Genetic Evolution Analysis and Host Characteristics of Hantavirus in Yunnan Province, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
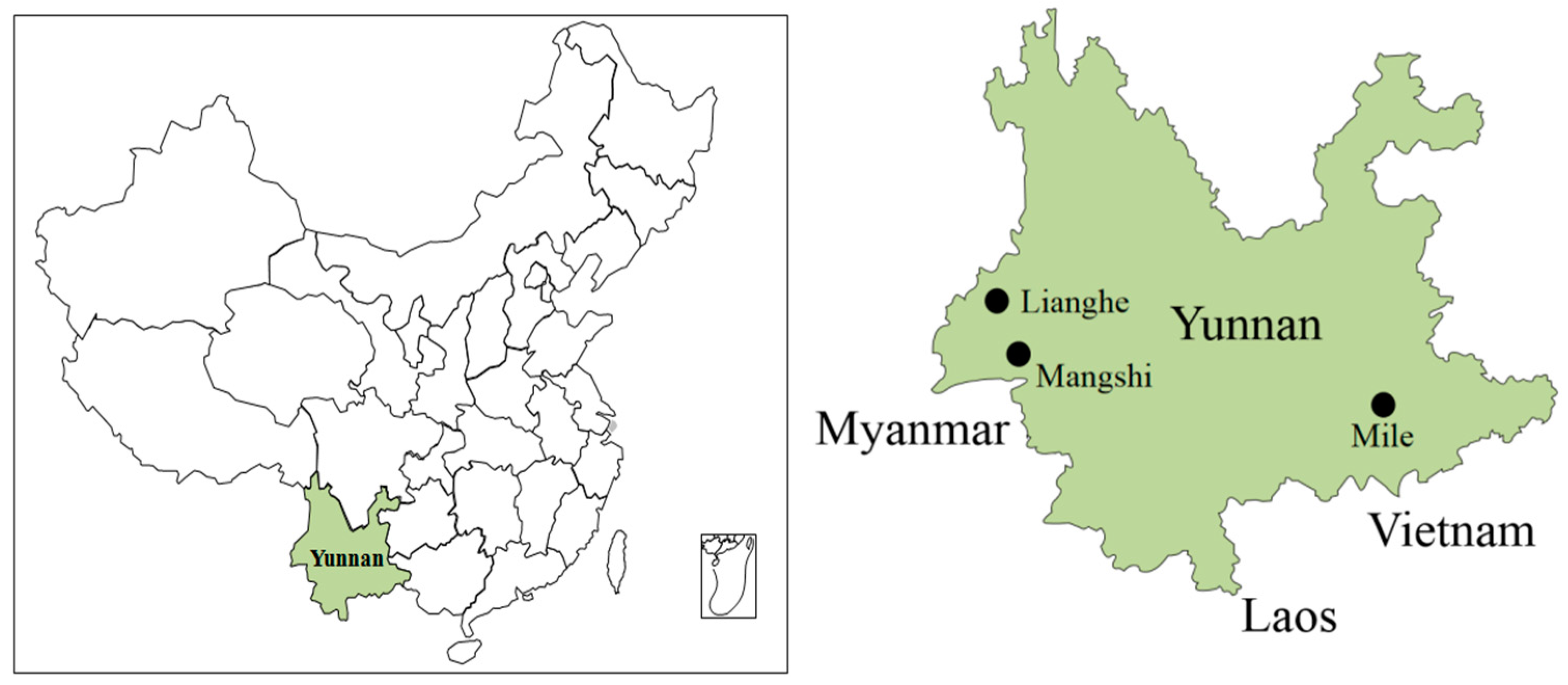
2.2. Sample Collection
2.3. Extraction of Nucleic Acid and HV Detection
2.4. Amplification and Sequencing of S-Segment Gene of SEOV
2.5. Genetic Characterization and Phylogenetic Analysis
2.6. Nucleotide Sequence Accession Numbers
3. Results
3.1. HV Infection in Murine-Shaped Animals
3.2. Sequence Homology Analysis
3.3. Genetic Evolution Analysis
4. Discussion
4.1. Expansion of the Monitoring Scope of Host Animal Species
4.2. Focusing on Genetic Variation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hsieh, C.J.; Li, C.W.; Cheng, C.A.; Wu, D.C.; Wu, W.C.; Lin, F.H.; Chou, Y.C.; Yu, C.P. Epidemiologic characteristics of domestic patients with hemorrhagic fever with renal syndrome in Taiwan: A 19-year retrospective study. Int. J. Environ. Res. Public Health 2020, 17, 5291. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.X.; Chen, M.J.; Sun, L. Haemorrhagic fever with renal syndrome: Literature review and distribution analysis in China. Int. J. Infect. Dis. 2016, 43, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Avsic-Zupanc, T.; Saksida, A.; Korva, M. Hantavirus infections. Clin. Microbiol. Infec. 2019, 21 (Suppl.), E6–E16. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Numan, A.; Sharma, D.; Shukla, R.; Alexander, A.; Jain, G.K.; Ahmad, F.J.; Kesharwani, P. Epidemiology, virology and clinical aspects of hantavirus infections: An overview. Int. J. Environ. Health Res. 2022, 32, 1815–1826. [Google Scholar] [CrossRef] [PubMed]
- Brocato, R.L.; Hooper, J.W. Progress on the prevention and treatment of hantavirus disease. Viruses 2019, 11, 610. [Google Scholar] [CrossRef]
- Figueiredo, L.T.M.; de Souza, W.M.; Ferres, M.; Enria, D.A. Hantaviruses and cardiopulmonary syndrome in South America. Virus Res. 2014, 187, 43–54. [Google Scholar] [CrossRef]
- Mittler, E.; Dieterle, M.E.; Kleinfelter, L.M.; Slough, M.M.; Chandran, K.; Jangra, R.K. Hantavirus entry: Perspectives and recent advances. Adv. Virus Res. 2019, 104, 185–224. [Google Scholar] [CrossRef]
- He, J.; Wang, Y.; Mu, D.; Xu, Z.; Qian, Q.; Chen, G.; Wen, L.; Yin, W.; Li, S.; Zhang, W.; et al. The impacts of climatic factors and vegetation on hemorrhagic fever with renal syndrome transmission in China: A study of 109 Counties. Int. J. Environ. Res. Public Health 2019, 16, 3434. [Google Scholar] [CrossRef]
- Bai, X.H.; Peng, C.; Jiang, T.; Hu, Z.M.; Huang, D.S.; Guan, P. Distribution of geographical scale, data aggregation unit and period in the correlation analysis between temperature and incidence of HFRS in mainland China: A systematic review of 27 ecological studies. PLoS Negl. Trop. Dis. 2019, 13, e0007688. [Google Scholar] [CrossRef]
- He, W.; Fu, J.; Wen, Y.; Cheng, M.; Mo, Y.; Chen, Q. Detection and genetic characterization of Seoul virus in liver tissue samples from Rattus norvegicus and Rattus tanezumi in Urban Areas of Southern China. Front. Vet. Sci. 2021, 8, 748232. [Google Scholar] [CrossRef]
- Wang, Q.W.; Tao, L.; Lu, S.Y.; Zhu, C.Q.; Ai, L.L.; Luo, Y.; Yu, R.B.; Lv, H.; Zhang, Y.; Wang, C.C.; et al. Genetic and hosts characterization of hantaviruses in port areas in Hainan Province, P.R. China. PLoS ONE 2022, 17, e0264859. [Google Scholar] [CrossRef]
- Arai, S.; Yanagihara, R. Genetic diversity and geographic distribution of bat-borne hantaviruses. In Bat-Borne Viruses; Corrales-Aguilar, E., Schwemmle, M., Eds.; Caister Academic Press: Poole, UK, 2020. [Google Scholar]
- Witkowski, P.T.; Drexler, J.F.; Kallies, R.; Lickova, M.; Bokorova, S.; Mananga, G.D.; Szemes, T.; Leroy, E.M.; Kruger, D.H.; Drosten, C.; et al. Phylogenetic analysis of a newfound bat-borne hantavirus supports a laurasiatherian host association for ancestral mammalian hantaviruses. Infect. Genet. Evol. 2016, 41, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yin, J.X. Epidemic process and influencing factors of hemorrhagic fever with renal syndrome: A review. Chin. J. Schistosomiasis Control. 2022, 34, 200–203. [Google Scholar] [CrossRef]
- Dheerasekara, K.; Sumathipala, S.; Muthugala, R. Hantavirus infections-Treatment and prevention. Curr. Treat. Options Infect. Dis. 2020, 12, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Guterres, A.; de Lemos, E.R.S. Hantaviruses and a neglected environmental determinant. One Health 2018, 5, 27–33. [Google Scholar] [CrossRef]
- Shang, C.; Yang, L.F.; Du, S.S.; Joseph, O.S.; Zhou, J.H.; Huang, X.X.; Han, Q.; Li, C.; Li, A.A.; Wang, Q.; et al. Genotyping of hantaviruses from Rodents in Yunnan Province, China, 2018-2019. Chin. J. Virol. 2022, 38, 149–155. [Google Scholar] [CrossRef]
- Pang, Z. Establishment of Multiplex Real Time RT-PCR Assays for Hemorrhagic Fever Viruses Detection; Chinese Center for Disease Control and Prevention: Beijing, China, 2013. [Google Scholar]
- Zhang, Y.H.; Ge, L.; Liu, L.; Huo, X.X.; Xiong, H.R.; Liu, Y.Y.; Liu, D.Y.; Luo, F.; Li, J.L.; Ling, J.X.; et al. The epidemic characteristics and changing trend of hemorrhagic fever with renal syndrome in Hubei Province, China. PLoS ONE 2014, 9, e92700. [Google Scholar] [CrossRef]
- Wang, Q.; Li, J.D.; Zhang, Q.F.; Qu, J.; Wang, S.W. Surveillance for hemorrhagic fever with renal syndrome in China, 2014. Dis. Surveill. 2016, 31, 192–199. [Google Scholar] [CrossRef]
- Yin, J.X.; Geater, A.; Chongsuvivatwong, V.; Dong, X.Q.; Du, C.H.; Zhong, Y.H.; McNeil, E. Predictors for presence and abundance of small mammals in households of villages endemic for commensal rodent plague in Yunnan Province, China. BMC Ecol. 2008, 8, 18. [Google Scholar] [CrossRef]
- Yin, J.X.; Geater, A.; Chongsuvivatwong, V.; Dong, X.Q.; Du, C.H.; Zhong, Y.H. Predictors for abundance of host flea and floor flea in households of villages with endemic commensal rodent plague, Yunnan Province, China. PLoS Neglect. Trop. D. 2011, 5, e997. [Google Scholar] [CrossRef]
- Zhang, Y. Investigation on the Infection of Hepatitis E in Rodents from Three Counties of Plague Natural Foci in Yunnan Province; Dali University: Dali, China, 2021. [Google Scholar] [CrossRef]
- Lledó, L.; Giménez-Pardo, C.; Gegúndez, M.I. Screening of forestry workers in Guadalajara Province (Spain) for antibodies to lymphocytic Choriomeningitis virus, hantavirus, rickettsia spp. and borrelia burgdorferi. Int. J. Environ. Res. Public Health 2019, 16, 4500. [Google Scholar] [CrossRef]
- Yashina, L.N.; Abramov, S.A.; Zhigalin, A.V.; Smetannikova, N.A.; Dupal, T.A.; Krivopalov, A.V.; Kikuchi, F.; Senoo, K.; Arai, S.; Mizutani, T.; et al. Geographic distribution and phylogeny of soricine shrew-borne Seewis virus and Altai virus in Russia. Viruses 2021, 13, 1286. [Google Scholar] [CrossRef] [PubMed]
- Clement, J.; LeDuc, J.W.; Lloyd, G.; Reynes, J.M.; McElhinney, L.; Van Ranst, M.; Lee, H.W. Wild rats, laboratory rats, pet rats: Global Seoul hantavirus disease revisited. Viruses 2019, 11, 652. [Google Scholar] [CrossRef] [PubMed]
- Cuperus, T.; de Vries, A.; Hoornweg, T.E.; Fonville, M.; Jaarsma, R.I.; Opsteegh, M.; Maas, M. Seoul virus in pet and feeder rats in the Netherlands. Viruses 2021, 13, 443. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, M.R.; Lin, X.D.; Guo, W.P.; Li, M.H.; Mei, S.H.; Li, Z.M.; Cong, M.L.; Jiang, R.L.; Zhou, R.H.; et al. Ongoing spillover of Hantaan and Gou hantaviruses from rodents is associated with hemorrhagic fever with renal syndrome (HFRS) in China. PLoS Neglect. Trop. D 2013, 7, e2484. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Yuan, J.F.; Yang, X.L.; Zhou, J.H.; Yang, W.H.; Peng, C.; Zhang, H.L.; Shi, Z.L. A novel hantavirus detected in Yunnan red-backed vole (Eothenomys miletus) in China. J. Gen. Virol. 2011, 92, 1454–1457. [Google Scholar] [CrossRef]
- Ge, X.Y.; Yang, W.H.; Pan, H.; Zhou, J.H.; Han, X.; Zhu, G.J.; Desmond, J.S.; Daszak, P.; Shi, Z.L.; Zhang, Y.Z. Fugong virus, a novel hantavirus harbored by the small oriental vole (Eothenomys Eleusis) in China. Virol. J. 2016, 13, 1–7. [Google Scholar] [CrossRef]
- Wang, J.L.; Zhang, H.L.; Zhou, J.H.; Yang, W.H.; Zhang, Y.Z.; Mi, Z.Q.; Luo, D.R.; Yang, L.G.; Yang, J.; Zhao, W.S.; et al. First detection of Tula-like Hantanvirus from Eothenomys milelus in China. Chin. J. Zoonoses 2010, 26, 408–412. [Google Scholar] [CrossRef]
- Deng, H.Y.; Wang, J.L.; Li, L.L.; Xin, Y.Y.; Liu, M.M.; Wang, Y.; Duan, Z.J. Genomic sequence analysis of a hantavirus found in Yunnan bat. Chin. J. Exp. Clin. Virol. 2018, 32, 140–144. [Google Scholar] [CrossRef]
- Klempa, B. Dobrava and Tula Hantaviruses from Central Europe: Molecular Evolution and Pathogenic Relevance. Ph.D. Thesis, Humboldt University of Berlin, Berlin, Germany, 2004. [Google Scholar]
- Plyusnina, A.; Heyman, P.; Baert, K.; Stuyck, J.; Cochez, C.; Plyusnin, A. Genetic characterization of seoul hantavirus originated from norway rats (Rattus norvegicus) captured in Belgium. J. Med. Virol. 2012, 84, 1298–1303. [Google Scholar] [CrossRef]
- Lin, X.D.; Guo, W.P.; Wang, W.; Zou, Y.; Hao, Z.Y.; Zhou, D.J.; Dong, X.; Qu, Y.G.; Li, M.H.; Tian, H.F.; et al. Migration of Norway rats resulted in the worldwide distribution of Seoul hantavirus today. J. Virol. 2012, 86, 972–981. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 2001, 11, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Schmaljohn, C.S.; Hasty, S.E.; Dalrymple, J.M.; LeDuc, J.W.; Lee, H.W.; von Bonsdorff, C.-H.; Brummer-Korvenkontio, M.; Vaheri, A.; Tsai, T.F.; Regnery, H.L.; et al. Antigenic and genetic properties of viruses linked to hemorrhagic fever with renal syndrome. Science 1985, 227, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, C.B.; Figueiredo, L.T.M.; Vapalahti, O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 2010, 23, 412–441. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.L.; Frias-Staheli, N.; Garcia-Sastre, A.; Schmaljohn, C.S. Hantaan virus nucleocapsid protein binds to importin α proteins and inhibits tumor necrosis factor Alpha-induced activation of nuclear factor kappa B. J. Virol. 2009, 83, 1271–1279. [Google Scholar] [CrossRef]
- Li, P.Y.; Bai, W.T. Research progress of hantavirus vaccine. Int. J. Virol. 2010, 17, 54–58. [Google Scholar] [CrossRef]


| Species | Region (Positive/Total) | Total (Positive/Total) | ||
|---|---|---|---|---|
| Mile | Mangshi | Lianghe | ||
| Rattus tanezumi | 1/29 (3.45% a) | 2/100 (2.00% a) | 5/115 (4.35% a) | 8/244 (3.28% a) |
| Suncus murinus | 0/5 | 1/71 (1.41% a) | 1/74 (1.35% a) | 2/150 (1.33% a) |
| Rattus rattus/sladeni | 0/3 | 0/4 | 0/7 | 0/14 |
| Mus pahari | 0/5 | 0/1 | 0/7 | 0/13 |
| Tupaia belangeri | 0/5 | 0/3 | 0/5 | 0/13 |
| Hylomys suillus | 0 | 0/1 | 0/11 | 0/12 |
| Mus caroli | 0/11 | 0/1 | 0 | 0/12 |
| Crocidura attenuate | 0/4 | 0/1 | 0/5 | 0/10 |
| Rattus nitidus | 0/4 | 0 | 2/2 (100% a) | 2/6 (33.33% a) |
| Niviventer confucianus | 0 | 0 | 0/4 | 0/4 |
| Niviventer fulvescens | 0 | 0/3 | 0 | 0/3 |
| Crocidura horsfieldi tadae | 0 | 0/1 | 0/2 | 0/3 |
| Dremomys rufigenis | 0 | 0/1 | 0 | 0/1 |
| Tamiops swinhoei | 0 | 0/1 | 0 | 0/1 |
| Squirrel | 0 | 0/1 | 0 | 0/1 |
| Berylmys bowersi | 0 | 0/1 | 0 | 0/1 |
| Total | 1/66 (1.52% a) | 3/190 (1.58% a) | 8/232 (3.45% a) | 12/488 (2.46% a) |
| Virus Strain | Type | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. YNLH-K40 | Unclassified | 99.3 | 86.0 | 86.5 | 84.3 | 86.7 | 88.5 | 85.2 | 66.3 | 41.0 | 41.1 | 65.5 | 48.1 | |
| 2. YNLH-K53 | Unclassified | 99.3 | 85.9 | 86.5 | 84.0 | 86.8 | 88.5 | 85.3 | 65.7 | 40.6 | 40.2 | 66.0 | 48.0 | |
| 3. AF288643 | SEOV-S1 | 85.3 | 84.9 | 96.6 | 96.2 | 96.2 | 88.8 | 95.3 | 70.8 | 64.1 | 48.7 | 70.9 | 49.1 | |
| 4. GU592948 | SEOV-S2 | 85.5 | 85.0 | 97.2 | 96.3 | 96.4 | 89.3 | 95.7 | 71.2 | 63.9 | 48.4 | 70.7 | 53.8 | |
| 5. AY766368 | SEOV-S3 | 84.1 | 83.6 | 95.9 | 95.9 | 96.1 | 88.4 | 95.0 | 70.5 | 63.1 | 49.5 | 70.5 | 54.3 | |
| 6. AY273791 | SEOV-S4 | 85.2 | 84.7 | 96.5 | 96.5 | 95.7 | 88.8 | 96.0 | 71.5 | 62.7 | 49.7 | 71.5 | 53.7 | |
| 7. JQ912777 | SEOV-S5 | 87.8 | 87.3 | 88.1 | 88.7 | 87.0 | 88.4 | 88.2 | 70.8 | 62.9 | 49.1 | 71.1 | 54.0 | |
| 8. AF329388 | SEOV-S6 | 84.4 | 83.9 | 95.8 | 96.4 | 95.4 | 96.4 | 88.3 | 71.2 | 63.7 | 48.6 | 70.8 | 53.3 | |
| 9. M14626 | HTNV | 70.0 | 70.0 | 72.8 | 72.9 | 72.5 | 73.0 | 71.9 | 73.2 | 60.8 | 50.4 | 72.2 | 53.0 | |
| 10. L11347 | PUUV | 65.2 | 65.0 | 65.2 | 65.6 | 64.1 | 64.8 | 65.5 | 64.9 | 60.4 | 68.7 | 61.8 | 68.0 | |
| 11. AF324902 | ANDV | 65.4 | 64.7 | 63.4 | 63.1 | 64.0 | 64.4 | 65.1 | 63.7 | 64.6 | 66.4 | 51.1 | 54.8 | |
| 12. L41916 | DOBV | 70.7 | 70.3 | 72.2 | 71.8 | 71.8 | 73.3 | 71.6 | 72.5 | 73.2 | 62.4 | 65.6 | 50.7 | |
| 13. L37904 | SNV | 63.8 | 64.4 | 65.8 | 65.1 | 64.1 | 65.4 | 65.5 | 64.5 | 63.9 | 66.5 | 74.2 | 63.9 |
| Virus | Region | Reference | ||
|---|---|---|---|---|
| Mile | Mangshi | Lianghe | ||
| HEV | 0.00% | 4.69% | 6.47% | Zhang [23] |
| HV | 1.52% | 1.58% | 3.45% | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Yin, J.-X.; Zhang, Y.; Wu, L.; Li, W.-H.; Luo, Y.-Y.; Li, R.; Li, Z.-W.; Liu, S.-Q. Genetic Evolution Analysis and Host Characteristics of Hantavirus in Yunnan Province, China. Int. J. Environ. Res. Public Health 2022, 19, 13433. https://doi.org/10.3390/ijerph192013433
Wang N, Yin J-X, Zhang Y, Wu L, Li W-H, Luo Y-Y, Li R, Li Z-W, Liu S-Q. Genetic Evolution Analysis and Host Characteristics of Hantavirus in Yunnan Province, China. International Journal of Environmental Research and Public Health. 2022; 19(20):13433. https://doi.org/10.3390/ijerph192013433
Chicago/Turabian StyleWang, Na, Jia-Xiang Yin, Yao Zhang, Li Wu, Wen-Hong Li, Yun-Yan Luo, Rui Li, Zi-Wei Li, and Shu-Qing Liu. 2022. "Genetic Evolution Analysis and Host Characteristics of Hantavirus in Yunnan Province, China" International Journal of Environmental Research and Public Health 19, no. 20: 13433. https://doi.org/10.3390/ijerph192013433
APA StyleWang, N., Yin, J.-X., Zhang, Y., Wu, L., Li, W.-H., Luo, Y.-Y., Li, R., Li, Z.-W., & Liu, S.-Q. (2022). Genetic Evolution Analysis and Host Characteristics of Hantavirus in Yunnan Province, China. International Journal of Environmental Research and Public Health, 19(20), 13433. https://doi.org/10.3390/ijerph192013433





