Impact of Integrating Machine Learning in Comparative Effectiveness Research of Oral Anticoagulants in Patients with Atrial Fibrillation
Abstract
:1. Introduction
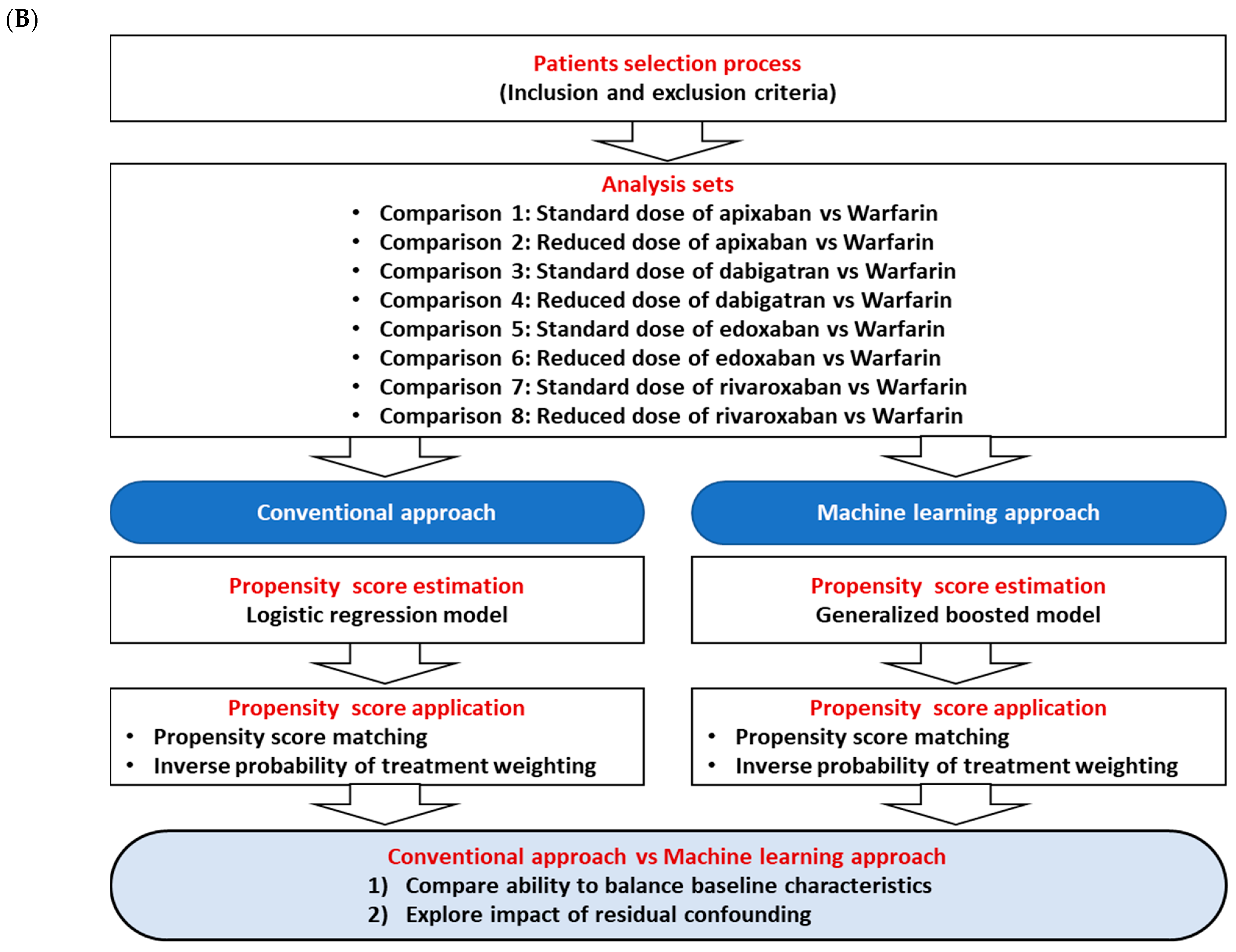
2. Materials and Methods
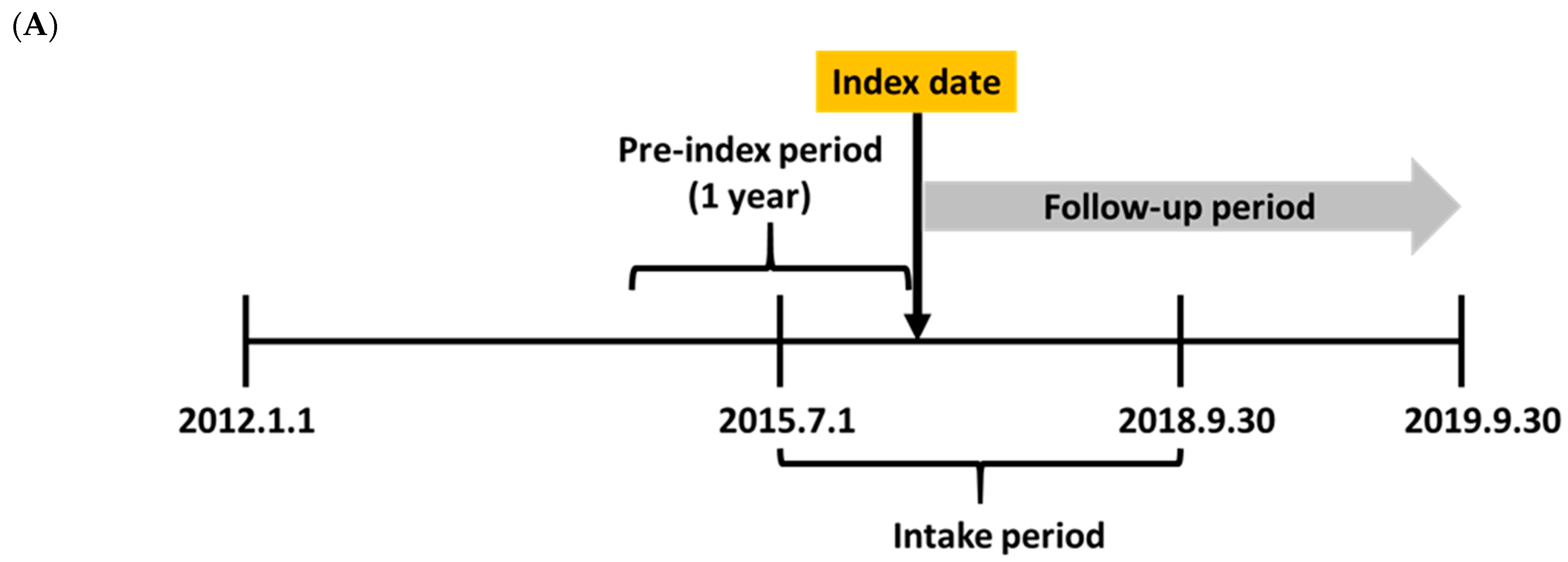
2.1. Study Scheme and Data Source
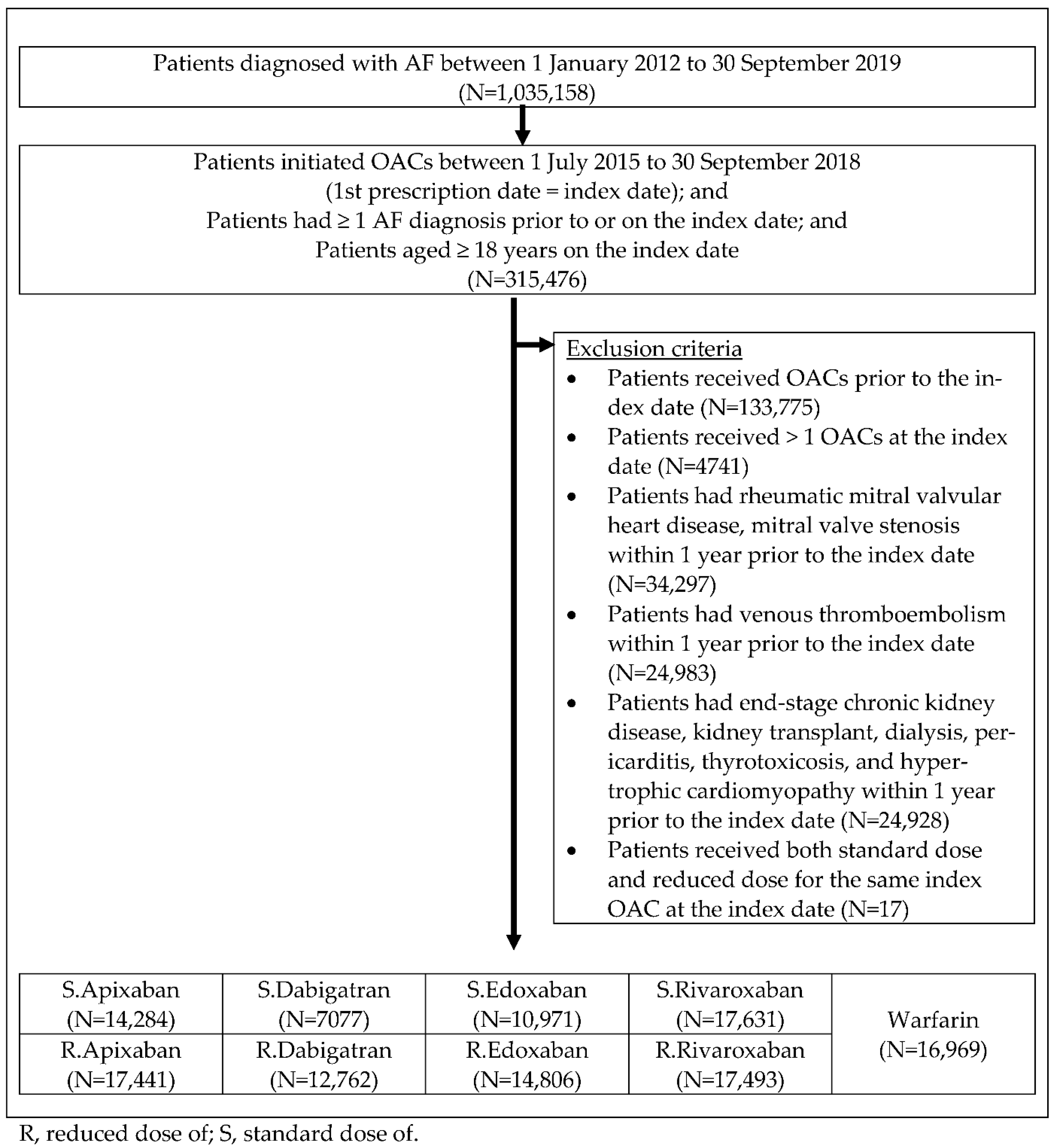
2.2. Study Population and Clinical Outcomes
2.3. Study Outcomes
2.4. Statistical Analyses—PS Estimation and Balance
2.5. Statistical Analyses—E-Value and Negative Control Outcome
3. Results
3.1. Baseline Characteristics and Balance
3.2. E-Value
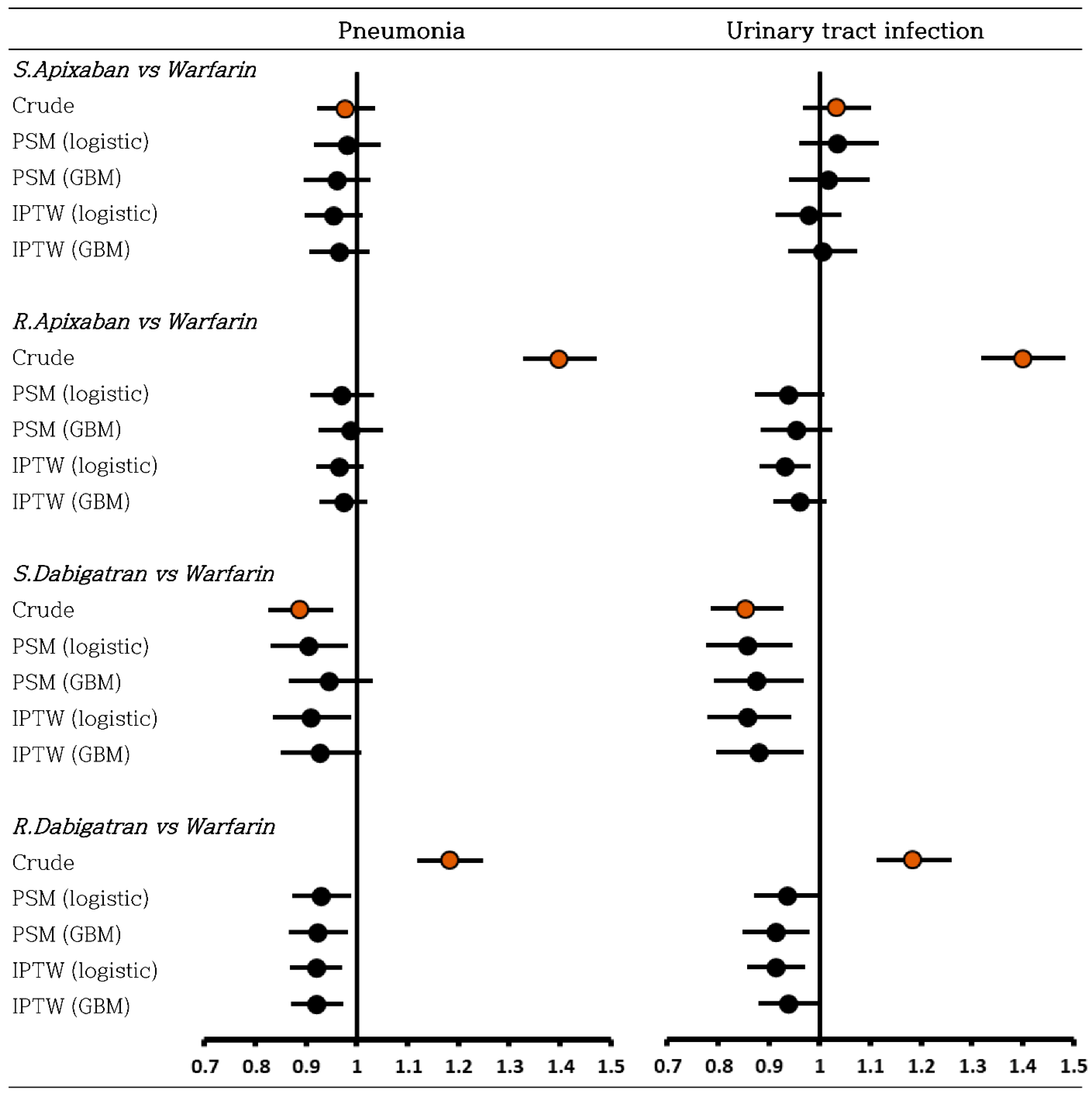
3.3. Negative Control Outcome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uddin, M.J.; Groenwold, R.H.; Ali, M.S.; de Boer, A.; Roes, K.C.; Chowdhury, M.A.; Klungel, O.H.J. Methods to control for unmeasured confounding in pharmacoepidemiology: An overview. Int. J. Clin. Pharm. 2016, 38, 714–723. [Google Scholar] [CrossRef]
- Monti, S.; Grosso, V.; Todoerti, M.; Caporali, R. Randomized controlled trials and real-world data: Differences and similarities to untangle literature data. Rheumatology 2018, 57, vii54–vii58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCaffrey, D.F.; Griffin, B.A.; Almirall, D.; Slaughter, M.E.; Ramchand, R.; Burgette, L.F. A tutorial on propensity score estimation for multiple treatments using generalized boosted models. Stat. Med. 2013, 32, 3388–3414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Austin, P.C. An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivar. Behav. Res. 2011, 46, 399–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, M.-J.; Choi, E.-K.; Han, K.-D.; Lee, S.-R.; Lim, W.-H.; Oh, S.; Lip, G.Y. Effectiveness and safety of non-vitamin K antagonist oral anticoagulants in Asian patients with atrial fibrillation. Stroke 2017, 48, 3040–3048. [Google Scholar] [CrossRef]
- Chan, Y.H.; See, L.C.; Tu, H.T.; Yeh, Y.H.; Chang, S.H.; Wu, L.S.; Lee, H.F.; Wang, C.L.; Kuo, C.F.; Kuo, C.T. Efficacy and safety of apixaban, dabigatran, rivaroxaban, and warfarin in Asians with nonvalvular atrial fibrillation. J. Am. Heart Assoc. 2018, 7, e008150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, T.B.; Skjøth, F.; Nielsen, P.B.; Kjældgaard, J.N.; Lip, G.Y. Comparative effectiveness and safety of non-vitamin K antagonist oral anticoagulants and warfarin in patients with atrial fibrillation: Propensity weighted nationwide cohort study. BMJ 2016, 353, i3189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Abraham, N.S.; Sangaralingham, L.R.; Bellolio, M.F.; McBane, R.D.; Shah, N.D.; Noseworthy, P.A. Effectiveness and safety of dabigatran, rivaroxaban, and apixaban versus warfarin in nonvalvular atrial fibrillation. J. Am. Heart Assoc. 2016, 5, e003725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, Y.-H.; Lee, H.-F.; Chao, T.-F.; Wu, C.-T.; Chang, S.-H.; Yeh, Y.-H.; See, L.-C.; Kuo, C.-T.; Chu, P.-H.; Wang, C.-L.; et al. Real-world comparisons of direct oral anticoagulants for stroke prevention in Asian patients with non-valvular atrial fibrillation: A systematic review and meta-analysis. Cardiovasc. Drugs Ther. 2019, 33, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.-L.; Lip, G.Y.; Lin, S.-J.; Chiang, C.-E. Non–vitamin K antagonist oral anticoagulants for stroke prevention in Asian patients with nonvalvular atrial fibrillation: Meta-analysis. Stroke 2015, 46, 2555–2561. [Google Scholar] [CrossRef]
- Bang, O.Y.; On, Y.K.; Lee, M.-Y.; Jang, S.-W.; Han, S.; Han, S.; Won, M.-M.; Park, Y.-J.; Lee, J.-M.; Choi, H.-Y.; et al. The risk of stroke/systemic embolism and major bleeding in Asian patients with non-valvular atrial fibrillation treated with non-vitamin K oral anticoagulants compared to warfarin: Results from a real-world data analysis. PLoS ONE 2020, 15, e0242922. [Google Scholar] [CrossRef]
- Kim, J.-A.; Yoon, S.; Kim, L.-Y.; Kim, D.-S. Towards actualizing the value potential of Korea Health Insurance Review and Assessment (HIRA) data as a resource for health research: Strengths, limitations, applications, and strategies for optimal use of HIRA data. J. Korean Med. Sci. 2017, 32, 718–728. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Health and Welfare. Production of Statistics of Quality of Health Care in 2015–16 [Korean]. 2017. Available online: http://www.google.co.kr/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&cad=rja&uact=8&ved=2ahUKEwjtrIK7gvPsAhW9L6YKHXR4AUAQFjAAegQIAhAC&url=http%3A%2F%2Fwww.mohw.go.kr%2Freact%2Fmodules%2Fdownload.jsp%3FBOARD_ID%3D60010%26CONT_SEQ%3D356240%26FILE_SEQ%3D296869&usg=AOvVaw1v2GXUM7OHTEc6mOB_fx1V (accessed on 18 October 2020).
- Joung, B.; Lee, J.M.; Lee, K.H.; Kim, T.-H.; Choi, E.-K.; Lim, W.-H.; Kang, K.-W.; Shim, J.; Lim, H.E.; Park, J. 2018 Korean guideline of atrial fibrillation management. Korean Circ. J. 2018, 48, 1033–1080. [Google Scholar] [CrossRef] [Green Version]
- McCaffrey, D.F.; Ridgeway, G.; Morral, A.R. Propensity score estimation with boosted regression for evaluating causal effects in observational studies. Psychol. Methods 2004, 9, 403–425. [Google Scholar] [CrossRef] [Green Version]
- Coffman, D.L.; Zhou, J.; Cai, X. Comparison of methods for handling covariate missingness in propensity score estimation with a binary exposure. BMC Med. Res. Methodol. 2020, 20, 168. [Google Scholar] [CrossRef] [PubMed]
- Allan, V.; Ramagopalan, S.V.; Mardekian, J.; Jenkins, A.; Li, X.; Pan, X.; Luo, X. Propensity score matching and inverse probability of treatment weighting to address confounding by indication in comparative effectiveness research of oral anticoagulants. J. Comp. Eff. Res. 2020, 9, 603–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferri-García, R.; Rueda, M.d.M. Propensity score adjustment using machine learning classification algorithms to control selection bias in online surveys. PLoS ONE 2020, 15, e0231500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, M.F. Causal Inference for the Brave and True. 11—Propensity Score. Available online: https://matheusfacure.github.io/python-causality-handbook/11-Propensity-Score.html# (accessed on 26 November 2021).
- Brookhart, M.A.; Schneeweiss, S.; Rothman, K.J.; Glynn, R.J.; Avorn, J.; Stürmer, T. Variable selection for propensity score models. Am. J. Epidemiol. 2006, 163, 1149–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connolly, S.J.; Ezekowitz, M.D.; Yusuf, S.; Eikelboom, J.; Oldgren, J.; Parekh, A.; Pogue, J.; Reilly, P.A.; Themeles, E.; Varrone, J.; et al. Dabigatran versus warfarin in patients with atrial fibrillation. N. Engl. J. Med. 2009, 361, 1139–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giugliano, R.P.; Ruff, C.T.; Braunwald, E.; Murphy, S.A.; Wiviott, S.D.; Halperin, J.L.; Waldo, A.L.; Ezekowitz, M.D.; Weitz, J.I.; Špinar, J. Edoxaban versus warfarin in patients with atrial fibrillation. N. Engl. J. Med. 2013, 369, 2093–2104. [Google Scholar] [CrossRef]
- Granger, C.B.; Alexander, J.H.; McMurray, J.J.; Lopes, R.D.; Hylek, E.M.; Hanna, M.; Al-Khalidi, H.R.; Ansell, J.; Atar, D.; Avezum, A. Apixaban versus warfarin in patients with atrial fibrillation. N. Engl. J. Med. 2011, 365, 981–992. [Google Scholar] [CrossRef] [Green Version]
- Noseworthy, P.A.; Yao, X.; Abraham, N.S.; Sangaralingham, L.R.; McBane, R.D.; Shah, N.D. Direct comparison of dabigatran, rivaroxaban, and apixaban for effectiveness and safety in nonvalvular atrial fibrillation. Chest 2016, 150, 1302–1312. [Google Scholar] [CrossRef]
- Patel, M.R.; Mahaffey, K.W.; Garg, J.; Pan, G.; Singer, D.E.; Hacke, W.; Breithardt, G.; Halperin, J.L.; Hankey, G.J.; Piccini, J.P. Rivaroxaban versus warfarin in nonvalvular atrial fibrillation. N. Engl. J. Med. 2011, 365, 883–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridgeway, G.; McCaffrey, D.; Morral, A.; Burgette, L.; Griffin, B.A. Toolkit for Weighting and Analysis of Nonequivalent Groups: A Tutorial for the Twang Package; RAND Corporation: Santa Monica, CA, USA, 2017. [Google Scholar]
- Parast, L.; McCaffrey, D.F.; Burgette, L.F.; de la Guardia, F.H.; Golinelli, D.; Miles, J.N.; Griffin, B.A.J. Optimizing variance-bias trade-off in the TWANG package for estimation of propensity scores. Health Serv. Outcomes Res. Methodol. 2017, 17, 175–197. [Google Scholar] [CrossRef]
- Austin, P.C. Using the standardized difference to compare the prevalence of a binary variable between two groups in observational research. Commun. Stat. Simul. Comput. 2009, 38, 1228–1234. [Google Scholar] [CrossRef]
- Cusson, A.; Infante-Rivard, C. Bias factor, maximum bias and the E-value: Insight and extended applications. Int. J. Epidemiol. 2020, 49, 1509–1516. [Google Scholar] [CrossRef]
- Blum, M.R.; Tan, Y.J.; Ioannidis, J.P.A. Use of E-values for addressing confounding in observational studies—An empirical assessment of the literature. Int. J. Epidemiol. 2020, 49, 1482–1494. [Google Scholar] [CrossRef]
- Mathur, M.B.; Ding, P.; Riddell, C.A.; VanderWeele, T.J. Website and R package for computing E-values. Epidemiology 2018, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- VanderWeele, T.J.; Ding, P. Sensitivity analysis in observational research: Introducing the E-value. Ann. Intern. Med. 2017, 167, 268–274. [Google Scholar] [CrossRef] [Green Version]
- Torp-Pedersen, C.; Goette, A.; Nielsen, P.B.; Potpara, T.; Fauchier, L.; John Camm, A.; Arbelo, E.; Boriani, G.; Skjoeth, F.; Rumsfeld, J.; et al. ‘Real-world’ observational studies in arrhythmia research: Data sources, methodology, and interpretation. A position document from European Heart Rhythm Association (EHRA), endorsed by Heart Rhythm Society (HRS), Asia-Pacific HRS (APHRS), and Latin America HRS (LAHRS). EP Eur. 2020, 22, 831–832. [Google Scholar]
- Lip, G.Y.H.; Skjøth, F.; Nielsen, P.B.; Kjældgaard, J.N.; Larsen, T.B. Effectiveness and Safety of Standard-Dose Nonvitamin K Antagonist Oral Anticoagulants and Warfarin Among Patients with Atrial Fibrillation with a Single Stroke Risk Factor: A Nationwide Cohort Study. JAMA Cardiol. 2017, 2, 872–881. [Google Scholar] [CrossRef]
- Alam, S.; Moodie, E.E.M.; Stephens, D.A. Should a propensity score model be super? The utility of ensemble procedures for causal adjustment. Stat. Med. 2019, 38, 1690–1702. [Google Scholar] [CrossRef]
- Elze, M.C.; Gregson, J.; Baber, U.; Williamson, E.; Sartori, S.; Mehran, R.; Nichols, M.; Stone, G.W.; Pocock, S.J. Comparison of propensity score methods and covariate adjustment: Evaluation in 4 cardiovascular studies. J. Am. Coll. Cardiol. 2017, 69, 345–357. [Google Scholar] [CrossRef]
- Harder, V.S.; Stuart, E.A.; Anthony, J.C. Propensity score techniques and the assessment of measured covariate balance to test causal associations in psychological research. Psychol. Methods 2010, 15, 234–249. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.K.; Lessler, J.; Stuart, E.A. Improving propensity score weighting using machine learning. Stat. Med. 2010, 29, 337–346. [Google Scholar] [CrossRef] [Green Version]
- Dehejia, R.H.; Wahba, S. Causal effects in nonexperimental studies: Reevaluating the evaluation of training programs. J. Am. Stat. Assoc. 1999, 94, 1053–1062. [Google Scholar] [CrossRef]
- Dehejia, R.H.; Wahba, S. Propensity score-matching methods for nonexperimental causal studies. Rev. Econ. Stat. 2002, 84, 151–161. [Google Scholar] [CrossRef] [Green Version]
- Rosenbaum, P.R.; Rubin, D.B. Reducing bias in observational studies using subclassification on the propensity score. J. Am. Stat. Assoc. 1984, 79, 516–524. [Google Scholar] [CrossRef]
- Ridgeway, G. Generalized Boosted Models: A Guide to the Gbm Package. Updated 2020; pp. 1–15. Available online: https://cran.r-project.org/web/packages/gbm/vignettes/gbm.pdf (accessed on 11 November 2021).
- Ding, P.; VanderWeele, T.J. Sensitivity Analysis Without Assumptions. Epidemiology 2016, 27, 368–377. [Google Scholar] [CrossRef] [Green Version]
- VanderWeele, T.J.; Mathur, M.B. Commentary: Developing best-practice guidelines for the reporting of E-values. Int. J. Epidemiol. 2020, 49, 1495–1497. [Google Scholar] [CrossRef]
- Madhavan, M.; Holmes, D.N.; Piccini, J.P.; Ansell, J.E.; Fonarow, G.C.; Hylek, E.M.; Kowey, P.R.; Mahaffey, K.W.; Thomas, L.; Peterson, E.D.; et al. Association of frailty and cognitive impairment with benefits of oral anticoagulation in patients with atrial fibrillation. Am. Heart J. 2019, 211, 77–89. [Google Scholar] [CrossRef]
- DeMaris, A. Combating unmeasured confounding in cross-sectional studies: Evaluating instrumental-variable and Heckman selection models. Psychol. Methods 2014, 19, 380. [Google Scholar] [CrossRef] [Green Version]
- Schneeweiss, S. Sensitivity analysis and external adjustment for unmeasured confounders in epidemiologic database studies of therapeutics. Pharmacoepidemiol. Drug Saf. 2006, 15, 291–303. [Google Scholar] [CrossRef]
- Hai, Q.; Ritchey, B.; Robinet, P.; Alzayed, A.M.; Brubaker, G.; Zhang, J.; Smith, J.D. Quantitative trait locus mapping of macrophage cholesterol metabolism and CRISPR/Cas9 editing implicate an ACAT1 truncation as a causal modifier variant. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 83–91. [Google Scholar] [CrossRef]
- Ritchey, B.; Hai, Q.; Han, J.; Barnard, J.; Smith, J.D. Genetic variant in 3′ untranslated region of the mouse pycard gene regulates inflammasome activity. eLife 2021, 10, e68203. [Google Scholar] [CrossRef]
- Hsu, J.; Gore-Panter, S.; Tchou, G.; Castel, L.; Lovano, B.; Moravec, C.S.; Pettersson, G.B.; Roselli, E.E.; Gillinov, A.M.; McCurry, K.R. Genetic control of left atrial gene expression yields insights into the genetic susceptibility for atrial fibrillation. Circ. Genom. Precis. Med. 2018, 11, e002107. [Google Scholar] [CrossRef] [Green Version]
- Austin, P.C. The use of propensity score methods with survival or time-to-event outcomes: Reporting measures of effect similar to those used in randomized experiments. Stat. Med. 2014, 33, 1242–1258. [Google Scholar] [CrossRef] [Green Version]
- Aljofey, A.; Rasool, A.; Jiang, Q.; Qu, Q. A Feature-Based Robust Method for Abnormal Contracts Detection in Ethereum Blockchain. Electronics 2022, 11, 2937. [Google Scholar] [CrossRef]
- Chadrasekhar, G. Time and Space Complexity of Machine Learning Models. Available online: https://pub.towardsai.net/time-and-space-complexity-of-machine-learning-models-df9b704e3e9c (accessed on 16 September 2022).
- Martindale, N.; Ismail, M.; Talbert, D.A. Ensemble-based online machine learning algorithms for network intrusion detection systems using streaming data. Information 2020, 11, 315. [Google Scholar] [CrossRef]

|
| Comparisons | E-value for the Hazard Ratios (1–3) (E-Value for the Limit of Confidence Interval Closest to the Null) | E-Value as an Anchor (4) | Meaningful Difference Δ (5) | Maximum Possible Coverage (%) (6) | |||
|---|---|---|---|---|---|---|---|
| PSM (Logistic) | PSM (GBM) | IPTW (Logistic) | IPTW (GBM) | IPTW (GBM) | |||
| Comparison 1 | 2.24 (1.92) | 2.23 (1.90) | 2.32 (2.04) | 2.35 (2.07) | 2.32 | 0.03 | - |
| Comparison 2 | 1.96 (1.66) | 2.01 (1.71) | 2.00 (1.78) | 2.08 (1.85) | 1.67 | 0.41 | 29 |
| Comparison 3 | 2.13 (1.76) | 2.19 (1.81) | 2.19 (1.82) | 2.30 (1.93) | 2.36 | 0.06 | - |
| Comparison 4 | 1.75 (1.47) | 1.89 (1.60) | 1.89 (1.64) | 1.93 (1.68) | 1.65 | 0.28 | 64 |
| Comparison 5 | 2.45 (2.05) | 2.27 (1.88) | 2.52 (2.13) | 2.43 (2.05) | 2.42 | 0.01 | - |
| Comparison 6 | 2.70 (2.32) | 2.67 (2.28) | 2.53 (2.21) | 2.56 (2.24) | 2.10 | 0.46 | 37 |
| Comparison 7 | 2.01 (1.73) | 2.04 (1.75) | 2.02 (1.78) | 2.04 (1.80) | 1.94 | 0.10 | 30 |
| Comparison 8 | 2.05 (1.76) | 2.08 (1.78) | 2.04 (1.80) | 2.10 (1.85) | 1.72 | 0.38 | 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.; Suh, H.S. Impact of Integrating Machine Learning in Comparative Effectiveness Research of Oral Anticoagulants in Patients with Atrial Fibrillation. Int. J. Environ. Res. Public Health 2022, 19, 12916. https://doi.org/10.3390/ijerph191912916
Han S, Suh HS. Impact of Integrating Machine Learning in Comparative Effectiveness Research of Oral Anticoagulants in Patients with Atrial Fibrillation. International Journal of Environmental Research and Public Health. 2022; 19(19):12916. https://doi.org/10.3390/ijerph191912916
Chicago/Turabian StyleHan, Sola, and Hae Sun Suh. 2022. "Impact of Integrating Machine Learning in Comparative Effectiveness Research of Oral Anticoagulants in Patients with Atrial Fibrillation" International Journal of Environmental Research and Public Health 19, no. 19: 12916. https://doi.org/10.3390/ijerph191912916
APA StyleHan, S., & Suh, H. S. (2022). Impact of Integrating Machine Learning in Comparative Effectiveness Research of Oral Anticoagulants in Patients with Atrial Fibrillation. International Journal of Environmental Research and Public Health, 19(19), 12916. https://doi.org/10.3390/ijerph191912916






