Gut Microbiome: The Interplay of an “Invisible Organ” with Herbal Medicine and Its Derived Compounds in Chronic Metabolic Disorders
Abstract
1. Introduction
2. The GM’s Interplay with Herbal Medicine, Altering Drugs’ Efficacy in Metabolic Disorders
2.1. Gut Microbial Metabolism Produces Ginsenosides from Ginseng Radix, Exerting Bioactivity
2.2. Gut Microbial Metabolism Produces Active Compounds from Puerariae
2.3. Gut Microbial Metabolism of Compounds from Coptidis Rhizoma Improves Their Absorption Rate
2.4. Gut Microbial Bioconversion of Compounds from Scutellaria Radix Improves Their Absorption Rate
| Herb Name | Microbial Metabolites | Treatment of Diseases | Study Design (In Vitro/In Vivo/Clinical Study) | Impact of Drug Efficacy | Ref. |
|---|---|---|---|---|---|
| Ginseng Radix | Compound K | Diabetes | In vivo (SD rats) In vitro (Caco-2 cell permeability) | Increased absorption | [73] |
| Compound K Ginsenoside Rh1 | NAFLD | In vivo (HFD-fed SD rats) In vitro (HSC-T6 cell) | Increased activity | [92] | |
| Compound K | Diabetes | In vivo (STZ and HFD-fed ICR mice) | Increased activity | [93] | |
| Puerariae Radix and Puerariae Flos | Irisolidone Tectorigenin | Estrogenic effect | In vitro (human fecal incubation, MCF-7 cells) | Increased activity (c-fos and pS2 gene) | [50] |
| Daidzein | Not indicated | In vitro (Caco-2 permeability) In vivo (hydrolyzation by rat microvilli) | Increased absorption | [81] | |
| Daidzein | Estrogenic effect | In vitro (human fecal incubation, MCF-7 cells) | Increased activity | [51] | |
| Equol | NAFLD | In vivo (HFD-fed mice) | Increased activity Changed bioactivity | [94] | |
| Coptidis Rhizoma | Oxyberberine | Colitis | In vivo (DSS-induced colitis Balb/C mice) | Increased activity | [53] |
| Dihydroberberine | Diabetes | In vivo (KK-Ay mice) | Increased absorption | [55] | |
| Berberrubine | Hypercholesterolemia | Clinical study (n = 12, moderate hypercholesterolemia) | Increased activity | [95] | |
| Scutellaria Radix | Baicalein | Not intended | In vivo (antibiotic-treated SD rats) | Increased absorption | [96] |
| Baicalein | Not intended | In vivo (germ-free Wistar rats) | Increased absorption | [56] | |
| Baicalein | Not intended | In vivo (bile-duct-ligated Wistar rats | Increased absorption | [89] | |
| Wogonin | Not intended | In vivo (antibiotic-treated SD rats) | Increased absorption | [91] | |
| Curcumae Radix | Tetrahydrocurcumin | Diabetes | In vivo (STZ-induced diabetic rats) | Increased activity | [97] |
| Tetrahydrocurcumin | Lipid accumulation | In vitro (THP-1 cells) | Decreased activity | [98] | |
| Mori folium, Bupleurum Radix, Houttuyniae Herba | Quercetin | Platelet activity | In vitro | Increased activity | [65] |
| Quercetin | Insulin resistance | In vitro (TNF-α-treated C2C12 cells) | Increased activity | [99] | |
| Glycyrrhizae Radix | Glycyrrhetic acid | Not indicated | In vivo (SD rats, Wistar germ-free rats) | Increased bioavailability | [100] |
| 18β-Glycyrrhetinic acid | Obesity | In vitro (3T3-L1) In vivo (HFD-fed C57/BL6 mice) | Not indicated | [101] | |
| 18β-Glycyrrhetinic acid | NASH | In vivo (MCD; C57/BL6 mice) | Increased bioactivity | [90] |
2.5. Gut Microbial Metabolism of Curcumin from Curcumae Radix Increases Its Bioavailability
2.6. Gut Microbial Bioconversion of Quercitrin from Several Herbs into Quercetin Increases Its Bioavailability
2.7. Glycyrrhizin from Glycyrrhizae Radix Requires Bacterial Transformation to Be Absorbed in the Intestine
| Herbal Medicine | Raw Compound | Properties of Raw Compound | Properties of Metabolite | Metabolite |
|---|---|---|---|---|
| Ginseng Radix | Ginsenoside Rb1 | PubChem CID | Ginsenoside Rd | |
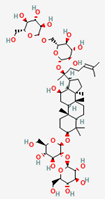 | 9898279 | 11679800 | 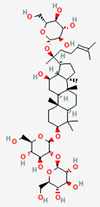 | |
| Molecular Weight | ||||
| 1109.3 | 963.30 | |||
| Bioavailability Score | ||||
| 0.17 | 0.17 | |||
| GI Absorption | ||||
| Low | Low | |||
| Lipinski’s Criteria | ||||
| No (3 violations) MW > 500, NorO > 10, NHorOH > 5 | No (3 violations) MW > 500, NorO > 10, NHorOH > 5 | |||
| Ginsenoside Rc | PubChem CID | Compound K | ||
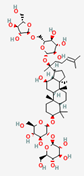 | 12855889 | 5481990 |  | |
| Molecular Weight | ||||
| 1079.3 | 653.8 | |||
| Bioavailability Score | ||||
| 0.17 | 0.55 | |||
| GI Absorption | ||||
| Low | High | |||
| Lipinski’s Criteria | ||||
| No (3 violations) MW > 500, NorO > 10, NHorOH > 5 | Yes (1 violation) MW > 500 | |||
| Puerariae Radix and Puerariae Flos | Daidzin | PubChem CID | Daidzein | |
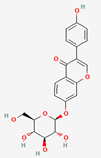 | 107971 | 5281708 | 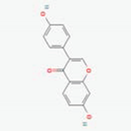 | |
| Molecular Weight | ||||
| 416.41 | 254.24 | |||
| Bioavailability Score | ||||
| 0.55 | 0.55 | |||
| GI Absorption | ||||
| Low | High | |||
| Lipinski’s Criteria | ||||
| Yes (0 violations) | Yes (0 violations) | |||
| Coptidis Rhizoma | Berberine | PubChem CID | Oxyberberine | |
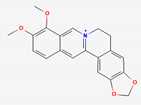 | 2353 | 11066 | 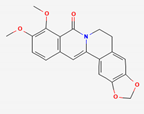 | |
| Molecular Weight | ||||
| 336.4 | 351.4 | |||
| Bioavailability Score | ||||
| 0.55 | 0.55 | |||
| GI Absorption | ||||
| High | High | |||
| Lipinski’s Criteria | ||||
| Yes (0 violations) | Yes (0 violations) | |||
| Scutellaria Radix | Baicalin | PubChem CID | Baicalein | |
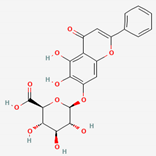 | 64982 | 5281605 | 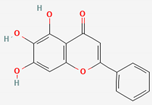 | |
| Molecular Weight | ||||
| 446.4 | 270.24 | |||
| Bioavailability Score | ||||
| 0.11 | 0.55 | |||
| GI Absorption | ||||
| Low | High | |||
| Lipinski’s Criteria | ||||
| No (2 violations) NorO > 10, NHorOH > 5 | Yes (0 violations) | |||
| Curcumae Radix | Curcumin | PubChem CID | Dihydrocurcumin | |
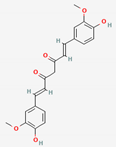 | 969516 | 10429233 | 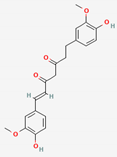 | |
| Molecular Weight | ||||
| 368.4 | 370.4 | |||
| Bioavailability Score | ||||
| 0.55 | 0.55 | |||
| GI Absorption | ||||
| High | High | |||
| Lipinski’s Criteria | ||||
| Yes (0 violations) | Yes (0 violations) | |||
| Mori folium/ Bupleurum Radix/Houttuyniae Herba | Quercitrin | PubChem CID | Quercetin | |
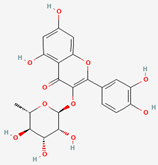 | 5280459 | 5280343 | 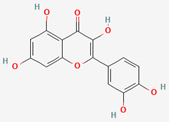 | |
| Molecular Weight | ||||
| 448.4 | 302.23 | |||
| Bioavailability Score | ||||
| 0.17 | 0.55 | |||
| GI Absorption | ||||
| Low | High | |||
| Lipinski’s Criteria | ||||
| No (2 violations) NorO > 10, NHorOH > 5 | Yes (0 violations) | |||
| Glycyrrhizae Radix | Glycyrrhizin (Glycyrrhizic Acid) | PubChem CID | 18-β-Glycyrrhetinic Acid (Glycyrrhetic Acid) | |
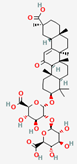 | 14982 | 10114 | 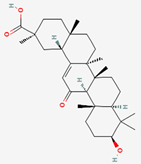 | |
| Molecular Weight | ||||
| 822.9 | 470.7 | |||
| Bioavailability Score | ||||
| 0.11 | 0.85 | |||
| GI Absorption | ||||
| Low | High | |||
| Lipinski’s Criteria | ||||
| No (3 violations) MW > 500, NorO > 10, NHorOH > 5 | Yes (1 violation) MLOGP > 4.15 | |||
3. Current Status and Future Perspectives
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davenport, E.R.; Sanders, J.G.; Song, S.J.; Amato, K.R.; Clark, A.G.; Knight, R. The human microbiome in evolution. BMC Biol. 2017, 15, 127. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Godoy-Vitorino, F.; Knight, R.; Blaser, M.J. Role of the microbiome in human development. Gut 2019, 68, 1108–1114. [Google Scholar] [CrossRef]
- Farré-Maduell, E.; Casals-Pascual, C. The origins of gut microbiome research in Europe: From Escherich to Nissle. Hum. Microbiome J. 2019, 14, 100065. [Google Scholar] [CrossRef]
- Satoor, S.N.; Patil, D.P.; Kristensen, H.D.; Joglekar, M.V.; Shouche, Y.; Hardikar, A.A. Manipulation and assessment of gut microbiome for metabolic studies. Methods Mol. Biol. 2014, 1194, 449–469. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Wang, X.; Li, L. Human gut microbiome: The second genome of human body. Protein Cell 2010, 1, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- D’Argenio, V.; Salvatore, F. The role of the gut microbiome in the healthy adult status. Clin. Chim. Acta 2015, 451, 97–102. [Google Scholar] [CrossRef]
- Dzherieva, I.; Volkova, N.; Panfilova, N. Depressive disorders in males with metabolic syndrome. J. Biomed. Clin. Res. 2011, 4, 46–49. [Google Scholar]
- Im, H.J.; Ahn, Y.C.; Wang, J.H.; Lee, M.M.; Son, C.G. Systematic review on the prevalence of nonalcoholic fatty liver disease in South Korea. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101526. [Google Scholar] [CrossRef]
- Karalliedde, J.; Gnudi, L. Diabetes mellitus, a complex and heterogeneous disease, and the role of insulin resistance as a determinant of diabetic kidney disease. Nephrol. Dial. Transplant. 2014, 31, 206–213. [Google Scholar] [CrossRef]
- Lê, K.-A.; Bortolotti, M. Role of dietary carbohydrates and macronutrients in the pathogenesis of nonalcoholic fatty liver disease. Curr. Opin. Clin. Nutr. Metab. Care 2008, 11, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Phelan, S.; Wadden, T.; Berkowitz, R.; Sarwer, D.; Womble, L.; Cato, R.; Rothman, R. Impact of weight loss on the metabolic syndrome. Int. J. Obes. 2007, 31, 1442–1448. [Google Scholar] [CrossRef] [PubMed]
- Alley, D.E.; Chang, V.W. Metabolic syndrome and weight gain in adulthood. J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2010, 65, 111–117. [Google Scholar] [CrossRef]
- Franklin, S.S.; Barboza, M.G.; Pio, J.R.; Wong, N.D. Blood pressure categories, hypertensive subtypes, and the metabolic syndrome. J. Hypertens. 2006, 24, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Di Daniele, N.J.N. Association of Dietary Patterns with Metabolic Syndrome; Multidisciplinary Digital Publishing Institute: Basel, Switzerland, 2020; Volume 12, p. 2840. [Google Scholar]
- Halpern, A.; Mancini, M.C.; Magalhães, M.E.C.; Fisberg, M.; Radominski, R.; Bertolami, M.C.; Bertolami, A.; de Melo, M.E.; Zanella, M.T.; Queiroz, M.S.J.D.; et al. Metabolic syndrome, dyslipidemia, hypertension and type 2 diabetes in youth: From diagnosis to treatment. Diabetol. Metab. Syndr. 2010, 2, 1–20. [Google Scholar] [CrossRef]
- Finsterer, J.; Frank, M. Repurposed drugs in metabolic disorders. Curr. Top. Med. Chem. 2013, 13, 2386–2394. [Google Scholar] [CrossRef]
- Scotti, L.; Monteiro, A.F.M.; de Oliveira Viana, J.; Mendonça Junior, F.J.B.; Ishiki, H.M.; Tchouboun, E.N.; Santos, R.; Scotti, M.T. Multi-Target Drugs Against Metabolic Disorders. Endocr. Metab. Immune Disord. Drug Targets 2019, 19, 402–418. [Google Scholar] [CrossRef]
- Jang, S.; Jang, B.-H.; Ko, Y.; Sasaki, Y.; Park, J.-S.; Hwang, E.-H.; Song, Y.-K.; Shin, Y.-C.; Ko, S.-G. Herbal Medicines for Treating Metabolic Syndrome: A Systematic Review of Randomized Controlled Trials. Evid. Based Complement. Altern. Med. 2016, 2016, 5936402. [Google Scholar] [CrossRef]
- Almazroo, O.A.; Miah, M.K.; Venkataramanan, R. Drug Metabolism in the Liver. Clin. Liver Dis. 2017, 21, 1–20. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, J.; Li, M.; Zhang, Y.; Jiang, B.; Zhao, X.; Huai, C.; Shen, L.; Zhang, N.; He, L.; et al. Cytochrome P450 Enzymes and Drug Metabolism in Humans. Int. J. Mol. Sci. 2021, 22, 12808. [Google Scholar] [CrossRef]
- Rath, S.; Rud, T.; Karch, A.; Pieper, D.H.; Vital, M. Pathogenic functions of host microbiota. Microbiome 2018, 6, 174. [Google Scholar] [CrossRef]
- Qin, Y.; Havulinna, A.S.; Liu, Y.; Jousilahti, P.; Ritchie, S.C.; Tokolyi, A.; Sanders, J.G.; Valsta, L.; Brożyńska, M.; Zhu, Q. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat. Genet. 2022, 54, 134–142. [Google Scholar] [CrossRef]
- Kang, C.; Wang, B.; Kaliannan, K.; Wang, X.; Lang, H.; Hui, S.; Huang, L.; Zhang, Y.; Zhou, M.; Chen, M. Gut microbiota mediates the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by high-fat diet. MBio 2017, 8, e00470-17. [Google Scholar] [CrossRef]
- Daly, A.K. Genetic polymorphisms affecting drug metabolism: Recent advances and clinical aspects. Adv. Pharm. 2012, 63, 137–167. [Google Scholar] [CrossRef]
- Noh, K.; Kang, Y.R.; Nepal, M.R.; Shakya, R.; Kang, M.J.; Kang, W.; Lee, S.; Jeong, H.G.; Jeong, T.C. Impact of gut microbiota on drug metabolism: An update for safe and effective use of drugs. Arch. Pharm. Res. 2017, 40, 1345–1355. [Google Scholar] [CrossRef]
- Zgheib, N.K.; Branch, R.A. Drug metabolism and liver disease: A drug-gene-environment interaction. Drug Metab. Rev. 2017, 49, 35–55. [Google Scholar] [CrossRef]
- Belle, D.J.; Singh, H. Genetic factors in drug metabolism. Am. Fam. Physician 2008, 77, 1553–1560. [Google Scholar]
- Pant, A.; Maiti, T.K.; Mahajan, D.; Das, B. Human Gut Microbiota and Drug Metabolism. Microb. Ecol. 2022, 1–15. [Google Scholar] [CrossRef]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef]
- An, X.; Bao, Q.; Di, S.; Zhao, Y.; Zhao, S.; Zhang, H.; Lian, F.; Tong, X. The interaction between the gut Microbiota and herbal medicines. Biomed. Pharmacother. 2019, 118, 109252. [Google Scholar] [CrossRef]
- Koppel, N.; Maini Rekdal, V.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef]
- Hollman, A. Plants and cardiac glycosides. Br. Heart J. 1985, 54, 258–261. [Google Scholar] [CrossRef]
- Leisegang, K. Herbal pharmacognosy: An introduction. In Herbal Medicine in Andrology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 17–26. [Google Scholar]
- Hollman, P.C.; Bijsman, M.N.; Van Gameren, Y.; Cnossen, E.P.; De Vries, J.H.; Katan, M.B. The sugar moiety is a major determinant of the absorption of dietary flavonoid glycosides in man. Free Radic. Res. 1999, 31, 569–573. [Google Scholar] [CrossRef]
- Bhattacharya, A. High-Temperature Stress and Metabolism of Secondary Metabolites in Plants. In Effect of High Temperature on Crop Productivity and Metabolism of Macro Molecules; Elsevier: London, UK, 2019; pp. 391–484. [Google Scholar]
- Kumar, S.; Pandey, A.K. Chemistry and biological activities of flavonoids: An overview. Sci. World J. 2013, 2013, 162750. [Google Scholar] [CrossRef]
- Xu, J.; Chen, H.B.; Li, S.L. Understanding the Molecular Mechanisms of the Interplay Between Herbal Medicines and Gut Microbiota. Med. Res. Rev. 2017, 37, 1140–1185. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.D.; Nicholson, J.K. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 2017, 179, 204–222. [Google Scholar] [CrossRef]
- Feng, W.; Liu, J.; Ao, H.; Yue, S.; Peng, C. Targeting gut microbiota for precision medicine: Focusing on the efficacy and toxicity of drugs. Theranostics 2020, 10, 11278–11301. [Google Scholar] [CrossRef]
- Shi, Z.Y.; Zeng, J.Z.; Wong, A.S.T. Chemical Structures and Pharmacological Profiles of Ginseng Saponins. Molecules 2019, 24, 2443. [Google Scholar] [CrossRef]
- Kim, D.H. Gut microbiota-mediated pharmacokinetics of ginseng saponins. J. Ginseng Res. 2018, 42, 255–263. [Google Scholar] [CrossRef]
- Kim, H.; Lee, J.H.; Kim, J.E.; Kim, Y.S.; Ryu, C.H.; Lee, H.J.; Kim, H.M.; Jeon, H.; Won, H.J.; Lee, J.Y.; et al. Micro-/nano-sized delivery systems of ginsenosides for improved systemic bioavailability. J. Ginseng Res. 2018, 42, 361–369. [Google Scholar] [CrossRef]
- Kim, H.-K. Pharmacokinetics of ginsenoside Rb1 and its metabolite compound K after oral administration of Korean Red Ginseng extract. J. Ginseng Res. 2013, 37, 451. [Google Scholar] [CrossRef]
- Paek, I.B.; Moon, Y.; Kim, J.; Ji, H.Y.; Kim, S.A.; Sohn, D.H.; Kim, J.B.; Lee, H.S. Pharmacokinetics of a ginseng saponin metabolite compound K in rats. Biopharm. Drug Dispos. 2006, 27, 39–45. [Google Scholar] [CrossRef]
- Jung, I.H.; Lee, J.H.; Hyun, Y.J.; Kim, D.H. Metabolism of ginsenoside Rb1 by human intestinal microflora and cloning of its metabolizing beta-D-glucosidase from Bifidobacterium longum H-1. Biol. Pharm. Bull. 2012, 35, 573–581. [Google Scholar] [CrossRef]
- Park, S.Y.; Bae, E.A.; Sung, J.H.; Lee, S.K.; Kim, D.H. Purification and characterization of ginsenoside Rb1-metabolizing beta-glucosidase from Fusobacterium K-60, a human intestinal anaerobic bacterium. Biosci. Biotechnol. Biochem. 2001, 65, 1163–1169. [Google Scholar] [CrossRef]
- Quan, L.H.; Min, J.W.; Yang, D.U.; Kim, Y.J.; Yang, D.C. Enzymatic biotransformation of ginsenoside Rb1 to 20(S)-Rg3 by recombinant beta-glucosidase from Microbacterium esteraromaticum. Appl. Microbiol. Biotechnol. 2012, 94, 377–384. [Google Scholar] [CrossRef]
- Akao, T.; Kida, H.; Kanaoka, M.; Hattori, M.; Kobashi, K. Drug metabolism: Intestinal bacterial hydrolysis is required for the appearance of compound K in rat plasma after oral administration of ginsenoside Rb1 from Panax ginseng. J. Pharm. Pharmacol. 1998, 50, 1155–1160. [Google Scholar] [CrossRef]
- Qian, T.; Cai, Z.; Wong, R.N.; Mak, N.K.; Jiang, Z.H. In vivo rat metabolism and pharmacokinetic studies of ginsenoside Rg3. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005, 816, 223–232. [Google Scholar] [CrossRef]
- Bae, E.-A.; Choo, M.-K.; Park, E.-K.; Park, S.-Y.; Shin, H.-Y.; Kim, D.-H. Metabolism of ginsenoside Rc by human intestinal bacteria and its related antiallergic activity. Biol. Pharm. Bull. 2002, 25, 743–747. [Google Scholar] [CrossRef]
- Tawab, M.A.; Bahr, U.; Karas, M.; Wurglics, M.; Schubert-Zsilavecz, M. Degradation of ginsenosides in humans after oral administration. Drug Metab. Dispos. 2003, 31, 1065–1071. [Google Scholar] [CrossRef]
- Hasegawa, H. Anticarcinogenesis in mice by ginseng-hydrolyzing colonic bacteria. Microb. Ecol. Health Dis. 2000, 12, 85–91. [Google Scholar]
- Hasegawa, H.; Sung, J.H.; Benno, Y. Role of human intestinal Prevotella oris in hydrolyzing ginseng saponins. Planta Med. 1997, 63, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Prasain, J.K.; Peng, N.; Rajbhandari, R.; Wyss, J.M. The Chinese Pueraria root extract (Pueraria lobata) ameliorates impaired glucose and lipid metabolism in obese mice. Phytomedicine 2012, 20, 17–23. [Google Scholar] [CrossRef]
- Jung, H.W.; Kang, A.N.; Kang, S.Y.; Park, Y.K.; Song, M.Y. The Root Extract of Pueraria lobata and Its Main Compound, Puerarin, Prevent Obesity by Increasing the Energy Metabolism in Skeletal Muscle. Nutrients 2017, 9, 33. [Google Scholar] [CrossRef]
- Zhang, Z.; Lam, T.N.; Zuo, Z. Radix Puerariae: An overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J. Clin. Pharmacol. 2013, 53, 787–811. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.S.; Nishihata, T.; Kakiuchi, N.; Hattori, M. Biotransformation of C-glucosylisoflavone puerarin to estrogenic (3S)-equol in co-culture of two human intestinal bacteria. Biol. Pharm. Bull. 2008, 31, 1621–1625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Pan Siu, A.K.; Lin, G.; Zuo, Z. Intestinal absorbability of three Radix Puerariae isoflavones including daidzein, daidzin and puerarin. Chin. Med. 2011, 6, 41. [Google Scholar] [CrossRef]
- Park, E.K.; Shin, J.; Bae, E.A.; Lee, Y.C.; Kim, D.H. Intestinal bacteria activate estrogenic effect of main constituents puerarin and daidzin of Pueraria thunbergiana. Biol. Pharm. Bull. 2006, 29, 2432–2435. [Google Scholar] [CrossRef]
- Shin, J.E.; Bae, E.A.; Lee, Y.C.; Ma, J.Y.; Kim, D.H. Estrogenic effect of main components kakkalide and tectoridin of Puerariae Flos and their metabolites. Biol. Pharm. Bull. 2006, 29, 1202–1206. [Google Scholar] [CrossRef][Green Version]
- Zhang, G.; Gong, T.; Kano, Y.; Yuan, D. Screening for in vitro metabolites of kakkalide and irisolidone in human and rat intestinal bacteria by ultra-high performance liquid chromatography/quadrupole time-of-flight mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 947–948, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Ran, Q.; Wang, J.; Wang, L.; Zeng, H.-R.; Yang, X.-B.; Huang, Q.-W. Rhizoma coptidis as a Potential Treatment Agent for Type 2 Diabetes Mellitus and the Underlying Mechanisms: A Review. Front. Pharmacol. 2019, 10, 805. [Google Scholar] [CrossRef]
- Feng, X.; Wang, K.; Cao, S.; Ding, L.; Qiu, F. Pharmacokinetics and Excretion of Berberine and Its Nine Metabolites in Rats. Front. Pharmacol. 2020, 11, 594852. [Google Scholar] [CrossRef]
- Dey, P. Gut microbiota in phytopharmacology: A comprehensive overview of concepts, reciprocal interactions, biotransformations and mode of actions. Pharmacol. Res. 2019, 147, 104367. [Google Scholar] [CrossRef]
- Cheng, H.; Liu, J.; Tan, Y.; Feng, W.; Peng, C. Interactions between gut microbiota and berberine, a necessary procedure to understand the mechanisms of berberine. J. Pharm. Anal. 2021, 12, 541–555. [Google Scholar] [CrossRef]
- Zheng, Y.; Gou, X.; Zhang, L.; Gao, H.; Wei, Y.; Yu, X.; Pang, B.; Tian, J.; Tong, X.; Li, M. Interactions Between Gut Microbiota, Host, and Herbal Medicines: A Review of New Insights Into the Pathogenesis and Treatment of Type 2 Diabetes. Front. Cell. Infect. Microbiol. 2020, 10, 360. [Google Scholar] [CrossRef]
- Feng, R.; Shou, J.-W.; Zhao, Z.-X.; He, C.-Y.; Ma, C.; Huang, M.; Fu, J.; Tan, X.-S.; Li, X.-Y.; Wen, B.-Y. Transforming berberine into its intestine-absorbable form by the gut microbiota. Sci. Rep. 2015, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ai, G.; Wang, Y.; Lu, Q.; Luo, C.; Tan, L.; Lin, G.; Liu, Y.; Li, Y.; Zeng, H.; et al. Oxyberberine, a novel gut microbiota-mediated metabolite of berberine, possesses superior anti-colitis effect: Impact on intestinal epithelial barrier, gut microbiota profile and TLR4-MyD88-NF-kappaB pathway. Pharm. Res. 2020, 152, 104603. [Google Scholar] [CrossRef]
- Baradaran Rahimi, V.; Askari, V.R.; Hosseinzadeh, H. Promising influences of Scutellaria baicalensis and its two active constituents, baicalin, and baicalein, against metabolic syndrome: A review. Phytother. Res. 2021, 35, 3558–3574. [Google Scholar] [CrossRef]
- Kim, D.-H.; Jang, I.-S.; Lee, H.-K.; Jung, E.-A.; Lee, K.-Y. Metabolism of glycyrrhizin and baicalin by human intestinal bacteria. Arch. Pharmacal Res. 1996, 19, 292–296. [Google Scholar] [CrossRef]
- Noh, K.; Kang, Y.; Nepal, M.R.; Jeong, K.S.; Oh, D.G.; Kang, M.J.; Lee, S.; Kang, W.; Jeong, H.G.; Jeong, T.C. Role of Intestinal Microbiota in Baicalin-Induced Drug Interaction and Its Pharmacokinetics. Molecules 2016, 21, 337. [Google Scholar] [CrossRef]
- Akao, T.; Kawabata, K.; Yanagisawa, E.; Ishihara, K.; Mizuhara, Y.; Wakui, Y.; Sakashita, Y.; Kobashi, K. Balicalin, the predominant flavone glucuronide of scutellariae radix, is absorbed from the rat gastrointestinal tract as the aglycone and restored to its original form. J. Pharm. Pharmacol. 2000, 52, 1563–1568. [Google Scholar] [CrossRef]
- Taiming, L.; Xuehua, J. Investigation of the absorption mechanisms of baicalin and baicalein in rats. J. Pharm. Sci. 2006, 95, 1326–1333. [Google Scholar] [CrossRef]
- Bak, E.J.; Kim, J.; Choi, Y.H.; Kim, J.H.; Lee, D.E.; Woo, G.H.; Cha, J.H.; Yoo, Y.J. Wogonin ameliorates hyperglycemia and dyslipidemia via PPARalpha activation in db/db mice. Clin. Nutr. 2014, 33, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Wang, M.; Peng, Y.; Li, X. Effects of Intestinal Microecology on Metabolism and Pharmacokinetics of Oral Wogonoside and Baicalin. Nat. Prod. Commun. 2017, 12, 509–514. [Google Scholar] [CrossRef]
- Ku, S.; Zheng, H.; Park, M.S.; Ji, G.E. Optimization of β-glucuronidase activity from Lactobacillus delbrueckii Rh2 and and its use for biotransformation of baicalin and wogonoside. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 275–280. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, J.Y.; Park, M.S.; Zheng, H.; Ji, G.E. Cloning and expression of beta-glucuronidase from Lactobacillus brevis in E. coli and application in the bioconversion of baicalin and wogonoside. J. Microbiol. Biotechnol. 2009, 19, 1650–1655. [Google Scholar] [CrossRef]
- Shao, W.; Yu, Z.; Chiang, Y.; Yang, Y.; Chai, T.; Foltz, W.; Lu, H.; Fantus, I.G.; Jin, T. Curcumin prevents high fat diet induced insulin resistance and obesity via attenuating lipogenesis in liver and inflammatory pathway in adipocytes. PLoS ONE 2012, 7, e28784. [Google Scholar] [CrossRef]
- Feng, D.; Zou, J.; Su, D.; Mai, H.; Zhang, S.; Li, P.; Zheng, X. Curcumin prevents high-fat diet-induced hepatic steatosis in ApoE(-/-) mice by improving intestinal barrier function and reducing endotoxin and liver TLR4/NF-kappaB inflammation. Nutr. Metab. 2019, 16, 79. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Deb, L.; Prasad, S. Curcumin differs from tetrahydrocurcumin for molecular targets, signaling pathways and cellular responses. Molecules 2014, 20, 185–205. [Google Scholar] [CrossRef]
- Lopresti, A.L. The Problem of Curcumin and Its Bioavailability: Could Its Gastrointestinal Influence Contribute to Its Overall Health-Enhancing Effects? Adv. Nutr. 2018, 9, 41–50. [Google Scholar] [CrossRef]
- Burapan, S.; Kim, M.; Han, J. Curcuminoid demethylation as an alternative metabolism by human intestinal microbiota. J. Agric. Food Chem. 2017, 65, 3305–3310. [Google Scholar] [CrossRef]
- Tan, S.; Rupasinghe, T.W.; Tull, D.L.; Boughton, B.; Oliver, C.; McSweeny, C.; Gras, S.L.; Augustin, M.A. Degradation of curcuminoids by in vitro pure culture fermentation. J. Agric. Food Chem. 2014, 62, 11005–11015. [Google Scholar] [CrossRef]
- Dai, X.; Ding, Y.; Zhang, Z.; Cai, X.; Li, Y. Quercetin and quercitrin protect against cytokineinduced injuries in RINm5F beta-cells via the mitochondrial pathway and NF-kappaB signaling. Int. J. Mol. Med. 2013, 31, 265–271. [Google Scholar] [CrossRef]
- Babaei, F.; Mirzababaei, M.; Nassiri-Asl, M. Quercetin in Food: Possible Mechanisms of Its Effect on Memory. J. Food Sci. 2018, 83, 2280–2287. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, S.Y.; Park, S.Y.; Han, M.J. Metabolism of quercitrin by human intestinal bacteria and its relation to some biological activities. Biol. Pharm. Bull. 1999, 22, 749–751. [Google Scholar] [CrossRef] [PubMed]
- Enkhmaa, B.; Shiwaku, K.; Katsube, T.; Kitajima, K.; Anuurad, E.; Yamasaki, M.; Yamane, Y. Mulberry (Morus alba L.) leaves and their major flavonol quercetin 3-(6-malonylglucoside) attenuate atherosclerotic lesion development in LDL receptor-deficient mice. J. Nutr. 2005, 135, 729–734. [Google Scholar] [CrossRef]
- Hirooka, K.; Fujita, Y. Excess production of Bacillus subtilis quercetin 2,3-dioxygenase affects cell viability in the presence of quercetin. Biosci. Biotechnol. Biochem. 2010, 74, 1030–1038. [Google Scholar] [CrossRef]
- PARK, S.-Y.; KIM, J.-H.; KIM, D.-H. Purification and characterization of quercitrin-hydrolyzing α-L-rhamnosidase from Fusobacterium K-60, a human intestinal bacterium. J. Microbiol. Biotechnol. 2005, 15, 519–524. [Google Scholar]
- Hosseini, A.; Razavi, B.M.; Banach, M.; Hosseinzadeh, H. Quercetin and metabolic syndrome: A review. Phytother. Res. 2021, 35, 5352–5364. [Google Scholar] [CrossRef]
- Comalada, M.; Camuesco, D.; Sierra, S.; Ballester, I.; Xaus, J.; Galvez, J.; Zarzuelo, A. In vivo quercitrin anti-inflammatory effect involves release of quercetin, which inhibits inflammation through down-regulation of the NF-kappaB pathway. Eur. J. Immunol. 2005, 35, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Graebin, C.S. The pharmacological activities of glycyrrhizinic acid (“glycyrrhizin”) and glycyrrhetinic acid. Sweeteners 2018, 245. [Google Scholar]
- Kwon, Y.J.; Son, D.H.; Chung, T.H.; Lee, Y.J. A Review of the Pharmacological Efficacy and Safety of Licorice Root from Corroborative Clinical Trial Findings. J. Med. Food 2020, 23, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, Y.; Santa, T.; Kotaki, H.; Uchino, K.; Sawada, Y.; Iga, T. Administration-route dependency of absorption of glycyrrhizin in rats: Intraperitoneal administration dramatically enhanced bioavailability. Biol. Pharm. Bull. 1995, 18, 337–341. [Google Scholar] [CrossRef]
- Akao, T. Differences in the metabolism of glycyrrhizin, glycyrrhetic acid and glycyrrhetic acid monoglucuronide by human intestinal flora. Biol. Pharm. Bull. 2000, 23, 1418–1423. [Google Scholar] [CrossRef]
- Shin, H.Y.; Lee, J.H.; Lee, J.Y.; Han, Y.O.; Han, M.J.; Kim, D.H. Purification and characterization of ginsenoside Ra-hydrolyzing beta-D-xylosidase from Bifidobacterium breve K-110, a human intestinal anaerobic bacterium. Biol. Pharm. Bull. 2003, 26, 1170–1173. [Google Scholar] [CrossRef]
- Nakamura, K.; Zhu, S.; Komatsu, K.; Hattori, M.; Iwashima, M. Deglycosylation of the Isoflavone C-Glucoside Puerarin by a Combination of Two Recombinant Bacterial Enzymes and 3-Oxo-Glucose. Appl. Env. Microbiol. 2020, 86, e00607-20. [Google Scholar] [CrossRef]
- Matthies, A.; Blaut, M.; Braune, A. Isolation of a human intestinal bacterium capable of daidzein and genistein conversion. Appl. Environ. Microbiol. 2009, 75, 1740–1744. [Google Scholar] [CrossRef]
- Tan, X.S.; Ma, J.Y.; Feng, R.; Ma, C.; Chen, W.J.; Sun, Y.P.; Fu, J.; Huang, M.; He, C.Y.; Shou, J.W.; et al. Tissue distribution of berberine and its metabolites after oral administration in rats. PLoS ONE 2013, 8, e77969. [Google Scholar] [CrossRef] [PubMed]
- Hassaninasab, A.; Hashimoto, Y.; Tomita-Yokotani, K.; Kobayashi, M. Discovery of the curcumin metabolic pathway involving a unique enzyme in an intestinal microorganism. Proc. Natl. Acad. Sci. USA 2011, 108, 6615–6620. [Google Scholar] [CrossRef]
- Akao, T. Purification and characterization of glycyrrhetic acid mono-glucuronide beta-D-glucuronidase in Eubacterium sp. GLH. Biol. Pharm. Bull. 1999, 22, 80–82. [Google Scholar] [CrossRef]
- Yim, J.S.; Kim, Y.S.; Moon, S.K.; Cho, K.H.; Bae, H.S.; Kim, J.J.; Park, E.K.; Kim, D.H. Metabolic activities of ginsenoside Rb1, baicalin, glycyrrhizin and geniposide to their bioactive compounds by human intestinal microflora. Biol. Pharm. Bull. 2004, 27, 1580–1583. [Google Scholar] [CrossRef] [PubMed]
- Akao, T. Influence of various bile acids on the metabolism of glycyrrhizin and glycyrrhetic acid by Ruminococcus sp. PO1-3 of human intestinal bacteria. Biol. Pharm. Bull. 1999, 22, 787–793. [Google Scholar] [CrossRef]
- Chen, X.J.; Liu, W.J.; Wen, M.L.; Liang, H.; Wu, S.M.; Zhu, Y.Z.; Zhao, J.Y.; Dong, X.Q.; Li, M.G.; Bian, L.; et al. Ameliorative effects of Compound K and ginsenoside Rh1 on nonalcoholic fatty liver disease in rats. Sci. Rep. 2017, 7, 41144. [Google Scholar] [CrossRef]
- Li, W.; Zhang, M.; Gu, J.; Meng, Z.J.; Zhao, L.C.; Zheng, Y.N.; Chen, L.; Yang, G.L. Hypoglycemic effect of protopanaxadiol-type ginsenosides and compound K on Type 2 diabetes mice induced by high-fat diet combining with streptozotocin via suppression of hepatic gluconeogenesis. Fitoterapia 2012, 83, 192–198. [Google Scholar] [CrossRef]
- Kim, M.H.; Park, J.S.; Jung, J.W.; Byun, K.W.; Kang, K.S.; Lee, Y.S. Daidzein supplementation prevents nonalcoholic fatty liver disease through alternation of hepatic gene expression profiles and adipocyte metabolism. Int. J. Obes. 2011, 35, 1019–1030. [Google Scholar] [CrossRef]
- Spinozzi, S.; Colliva, C.; Camborata, C.; Roberti, M.; Ianni, C.; Neri, F.; Calvarese, C.; Lisotti, A.; Mazzella, G.; Roda, A. Berberine and its metabolites: Relationship between physicochemical properties and plasma levels after administration to human subjects. J. Nat. Prod. 2014, 77, 766–772. [Google Scholar] [CrossRef]
- Kang, M.J.; Ko, G.S.; Oh, D.G.; Kim, J.S.; Noh, K.; Kang, W.; Yoon, W.K.; Kim, H.C.; Jeong, H.G.; Jeong, T.C. Role of metabolism by intestinal microbiota in pharmacokinetics of oral baicalin. Arch. Pharm. Res. 2014, 37, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Murugan, P.; Pari, L. Influence of tetrahydrocurcumin on erythrocyte membrane bound enzymes and antioxidant status in experimental type 2 diabetic rats. J. Ethnopharmacol. 2007, 113, 479–486. [Google Scholar] [CrossRef]
- Nakagawa, K.; Zingg, J.M.; Kim, S.H.; Thomas, M.J.; Dolnikowski, G.G.; Azzi, A.; Miyazawa, T.; Meydani, M. Differential cellular uptake and metabolism of curcuminoids in monocytes/macrophages: Regulatory effects on lipid accumulation. Br. J. Nutr. 2014, 112, 8–14. [Google Scholar] [CrossRef]
- Dai, X.; Ding, Y.; Zhang, Z.; Cai, X.; Bao, L.; Li, Y. Quercetin but not quercitrin ameliorates tumor necrosis factor-alpha-induced insulin resistance in C2C12 skeletal muscle cells. Biol. Pharm. Bull. 2013, 36, 788–795. [Google Scholar] [CrossRef]
- Takeda, S.; Ishthara, K.; Wakui, Y.; Amagaya, S.; Maruno, M.; Akao, T.; Kobashi, K. Bioavailability study of glycyrrhetic acid after oral administration of glycyrrhizin in rats; relevance to the intestinal bacterial hydrolysis. J. Pharm. Pharmacol. 1996, 48, 902–905. [Google Scholar] [CrossRef]
- Park, M.; Lee, J.H.; Choi, J.K.; Hong, Y.D.; Bae, I.H.; Lim, K.M.; Park, Y.H.; Ha, H. 18β-glycyrrhetinic acid attenuates anandamide-induced adiposity and high-fat diet induced obesity. Mol. Nutr. Food Res. 2014, 58, 1436–1446. [Google Scholar] [CrossRef]
- Yan, T.; Wang, H.; Cao, L.; Wang, Q.; Takahashi, S.; Yagai, T.; Li, G.; Krausz, K.W.; Wang, G.; Gonzalez, F.J.; et al. Glycyrrhizin Alleviates Nonalcoholic Steatohepatitis via Modulating Bile Acids and Meta-Inflammation. Drug Metab. Dispos. 2018, 46, 1310–1319. [Google Scholar] [CrossRef]
- Kim, K.A.; Jung, I.H.; Park, S.H.; Ahn, Y.T.; Huh, C.S.; Kim, D.H. Comparative analysis of the gut microbiota in people with different levels of ginsenoside Rb1 degradation to compound K. PLoS ONE 2013, 8, e62409. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Ishige, A.; Yazawa, Y.; Kondo, M.; Muramatsu, K.; Watanabe, K. Promotion of intestinal peristalsis by Bifidobacterium spp. capable of hydrolysing sennosides in mice. PLoS ONE 2012, 7, e31700. [Google Scholar] [CrossRef]
- Brooks, A.W.; Priya, S.; Blekhman, R.; Bordenstein, S.R. Gut microbiota diversity across ethnicities in the United States. PLoS Biol 2018, 16, e2006842. [Google Scholar] [CrossRef]
- Kim, C.Y.; Lee, M.; Yang, S.; Kim, K.; Yong, D.; Kim, H.R.; Lee, I. Human reference gut microbiome catalog including newly assembled genomes from under-represented Asian metagenomes. Genome Med. 2021, 13, 1–20. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Y.; Zhang, M.; Pang, X.; Xu, J.; Kang, C.; Li, M.; Zhang, C.; Zhang, Z.; Zhang, Y. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS ONE 2012, 7, e42529. [Google Scholar] [CrossRef]
- Zhang, B.; Sun, W.; Yu, N.; Sun, J.; Yu, X.; Li, X.; Xing, Y.; Yan, D.; Ding, Q.; Xiu, Z.; et al. Anti-diabetic effect of baicalein is associated with the modulation of gut microbiota in streptozotocin and high-fat-diet induced diabetic rats. J. Funct. Foods 2018, 46, 256–267. [Google Scholar] [CrossRef]
- Cani, P.D.; Delzenne, N.M. The role of the gut microbiota in energy metabolism and metabolic disease. Curr. Pharm. Des. 2009, 15, 1546–1558. [Google Scholar] [CrossRef]
- Ansari, M.H.R.; Saher, S.; Parveen, R.; Khan, W.; Khan, I.A.; Ahmad, S. Role of gut microbiota metabolism and biotransformation on dietary natural products to human health implications with special reference to biochemoinformatics approach. J. Tradit. Complement. Med. 2022. [Google Scholar] [CrossRef]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their Impact on host health. Microbiome 2019, 7, 1–15. [Google Scholar] [CrossRef]
- McCoubrey, L.E.; Thomaidou, S.; Elbadawi, M.; Gaisford, S.; Orlu, M.; Basit, A.W. Machine Learning Predicts Drug Metabolism and Bioaccumulation by Intestinal Microbiota. Pharmaceutics 2021, 13, 2001. [Google Scholar] [CrossRef]
| Herbal Medicine | Compound | Related Microbiota | Microbial Metabolites | Mechanisms | Ref. |
|---|---|---|---|---|---|
| Ginseng Radix | Ginsenoside Rb1 | Bifidobacterium longum H-1 | Ginsenoside Rd Compound K | β-D-glucosidase | [41] |
| Ginsenoside Rb1 | Fusobacterium K-60 | Compound K | β-Glucosidase | [42] | |
| Ginsenosides Ra1 and Ra2 | Bifidobacterium breve K-110 | Ginsenosides Rb2, Rc | β-D-Xylosidase | [43] | |
| Ginsenoside Rb1 | Microbacterium esteraromaticum | Ginsenoside Rd Ginsenoside 20(S)-Rg3 | β-Glucosidase | [44] | |
| Ginsenoside Rb1 | Eubacterium sp. A-44 | Ginsenoside Rd Ginsenoside F2 Compound K | β-D-glucosidase | [45] | |
| Ginsenoside Rc | Bifidobacterium K-103 Eubacterium A-44 | Ginsenoside Rd (intermediate) Compound K | Hydrolysis | [46] | |
| Bacteriodes HJ-15 Bifidobacterium K-506 | Ginsenoside Mb (intermediate) Compound K | Hydrolysis | |||
| Ginsenoside Rb1 | Prevotella oris | 20-O-/J-o-glucopyranosyl-20(S)-protopanaxadiol | β-Glucosidase hydrolysis | [47] | |
| Puerariae Radix And Puerariae Flos | Puerarin | Dorea longicatena PUE | Daidzein | Deglycosylation | [48] |
| Daidzein | Slackia isoflavoniconvertens. | Equol | Not identified | [49] | |
| Kakkalide Tectoridin | Bifidobacterium breve K-110 | Irisolidone tectorigenin | β-D-Xylosidase | [50] | |
| Puerarin Daidzin | Bacteroides sterocoris HJ-15 Bifidobacterium longum H-1 Eubacterium rectale A-44 Streptococcus faecium S-9 | Daidzein | Hydrolysis | [51] | |
| Kakkalide Irisolidone | Not identified | Irisolidone Biochanin A | Hydrolysis Dehydroxylation Demethoxylation Demethylation Hydroxylation Decarbonylation Reduction | [52] | |
| Coptidis Rhizoma | Berberine | Escherichia coli Streptococcus faecalis Lactobacillus acidophilus | Oxyberberine | Oxidation | [53] |
| Berberine | Not identified | Thalifendine Berberrubine Jatrorrhizine | Not identified | [54] | |
| Berberine | Enterobacter cloacae Enterococcus faecium | Dihydroberberine | Nitroreductase | [55] | |
| Scutellaria Radix | Baicalin | Not identified | Baicalein | Not identified | [56] |
| Baicalin | Escherichia coli | Baicalein | Beta-D-glucuronidase | [57] | |
| Baicalin Wogonoside | Lactobacillus delbrueckii Rh2 | Baicalein Wogonin | β-glucuronidase | [58] | |
| Baicalin Wogonoside | Lactobacillus brevis RO1 | Baicalein Wogonin | β-glucuronidase | [59] | |
| Curcumae Radix | Curcumin Demethoxycurcumin Bis-demethoxycurcumin | Escherichia fergusonii Escherichia coli ATCC 8739 Escherichia coli DH10B | Dihydrocurcumin Tetrahydrocurcumin Ferulic acid | Reduction (CurA) | [60] |
| Curcumin | E. Coli strain DH10B | Dihydrocurcumin Tetrahydrocurcumin | Reduction (CurA) | [61] | |
| Curcumin (1) Demethoxycurcumin (2) Bisdemethoxycurcumin (3) | Blautia sp. MRG-PMF1 | Dimethylcurcumin (from 1) Bisdemethylcurcumin (from 1) Demethyldemethoxycurcumin (from 2) | Reduction | [62] | |
| Mori folium/ Bupleurum Radix/ Houttuyniae Herba | Quercitrin | Bacillus subtilis | Quercetin | Dioxygenase (C-ring cleavage) | [63] |
| Quercitrin | Fusobacterium K-60 | Quercetin | Hydrolysis (α-L-Rhamnosidase) | [64] | |
| Quercitrin | Fusobacterium K-60 | Quercetin 3,4-Dihydroxyphenylacetic acid 4-Hydroxylphenylacetic acid | Not identified | [65] | |
| Glycyrrhizae Radix | Glycyrrhizin | Eubacterium sp. GLH | 18β-Glycyrrhetinic acid monoglucuronide 18β-Glycyrrhetinic acid | Deglycosylation | [66] |
| Glycyrrhizin | Not indicated (human feces sample) | 18β-Glycyrrhetic acid | b-D-glucuronidases | [67] | |
| Glycyrrhizin | Ruminococcus sp. PO1-3 | 18β-Glycyrrhetic acid 3-Oxo-glycyrrhetic acid | b-D-glucuronidases 3β-Hydroxysteroid dehydrogenase | [68] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, D.-W.; Wang, J.-H. Gut Microbiome: The Interplay of an “Invisible Organ” with Herbal Medicine and Its Derived Compounds in Chronic Metabolic Disorders. Int. J. Environ. Res. Public Health 2022, 19, 13076. https://doi.org/10.3390/ijerph192013076
Lim D-W, Wang J-H. Gut Microbiome: The Interplay of an “Invisible Organ” with Herbal Medicine and Its Derived Compounds in Chronic Metabolic Disorders. International Journal of Environmental Research and Public Health. 2022; 19(20):13076. https://doi.org/10.3390/ijerph192013076
Chicago/Turabian StyleLim, Dong-Woo, and Jing-Hua Wang. 2022. "Gut Microbiome: The Interplay of an “Invisible Organ” with Herbal Medicine and Its Derived Compounds in Chronic Metabolic Disorders" International Journal of Environmental Research and Public Health 19, no. 20: 13076. https://doi.org/10.3390/ijerph192013076
APA StyleLim, D.-W., & Wang, J.-H. (2022). Gut Microbiome: The Interplay of an “Invisible Organ” with Herbal Medicine and Its Derived Compounds in Chronic Metabolic Disorders. International Journal of Environmental Research and Public Health, 19(20), 13076. https://doi.org/10.3390/ijerph192013076







