with an azo-dye-feeding cathode. Appl. Microbiol. Biotechnol. 2009, 85, 175–183. [Google Scholar] [CrossRef]
- Miran, W.; Nawaz, M.; Kadam, A.; Shin, S.; Heo, J.; Jang, J.; Lee, D.S. Microbial community structure in a dual chamber microbial fuel cell fed with brewery waste for azo dye degradation and electricity generation. Environ. Sci. Pollut. Res. 2015, 22, 13477–13485. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Zhang, S.; Liu, H.; Huang, J.; Li, L. Sulfide-mediated azo dye degradation and microbial community analysis in a single-chamber air cathode microbial fuel cell. Bioelectrochemistry 2020, 131, 107349. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.D.; Abdulateif, H.; Ismail, I.M.; Sabir, S.; Khan, M.Z. Bioelectricity generation and bioremediation of an azo-dye in a microbial fuel cell coupled activated sludge process. PLoS ONE 2015, 10, e0138448. [Google Scholar] [CrossRef] [PubMed]


| Enzyme Class Involved | Culture | Dyes | % Decolorization | References |
|---|---|---|---|---|
| Laccases | Marasmius scorodonius | Congo Red | 90% | [101] |
| Malachite Green | 82% | |||
| Crystal Violet | 69% | |||
| Methylene Green | 63% | |||
| Reactive Orange 16 | 48% | |||
| (+ 1-hydroxybenzotriazole) | ||||
| Remazol Brilliant Blue R | 61% | |||
| (+ 1-hydroxybenzotriazole) | ||||
| Myceliopthora thermophila | Acid Blue 74 | 15.20% | [97] | |
| Acid Blue 25 | 53.30% | |||
| Acid Green 27 | 67% | |||
| Reactive Blue 19 | 31.20% | |||
| Direct Red 28 | 9.60% | |||
| Trametes versicolor | Acid Blue 74 | 88.40% | [97] | |
| Acid Blue 25 | 66.00% | |||
| Acid Green 27 | 76.00% | |||
| Reactive Blue 19 | 64.50% | |||
| Direct Red 28 | 11.90% | |||
| Aspergillus ochraceus NCIM 1146 | Reactive Navy Blue HER | 90.00% | [102] | |
| Reactive Golden Yellow HER | 90.00% | |||
| Methyl Orange | ||||
| 56.00% | ||||
| Lignin peroxidases | Phanerochaete chrysosporium (Crude lignin peroxidases with 2 mM veratryl alcohol) | Bromophenol Blue | 93% | [103] |
| Congo Red | 54% | |||
| Methylene Blue | ~85% | |||
| Methyl Green | ~85% | |||
| Methyl Orange | ~85% | |||
| Remazol Brilliant Blue R | ~70% | |||
| Toluidine Blue | 80% | |||
| Poly R-478 | 46% | |||
| Poly S-119 | 80% | |||
| Poly T-128 | 48% | |||
| Ganoderma lucidum IBL-05 (with 4 mM veratryl alcohol) | Sandal-fix Red C4BLN | 66% | [104] | |
| Sandal-fix Turq Blue GWF | 59% | |||
| Sandal-fix Foron Blue E2BLN | 52% | |||
| Sandal-fix Black CKF | 40% | |||
| Sandal-fix Golden Yellow CRL | 48% | |||
| Bjerkandera adusta CX-9 | Acid Blue 158 | ~40% | [105] | |
| Cibacet Brilliant Blue BG | 25% | |||
| Poly R-478 | ~30% | |||
| Methyl Green | 75% | |||
| Indigo Carmine | 50% | |||
| Remazol Brilliant Blue R | ~90% | |||
| Remazol Brilliant Violet 5R | <20% | |||
| Manganese peroxidase | Bjerkandera adusta CX-9 | Acid Blue 158 | 91% | [105] |
| Cibacet Brilliant Blue BG | 70% | |||
| Poly R-478 | 80% | |||
| Methyl Green | <20% | |||
| Indigo Carmine | ~45% | |||
| Remazol Brilliant Blue R | ~40% | |||
| Remazol Brilliant Violet 5R | 70% | |||
| Cerrena unicolor BBP6 | Congo Red | 54% | [106] | |
| Methyl Orange | 78% | |||
| Remazol Brilliant Blue R | 81% | |||
| Bromophenol Blue | 62% | |||
| Crystal Violet | 81% | |||
| Azure Blue (+gallic acid) | 63% |
| Culture | Dyes | % Decolorization | References |
|---|---|---|---|
| (Time of Incubation) | |||
| Bacillus sp. AK1 | Metanil Yellow | 99% (24 h) | [125] |
| Sphingomonas paucimobilis | Methyl Red | 99.6% (10 h) | [126] |
| Proteus mirabilis | Red RBN | 95% (20 h) | [127] |
| Aeromonas hydrophila | Red RBN | 90% (8 days) | [128] |
| Brevibacterium sp. VN-15 | Reactive Yellow 107 | 98% (96 h) | [129] |
| Reactive Black 5 | 95% (144 h) | ||
| Reactive Red 198 | 97% (120 h) | ||
| Direct Blue 71 | 94% (168 h) | ||
| Acinetobacter calcoaceticus NCIM 2890 | Amaranth | 93% (48 h) | [93] |
| Methyl Red | 95% (24 h) | ||
| Amido Black 10 B | 87% (72 h) | ||
| Congo Red | 17% (72 h) | ||
| Bacillus firmus H4 | Novacron Red | 80–89% (24 h) | [130] |
| Bacillus filamentosus T13 | Novacron Red | 80–89% (24 h) | [130] |
| Bacillus subterraneus A36 | Novacron Red | 80–89% (24 h) | [130] |
| Micrococcus luteus 24M | Congo Red | 99% (11 days) | [131] |
| Pseudomonas sp. SUK1 | Red BLI | 99% (1 h) | [132] |
| Pseudomonas sp. SUK1 | Reactive Red 2 | >80% (48 h to 72 h) | [133] |
| Shewanella putrefaciens | Acid Red 88 | 100% (4 h) | [134] |
| Direct Red 81 | 100% (4 h) | ||
| Reactive Black 5 | 100% (6 h) | ||
| Disperse Orange 3 | 100% (8 h) | ||
| Kocuria indica DP-K7 | Methyl Red | 68% (160 h) | [135] |
| Arthrobacter bambusae DP-A9 | Methyl Red | 100% (24 h) | [136] |
| Brilliant Black | 100% (24 h) | ||
| Leifsonia shinshuensis DP-L11 | Methyl Red | 53% (24 h) | [136] |
| Brilliant Black | 85% (24 h) | ||
| Dermacoccus nishinomiyaensis DP-D10 | Methyl Red | 84% (24 h) | [136] |
| Brilliant Black | 100% (24 h) | ||
| Paraburkholderia sp. DP-P12 | Methyl Red | 58% (24 h) | [136] |
| Brilliant Black | 62.5% (24 h) | ||
| Rhodococcus sp. UCC 0008 | Methyl Red | 100% (72 h) | [137] |
| Rhodococcus sp. UCC 0016 | Methyl Red | 100% (24 h) | [137] |
| Staphylococcus sp. EY-3 | Congo Red | >96% (48 h) | [138] |
| Kocuria rosea MTCC 1532 | Methyl Orange | 100% (72 h) | [139] |
| Citrobacter sp. CK3 | Reactive Red 180 | 95% (36 h) | [140] |
| Bacillus sp. YZU1 | Reactive Black 5 | 95% (120 h) | [141] |
| Culture | Dyes | % Decolorization (Time of Incubation) | References | |
|---|---|---|---|---|
| Bacterial consortium | Bacillus circulans BPB8 | Textile effluents with mixed azo dyes (Reactive Red, Reactive Brown, Reactive Black) and Cr(VI) | 82% (5 days) | [142] |
| Bacillus circulans HQB947 | ||||
| Bacillis subtilis | ||||
| Terribacillus gorriensis | ||||
| Fungal–bacterial consortium | White Rot fungus 8-4* Pseudomonas | Direct Fast Scarlet 4BS (Sole Carbon Source) | 100% (30 h) | [143] |
| Bacterial consortium | Pseudomonas sp. ARa | Reactive Red 195 (Maltose and Proteose Peptone) | 100% (14 h) | [144] |
| Bacillus sp. ARc | ||||
| Bacillus sp. ARd | ||||
| Ochrobactrum sp. ARf | ||||
| Bacterial consortium | Bacillus cereus BN-7 | Acid Red 88 | 100% (24 h) | [145] |
| Pseudomonas putida BN-4 | ||||
| Pseudomonas fluorescence BN-5 | ||||
| Stenotrophomonas acidaminiphila BN-3 | ||||
| Fungal–bacterial consortium | Brevibacillus laterosporus Galactomyces geotrichum | Reactive Red 198 | 92% (18 h) | [146] |
| Fungal–bacterial consortium | Aspergillus ochraceous NCIM-1146 | Rubine GFL | 95% (30 h) | [147] |
| Pseudomonas sp. SUK1 | Textile effluent | 98% (35 h) | ||
| Bacterial consortium | Bacillus sp. AK1 | Ponceau 4R | 100% (18 h) | [148] |
| Lysinibacillus sp. AK2 | ||||
| Kerstersia sp. VKY1 | ||||
| Bacterial consortium | Paenibacillus polymyxa | Reactive Violet 5R | 100% (36 h) | [149] |
| Micrococcus luteus | ||||
| Micrococcus sp. | ||||
| Bacterial consortium | Enterobacter dissolvens AGYP1 | Acid Maroon V | 93% (20 h) | [150] |
| Pseudomonas aeruginosa AGYP2 | ||||
| Bacterial consortium | Bacillus odyssey SUK3 | Red HE3B | 97% (24 h) | [151] |
| Morganella morganii SUK5 | ||||
| Proteus sp. SUK7 | ||||
| Bacterial consortium | Providencia sp. SDS | Red HE3B | 100% (1 h) | [152] |
| Pseudomonas aeruginosa BCH | ||||
| Bacterial consortium | Proteus vulgaris NCIM-2027 (PV) | Scarlet Red Dye Mixture (Scarlet R, Navy Blue HER, Red HE7B, Green HE4BD, Orange HE2R, Navy Blue G, Red HE3B, Navy Blue HE2R, Golden Yellow 24D, Brilliant Blue G, Direct Brown MR, Direct Blue GLL) | 100% (3 h) | [153] |
| Micrococcus glutamicus NCIM-2168 (MG) | 88% (72 h) | |||
| Bacterial consortium | Bacillus subtilis WGI3 | Direct Red 23 | 70% (48 h) | [154] |
| Bacillus subtilis WGI4 | Direct Yellow 12 | 84% (48 h) | ||
| Bacillus cereus WGI9 | Direct Blue 15 | 66% (48 h) | ||
| Dye Mixture | 75% (48 h) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngo, A.C.R.; Tischler, D. Microbial Degradation of Azo Dyes: Approaches and Prospects for a Hazard-Free Conversion by Microorganisms. Int. J. Environ. Res. Public Health 2022, 19, 4740. https://doi.org/10.3390/ijerph19084740
Ngo ACR, Tischler D. Microbial Degradation of Azo Dyes: Approaches and Prospects for a Hazard-Free Conversion by Microorganisms. International Journal of Environmental Research and Public Health. 2022; 19(8):4740. https://doi.org/10.3390/ijerph19084740
Chicago/Turabian StyleNgo, Anna Christina R., and Dirk Tischler. 2022. "Microbial Degradation of Azo Dyes: Approaches and Prospects for a Hazard-Free Conversion by Microorganisms" International Journal of Environmental Research and Public Health 19, no. 8: 4740. https://doi.org/10.3390/ijerph19084740
APA StyleNgo, A. C. R., & Tischler, D. (2022). Microbial Degradation of Azo Dyes: Approaches and Prospects for a Hazard-Free Conversion by Microorganisms. International Journal of Environmental Research and Public Health, 19(8), 4740. https://doi.org/10.3390/ijerph19084740







