The Effects of Paroxetine on Benthic Microbial Food Web and Nitrogen Transformation in River Sediments
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collection
2.2. Microcosm Experiments and Environmental Parameter Analysis
2.3. eDNA Extraction, PCR Amplification, and Sequencing Analysis
2.4. Statistical Analysis
3. Results and Discussion
3.1. Effect of Paroxetine on Different Forms of Nitrogen in Sediments
3.2. Effect of Paroxetine on Bacterial Communities in Sediment
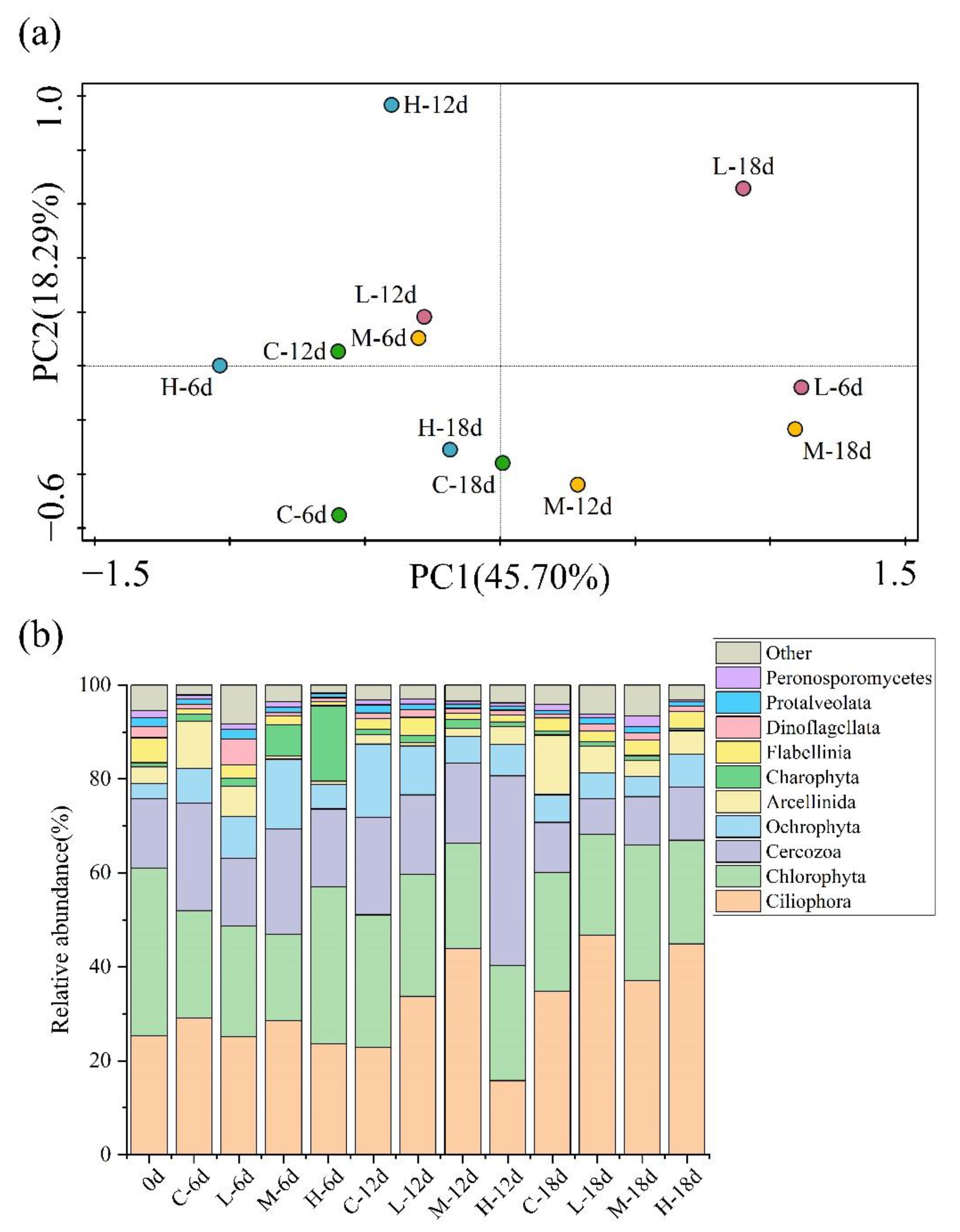
3.3. Effect of Paroxetine on Eukaryotic Communities in Sediments
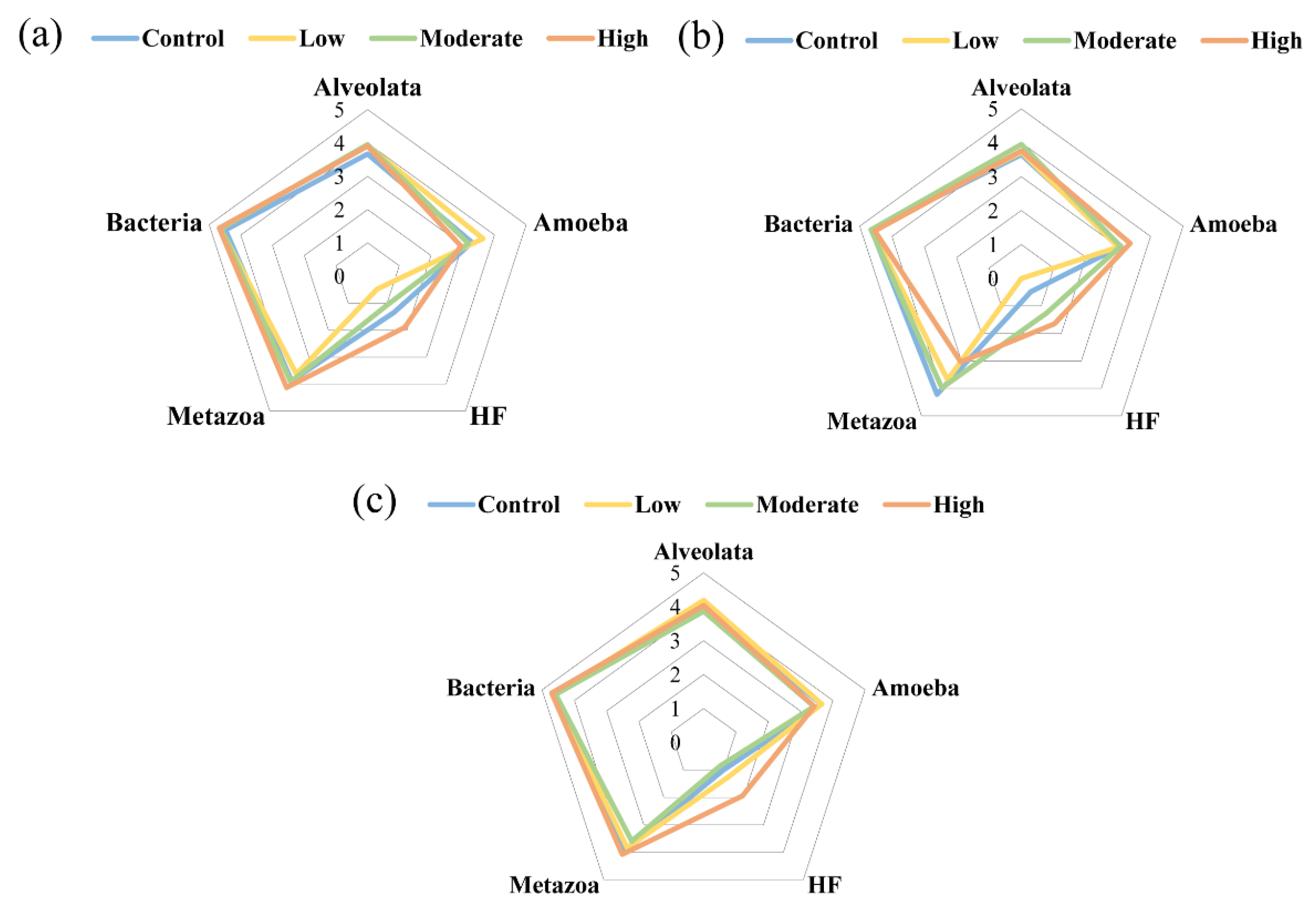
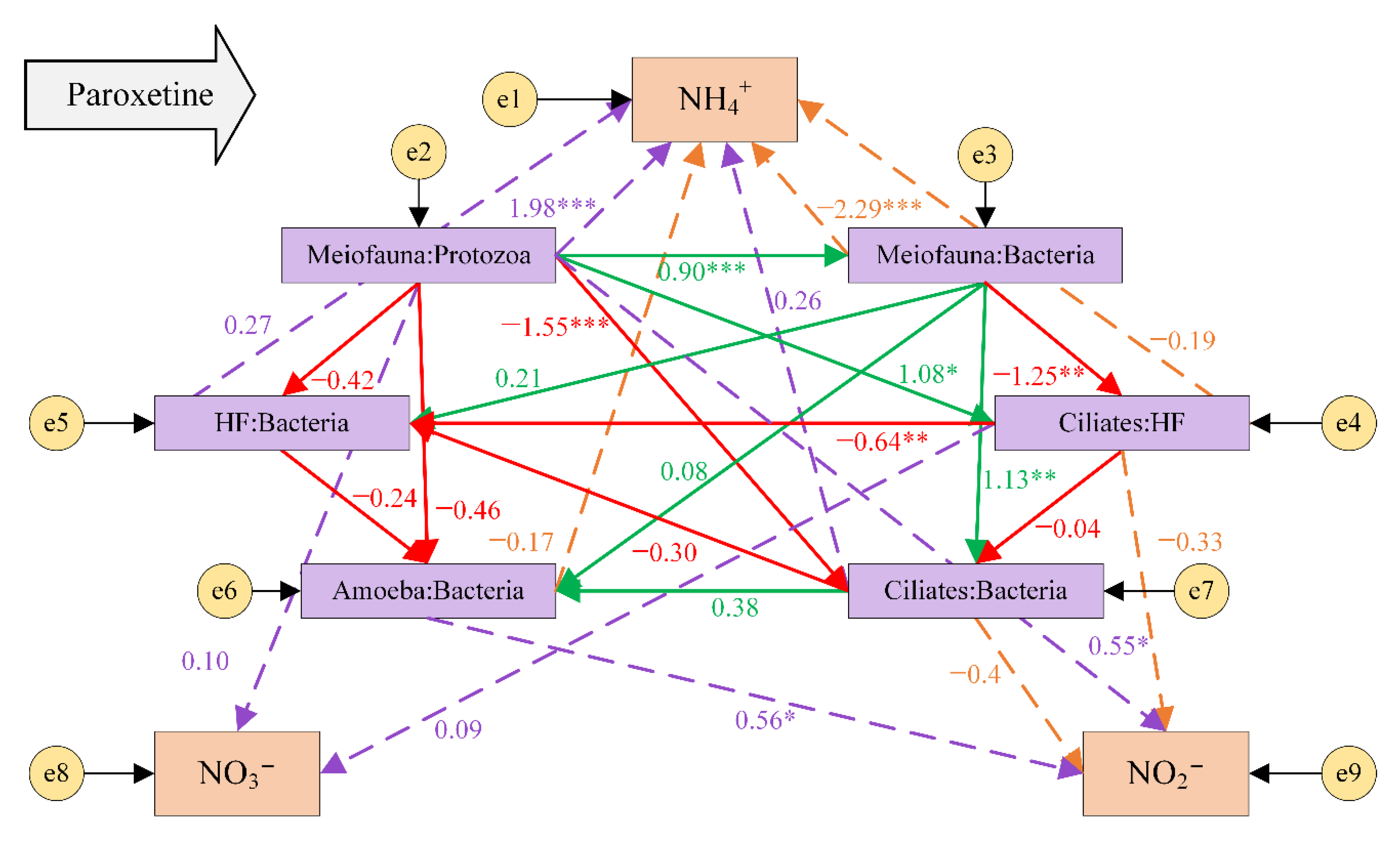
3.4. Top-Down Controls in the Microbial Food Web under the Influence of Paroxetine
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Smith, L.; Jacob, L.; Yakkundi, A.; McDermott, D.; Armstrong, N.C.; Barnett, Y.; López-Sánchez, G.F.; Martin, S.; Butler, L.; Tully, M.A. Correlates of Symptoms of Anxiety and Depression and Mental Wellbeing Associated with COVID-19: A Cross-Sectional Study of UK-Based Respondents. Psychiatry Res. 2020, 291, 113138. [Google Scholar] [CrossRef]
- Niederkrotenthaler, T.; Laido, Z.; Kirchner, S.; Braun, M.; Metzler, H.; Waldhör, T.; Strauss, M.J.; Garcia, D.; Till, B. Mental Health over Nine Months during the SARS-CoV2 Pandemic: Representative Cross-Sectional Survey in Twelve Waves between April and December 2020 in Austria. J. Affect. Disord. 2022, 296, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Robinson, E.; Sutin, A.R.; Daly, M.; Jones, A. A Systematic Review and Meta-Analysis of Longitudinal Cohort Studies Comparing Mental Health before versus during the COVID-19 Pandemic in 2020. J. Affect. Disord. 2022, 296, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Lopes, D.G.; Duarte, I.A.; Antunes, M.; Fonseca, V.F. Effects of Antidepressants in the Reproduction of Aquatic Organisms: A Meta-Analysis. Aquat. Toxicol. 2020, 227, 105569. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Camal, N.; Cardoso-Vera, J.D.; Islas-Flores, H.; Gómez-Oliván, L.M.; Mejía-García, A. Consumption and Ocurrence of Antidepressants (SSRIs) in Pre- and Post-COVID-19 Pandemic, Their Environmental Impact and Innovative Removal Methods: A Review. Sci. Total Environ. 2022, 829, 154656. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Kubec, J.; Guo, W.; Roje, S.; Ložek, F.; Grabicová, K.; Randák, T.; Kouba, A.; Buřič, M. A Combination of Six Psychoactive Pharmaceuticals at Environmental Concentrations Alter the Locomotory Behavior of Clonal Marbled Crayfish. Sci. Total Environ. 2021, 751, 141383. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Ohman, K.; Metcalfe, C.; Ikonomou, M.G.; Amatya, P.L.; Wilson, J. Pharmaceuticals and Endocrine Disruptors in Wastewater Treatment Effluents and in the Water Supply System of Calgary, Alberta, Canada. Water Qual. Res. J. 2006, 41, 351–364. [Google Scholar] [CrossRef]
- Yang, Z.; Lu, T.; Zhu, Y.; Zhang, Q.; Zhou, Z.; Pan, X.; Qian, H. Aquatic Ecotoxicity of an Antidepressant, Sertraline Hydrochloride, on Microbial Communities. Sci. Total Environ. 2019, 654, 129–134. [Google Scholar] [CrossRef]
- Li, Y.; Miao, Y.; Zhang, W.; Yang, N.; Niu, L.; Zhang, H.; Wang, L. Sertraline Inhibits Top-down Forces (Predation) in Microbial Food Web and Promotes Nitrification in Sediment. Environ. Pollut. 2020, 267, 115580. [Google Scholar] [CrossRef]
- Macaluso, M.; Preskorn, S.H. (Eds.) Antidepressants: From Biogenic Amines to New Mechanisms of Action. In Handbook of Experimental Pharmacology; Springer International Publishing: Cham, Switzerland, 2019; Volume 250, ISBN 978-3-030-10948-6. [Google Scholar]
- Vaclavik, J.; Sehonova, P.; Hodkovicova, N.; Vecerkova, L.; Blahova, J.; Franc, A.; Marsalek, P.; Mares, J.; Tichy, F.; Svobodova, Z.; et al. The Effect of Foodborne Sertraline on Rainbow Trout (Oncorhynchus Mykiss). Sci. Total Environ. 2020, 708, 135082. [Google Scholar] [CrossRef]
- Zhu, S.; Wu, G.; Gu, L.; Sun, Y.; Zhang, L.; Huang, Y.; Lyu, K.; Yang, Z. Antidepressant Sertraline Impairs the Induced Morphological Defense of Ceriodaphnia Cornuta in Response to Chaoborus Larvae Kairomone. Environ. Pollut. 2020, 266, 115092. [Google Scholar] [CrossRef]
- Zorita, S.; Mårtensson, L.; Mathiasson, L. Occurrence and Removal of Pharmaceuticals in a Municipal Sewage Treatment System in the South of Sweden. Sci. Total Environ. 2009, 407, 2760–2770. [Google Scholar] [CrossRef] [PubMed]
- Klaminder, J.; Brodin, T.; Sundelin, A.; Anderson, N.J.; Fahlman, J.; Jonsson, M.; Fick, J. Long-Term Persistence of an Anxiolytic Drug (Oxazepam) in a Large Freshwater Lake. Environ. Sci. Technol. 2015, 49, 10406–10412. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, X.; Miao, Y.; Li, Y.; Zhang, H.; Niu, L.; Wang, L. Determining the Effect of Sertraline on Nitrogen Transformation through the Microbial Food Web in Sediments Based on 15N-DNA-Stable Isotope Probing. Environ. Res. 2021, 199, 111347. [Google Scholar] [CrossRef] [PubMed]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the Nitrogen Cycle: Recent Trends, Questions, and Potential Solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef]
- Fowler, D.; Pyle, J.A.; Raven, J.A.; Sutton, M.A. The Global Nitrogen Cycle in the Twenty-First Century: Introduction. Phil. Trans. R. Soc. B 2013, 368, 20130165. [Google Scholar] [CrossRef]
- Bonaglia, S.; Nascimento, F.J.A.; Bartoli, M.; Klawonn, I.; Brüchert, V. Meiofauna Increases Bacterial Denitrification in Marine Sediments. Nat. Commun. 2014, 5, 5133. [Google Scholar] [CrossRef]
- Lotti, T.; Cordola, M.; Kleerebezem, R.; Caffaz, S.; Lubello, C.; van Loosdrecht, M.C.M. Inhibition Effect of Swine Wastewater Heavy Metals and Antibiotics on Anammox Activity. Water Sci. Technol. 2012, 66, 1519–1526. [Google Scholar] [CrossRef]
- Li, N.; Liu, Q.; Zhou, G.-Q.; Dai, M.-X.; Kong, Q. Contaminant Removal and Microorganism Response of Activated Sludge in Sulfamethazine Wastewater Treatment. Int. Biodeterior. Biodegrad. 2019, 143, 104705. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Y.; Zhang, H.; Wang, L.; Zhang, W.; Niu, L.; Wang, P.; Wang, C. The Responses of Bacterial Community and N2O Emission to Nitrogen Input in Lake Sediment: Estrogen as a Co-Pollutant. Environ. Res. 2019, 179, 108769. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Li, Y.; Wang, P.; Wang, C. Background Nutrients and Bacterial Community Evolution Determine 13C-17β-Estradiol Mineralization in Lake Sediment Microcosms. Sci. Total Environ. 2019, 651, 2304–2311. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, R.; Wang, L.; Niu, L.; Wang, C.; Hu, J.; Wu, H.; Zhang, W.; Wang, P. Silver Nanoparticles and Fe(III) Co-Regulate Microbial Community and N2O Emission in River Sediments. Sci. Total Environ. 2020, 706, 135712. [Google Scholar] [CrossRef] [PubMed]
- Damashek, J.; Francis, C.A. Microbial Nitrogen Cycling in Estuaries: From Genes to Ecosystem Processes. Estuaries Coasts 2018, 41, 626–660. [Google Scholar] [CrossRef]
- Delmont, T.O.; Quince, C.; Shaiber, A.; Esen, Ö.C.; Lee, S.T.; Rappé, M.S.; McLellan, S.L.; Lücker, S.; Eren, A.M. Nitrogen-Fixing Populations of Planctomycetes and Proteobacteria Are Abundant in Surface Ocean Metagenomes. Nat. Microbiol. 2018, 3, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Hüpeden, J.; Wemheuer, B.; Indenbirken, D.; Schulz, C.; Spieck, E. Taxonomic and Functional Profiling of Nitrifying Biofilms in Freshwater, Brackish and Marine RAS Biofilters. Aquac. Eng. 2020, 90, 102094. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete Nitrification by Nitrospira Bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef]
- Cotto, I.; Dai, Z.; Huo, L.; Anderson, C.L.; Vilardi, K.J.; Ijaz, U.; Khunjar, W.; Wilson, C.; De Clippeleir, H.; Gilmore, K.; et al. Long Solids Retention Times and Attached Growth Phase Favor Prevalence of Comammox Bacteria in Nitrogen Removal Systems. Water Res. 2020, 169, 115268. [Google Scholar] [CrossRef]
- Cunningham-Bussel, A.; Zhang, T.; Nathan, C.F. Nitrite Produced by Mycobacterium Tuberculosis in Human Macrophages in Physiologic Oxygen Impacts Bacterial ATP Consumption and Gene Expression. Proc. Natl. Acad. Sci. USA 2013, 110, E4256–E4265. [Google Scholar] [CrossRef]
- Malhotra, V.; Agrawal, R.; Duncan, T.R.; Saini, D.K.; Clark-Curtiss, J.E. Mycobacterium Tuberculosis Response Regulators, DevR and NarL, Interact in Vivo and Co-Regulate Gene Expression during Aerobic Nitrate Metabolism. J. Biol. Chem. 2015, 290, 8294–8309. [Google Scholar] [CrossRef]
- Moreno-Vivián, C.; Flores, E. Nitrate Assimilation in Bacteria. In Biology of the Nitrogen Cycle, 1st ed.; Elsevier: London, UK, 2007; pp. 263–282. ISBN 978-0-444-52857-5. [Google Scholar]
- Huang, Q.; Abdalla, A.E.; Xie, J. Phylogenomics of Mycobacterium Nitrate Reductase Operon. Curr. Microbiol. 2015, 71, 121–128. [Google Scholar] [CrossRef]
- Yang, N.; Li, Y.; Zhang, W.; Lin, L.; Qian, B.; Wang, L.; Niu, L.; Zhang, H. Cascade Dam Impoundments Restrain the Trophic Transfer Efficiencies in Benthic Microbial Food Web. Water Res. 2020, 170, 115351. [Google Scholar] [CrossRef] [PubMed]
- Neuparth, T.; Lopes, A.I.; Alves, N.; Oliveira, J.M.A.; Santos, M.M. Does the Antidepressant Sertraline Show Chronic Effects on Aquatic Invertebrates at Environmentally Relevant Concentrations? A Case Study with the Keystone Amphipod, Gammarus Locusta. Ecotoxicol. Environ. Saf. 2019, 183, 109486. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.W.; Wu, W.; Chung, C.-C.; Chiang, K.-P.; Gong, G.-C.; Hsieh, C. Predator and Prey Biodiversity Relationship and Its Consequences on Marine Ecosystem Functioning—Interplay between Nanoflagellates and Bacterioplankton. ISME J. 2018, 12, 1532–1542. [Google Scholar] [CrossRef] [PubMed]
- Weisse, T. Functional Diversity of Aquatic Ciliates. Eur. J. Protistol. 2017, 61, 331–358. [Google Scholar] [CrossRef] [PubMed]
- Prast, M.; Bischoff, A.; Waller, U.; Amann, R.; Berninger, U. Effect of Ciliates on Nitrification and Nitrifying Bacteria in Baltic Sea Sediments. Mar. Ecol. Prog. Ser. 2007, 350, 55–61. [Google Scholar] [CrossRef]
- Turley, C.; Newell, R.; Robins, D. Survival Strategies of Two Small Marine Ciliates and Their Role in Regulating Bacterial Community Structure under Experimental Conditions. Mar. Ecol. Prog. Ser. 1986, 33, 59–70. [Google Scholar] [CrossRef]
- Domingues, C.D.; da Silva, L.H.S.; Rangel, L.M.; de Magalhães, L.; de Melo Rocha, A.; Lobão, L.M.; Paiva, R.; Roland, F.; Sarmento, H. Microbial Food-Web Drivers in Tropical Reservoirs. Microb. Ecol. 2017, 73, 505–520. [Google Scholar] [CrossRef]



| Samples | Chao1 Index | ACE Index | Simpson Index | Shannon Index |
|---|---|---|---|---|
| C-6d | 1303.455 | 1308.979 | 0.954122 | 6.399949 |
| C-12d | 1259.007 | 1273.765 | 0.927629 | 5.935353 |
| C-18d | 1649.173 | 1690.546 | 0.966728 | 6.623691 |
| L-6d | 1482.061 | 1479.052 | 0.982247 | 7.328378 |
| L-12d | 1200.021 | 1221.052 | 0.962134 | 6.803168 |
| L-18d | 1435.575 | 1347.239 | 0.968264 | 6.634653 |
| M-6d | 1887.182 | 1908.239 | 0.971867 | 7.001973 |
| M-12d | 1335.679 | 1328.988 | 0.947955 | 6.202316 |
| M-18d | 1427.407 | 1386.27 | 0.983062 | 7.209895 |
| H-6d | 2115.781 | 2197.906 | 0.948932 | 6.364666 |
| H-12d | 2003.015 | 1936.051 | 0.98843 | 7.652237 |
| H-18d | 1477.005 | 1521.179 | 0.953361 | 6.300156 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Chen, X.; Wang, X.; Shang, J.; Niu, L.; Wang, L.; Zhang, H.; Zhang, W. The Effects of Paroxetine on Benthic Microbial Food Web and Nitrogen Transformation in River Sediments. Int. J. Environ. Res. Public Health 2022, 19, 14602. https://doi.org/10.3390/ijerph192114602
Li Y, Chen X, Wang X, Shang J, Niu L, Wang L, Zhang H, Zhang W. The Effects of Paroxetine on Benthic Microbial Food Web and Nitrogen Transformation in River Sediments. International Journal of Environmental Research and Public Health. 2022; 19(21):14602. https://doi.org/10.3390/ijerph192114602
Chicago/Turabian StyleLi, Yi, Xinqi Chen, Xinzi Wang, Jiahui Shang, Lihua Niu, Longfei Wang, Huanjun Zhang, and Wenlong Zhang. 2022. "The Effects of Paroxetine on Benthic Microbial Food Web and Nitrogen Transformation in River Sediments" International Journal of Environmental Research and Public Health 19, no. 21: 14602. https://doi.org/10.3390/ijerph192114602
APA StyleLi, Y., Chen, X., Wang, X., Shang, J., Niu, L., Wang, L., Zhang, H., & Zhang, W. (2022). The Effects of Paroxetine on Benthic Microbial Food Web and Nitrogen Transformation in River Sediments. International Journal of Environmental Research and Public Health, 19(21), 14602. https://doi.org/10.3390/ijerph192114602





