Antibiotic Resistance during COVID-19: A Systematic Review
Abstract
:1. Introduction
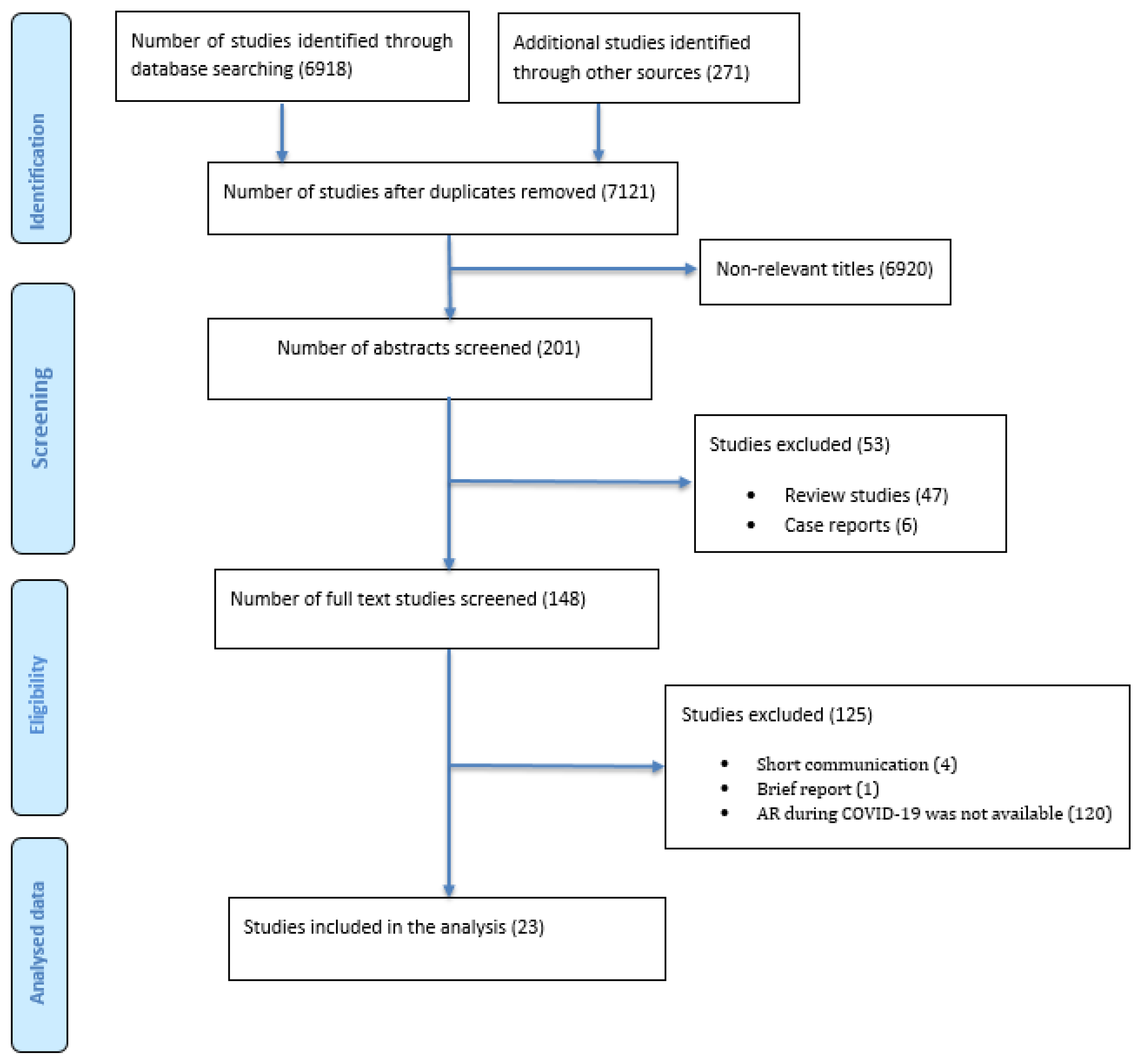
2. Materials and Methods
2.1. Inclusion Criteria
- Articles should be original studies.
- Studies should report data on at least these two variables: antibiotic resistance, and COVID-19.
- Studies should be written in English or at least their abstract should be in English.
- Studies should be published between 2019 (since announcing COVID-19 in the country where the included study conducted) and May 2022.
2.2. Exclusion Criteria
2.3. Search Strategy
2.4. Study Selection and Data Extraction
2.5. Quality Assessment
2.6. Data Analysis
3. Results
3.1. Study Characteristics
3.2. Antibiotic Resistance Findings during COVID-19
3.3. Nature of AR during COVID-19
3.4. Pattern of Resistant Bacteria to Tested Antibiotics during the COVID-19 Pandemic
3.5. Potential Risk Factors
4. Discussion
Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Director-General’s Remarks at the Media Briefing on 2019. 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020 (accessed on 9 April 2022).
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef]
- World Health Organization. Coronavirus Disease (COVID-19) Pandemic. 2021. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 10 April 2022).
- Getahun, H.; Smith, I.; Trivedi, K.; Paulin, S.; Balkhy, H.H. Tackling antimicrobial resistance in the COVID-19 pandemic. Bull. World Health Organ. 2020, 98, 442. [Google Scholar] [CrossRef]
- Lynch, C.; Mahida, N.; Gray, J. Antimicrobial stewardship: A COVID casualty? J. Hosp. Inf. 2020, 106, 401–403. [Google Scholar] [CrossRef]
- Zavala-Flores, E.; Salcedo-Matienzo, J. Medicación prehospitalaria en pacientes hospitalizados por COVID-19 en un hospital público de Lima-Perú. Acta Méd. Peru. 2020, 37, 393–395. [Google Scholar] [CrossRef]
- Sharifipour, E.; Shams, S.; Esmkhani, M.; Khodadadi, J.; Fotouhi-Ardakani, R.; Koohpaei, A.; Doosti, Z.; Golzari, S.E. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect. Dis. 2020, 20, 646. [Google Scholar] [CrossRef] [PubMed]
- Fattorini, L.; Creti, R.; Palma, C.; Pantosti, A. Bacterial coinfections in COVID-19: An underestimated adversary. Ann. Ist. Super. Sanit. 2020, 56, 359–364. [Google Scholar]
- Dhesi, Z.; Enne, V.I.; Brealey, D.; Livermore, D.M.; High, J.; Russell, C.; Colles, A.; Kandil, H.; Mack, D.; Martin, D.; et al. Organisms causing secondary pneumonias in COVID-19 patients at 5 UK ICUs as detected with the FilmArray test. medRxiv 2020. Available online: https://www.medrxiv.org/content/10.1101/2020.06.22.20131573v1 (accessed on 10 April 2022).
- Dudoignon, E.; Caméléna, F.; Deniau, B.; Habay, A.; Coutrot, M.; Ressaire, Q.; Plaud, B.; Berçot, B.; Dépret, F. Bacterial pneumonia in COVID-19 critically ill patients: A case series. Clin. Infect. Dis. 2021, 72, 905–906. [Google Scholar] [CrossRef]
- Rawson, T.M.; Moore, L.S.P.; Zhu, N.; Ranganathan, N.; Skolimowska, K.; Gilchrist, M.; Satta, G.; Cooke, G.; Holmes, A.H. Bacterial and Fungal Coinfection in Individuals With Coronavirus: A Rapid Review To Support COVID-19 Antimicrobial Prescribing. Clin. Infect. Dis. 2020, 71, 2459–2468. [Google Scholar] [CrossRef]
- Abelenda-Alonso, G.; Padullés, A.; Rombauts, A.; Gudiol, C.; Pujol, M.; Alvarez-Pouso, C.; Jodar, R.; Carratalà, J. Antibiotic prescription during the COVID-19 pandemic: A biphasic pattern. In Infection Control and Hospital Epidemiology; Cambridge University Press: Cambridge, UK, 2020; Volume 41, pp. 1371–1372. [Google Scholar]
- Aurilio, C.; Sansone, P.; Paladini, A.; Barbarisi, M.; Coppolino, F.; Pota, V.; Pace, M. Multidrug Resistence Prevalence in COVID Area. Life 2021, 11, 601. [Google Scholar] [CrossRef]
- Gaspar, G.G.; Ferreira, L.R.; Feliciano, C.S.; Júnior, C.P.C.; Molina, F.M.R.; Vendruscolo, A.C.S.; Bradan, G.M.A.; Lopes, N.A.P.; Martinez, R.; Bollela, V.R. Pre- and post-COVID-19 evaluation of antimicrobial susceptibility for healthcare-associated infections in the intensive care unit of a tertiary hospital. Rev. Soc. Bras. Med. Trop. 2021, 54, e00902021. [Google Scholar] [CrossRef]
- Fernández, P.; Moreno, L.; Yagüe, G.; Andreu, E.; Jara, R.; Segovia, M. Colonization by multidrug-resistant microorganisms in ICU patients during the COVID-19 pandemic. Med. Intensiva 2021, 45, 313–315. [Google Scholar] [CrossRef] [PubMed]
- Bork, J.T.; Leekha, S.; Claeys, K.; Seung, H.; Tripoli, M.; Amoroso, A.; Heil, E.L. Change in hospital antibiotic use and acquisition of multidrug-resistant gram-negative organisms after the onset of coronavirus disease 2019. Infect. Control Hosp. Epidemiol. 2020, 42, 1115–1117. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.A.; Clarke, M.; Rovers, M.; Riley, R.D.; Simmonds, M.; Stewart, G.; Tierney, J.F. Preferred reporting items for a systematic review and meta-analysis of individual participant data: The PRISMA-IPD statement. JAMA 2015, 313, 1657–1665. [Google Scholar] [CrossRef] [PubMed]
- Institute, J.B. Critical Appraisal Tools. 2020. Available online: https://jbi.global/critical-appraisal-tools (accessed on 20 April 2022).
- Akhtar, S.; Nasir, J.A.; Ali, A.; Asghar, M.; Majeed, R.; Sarwar, A. Prevalence of type-2 diabetes and prediabetes in Malaysia: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0263139. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.; Yang, Y.; Cai, P.; Cao, J.; Cai, X.; Zhang, Y. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: A retrospective analysis. Antimicrob. Resist. Infect. Control 2020, 9, 153. [Google Scholar] [CrossRef]
- Gomez-Simmonds, A.; Annavajhala, M.K.; McConville, T.H.; E Dietz, D.; Shoucri, S.M.; Laracy, J.C.; Rozenberg, F.D.; Nelson, B.; Greendyke, W.G.; Furuya, E.Y.; et al. Carbapenemase-producing Enterobacterales causing secondary infections during the COVID-19 crisis at a New York City hospital. J. Antimicrob. Chemother. 2020, 76, 380–384. [Google Scholar] [CrossRef]
- Sang, L.; Xi, Y.; Lin, Z.; Pan, Y.; Song, B.; Li, C.-A.; Zheng, X.; Zhong, M.; Jiang, L.; Pan, C.; et al. Secondary infection in severe and critical COVID-19 patients in China: A multicenter retrospective study. Ann. Palliat. Med. 2021, 10, 8557–8570. [Google Scholar] [CrossRef]
- Despotovic, A.; Milosevic, B.; Cirkovic, A.; Vujovic, A.; Cucanic, K.; Cucanic, T.; Stevanovic, G. The Impact of COVID-19 on the Profile of Hospital-Acquired Infections in Adult Intensive Care Units. Antibiotics 2021, 10, 1146. [Google Scholar] [CrossRef]
- Caruso, P.; Maiorino, M.I.; Macera, M.; Signoriello, G.; Castellano, L.; Scappaticcio, L.; Longo, M.; Gicchino, M.; Campitiello, F.; Bellastella, G.; et al. Antibiotic Resistance in Diabetic Foot Infection: How it Changed with COVID-19 Pandemic In A Tertiary Care Center. Diabetes Res. Clin. Pr. 2021, 175, 108797. [Google Scholar] [CrossRef]
- Wardoyo, E.H.; Suardana, I.W.; Yasa, I.W.P.S.; Sukrama, I.D.M. Antibiotics susceptibility of Escherichia coli isolates from clinical specimens before and during COVID-19 pandemic. Iran. J. Microbiol. 2021, 13, 156–160. [Google Scholar]
- Temperoni, C.; Caiazzo, L.; Barchiesi, F. High Prevalence of Antibiotic Resistance among Opportunistic Pathogens Isolated from Patients with COVID-19 under Mechanical Ventilation: Results of a Single-Center Study. Antibiotics 2021, 10, 1080. [Google Scholar] [CrossRef] [PubMed]
- Bazaid, A.S.; Barnawi, H.; Qanash, H.; Alsaif, G.; Aldarhami, A.; Gattan, H.; Alharbi, B.; Alrashidi, A.; Abu Al-Soud, W.; Moussa, S.; et al. Bacterial Coinfection and Antibiotic Resistance Profiles among Hospitalised COVID-19 Patients. Microorganisms 2022, 10, 495. [Google Scholar] [CrossRef] [PubMed]
- Pourajam, S.; Kalantari, E.; Talebzadeh, H.; Mellali, H.; Sami, R.; Soltaninejad, F.; Amra, B.; Sajadi, M.; Alenaseri, M.; Kalantari, F.; et al. Secondary Bacterial Infection and Clinical Characteristics in Patients With COVID-19 Admitted to Two Intensive Care Units of an Academic Hospital in Iran During the First Wave of the Pandemic. Front. Cell. Infect. Microbiol. 2022, 12, 141. [Google Scholar] [CrossRef] [PubMed]
- Gysin, M.; Acevedo, C.T.; Haldimann, K.; Bodendoerfer, E.; Imkamp, F.; Bulut, K.; Buehler, P.K.; Brugger, S.D.; Becker, K.; Hobbie, S.N. Antimicrobial susceptibility patterns of respiratory Gram-negative bacterial isolates from COVID-19 patients in Switzerland. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 64. [Google Scholar] [CrossRef]
- Polemis, M.; Mandilara, G.; Pappa, O.; Argyropoulou, A.; Perivolioti, E.; Koudoumnakis, N.; Pournaras, S.; Vasilakopoulou, A.; Vourli, S.; Katsifa, H.; et al. COVID-19 and Antimicrobial Resistance: Data from the Greek Electronic System for the Surveillance of Antimicrobial Resistance—WHONET-Greece (January 2018–March 2021). Life 2021, 11, 996. [Google Scholar] [CrossRef]
- Bahçe, Y.G.; Acer, Ö.; Özüdoğru, O. Evaluation of bacterial agents isolated from endotracheal aspirate cultures of Covid-19 general intensive care patients and their antibiotic resistance profiles compared to pre-pandemic conditions. Microb. Pathog. 2022, 164, 105409. [Google Scholar] [CrossRef]
- Karataş, M.; Yaşar-Duman, M.; Tünger, A.; Çilli, F.; Aydemir, Ş.; Özenci, V. Secondary bacterial infections and antimicrobial resistance in COVID-19: Comparative evaluation of pre-pandemic and pandemic-era, a retrospective single center study. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 51. [Google Scholar] [CrossRef]
- Tiri, B.; Sensi, E.; Marsiliani, V.; Cantarini, M.; Priante, G.; Vernelli, C.; Martella, L.A.; Costantini, M.; Mariottini, A.; Andreani, P.; et al. Antimicrobial Stewardship Program, COVID-19, and Infection Control: Spread of Carbapenem-Resistant Klebsiella Pneumoniae Colonization in ICU COVID-19 Patients. What Did Not Work? J. Clin. Med. 2020, 9, 2744. [Google Scholar] [CrossRef]
- Jamnani, A.N.; Montazeri, M.; Mirzakhani, M.; Moosazadeh, M.; Haghighi, M. Evaluation of Bacterial Coinfection and Antibiotic Resistance in Patients with COVID-19 under Mechanical Ventilation. SN Compr. Clin. Med. 2022, 4, 4–8. [Google Scholar] [CrossRef]
- Meawed, T.E.; Ahmed, S.M.; Mowafy, S.M.; Samir, G.M.; Anis, R.H. Bacterial and fungal ventilator associated pneumonia in critically ill COVID-19 patients during the second wave. J. Infect. Public Health 2021, 14, 1375–1380. [Google Scholar] [CrossRef]
- Saini, V.; Jain, C.; Singh, N.; Alsulimani, A.; Gupta, C.; Dar, S.; Haque, S.; Das, S. Paradigm shift in antimicrobial resistance pattern of bacterial isolates during the covid-19 pandemic. Antibiotics 2021, 10, 954. [Google Scholar] [CrossRef] [PubMed]
- Aldhwaihi, K.A.; Alsanad, S.M.; Almutiri, A.H.; Aldoihi, S. Assessment of Antibiotic Resistance Pattern of Bacteria Prevalent during COVID-19 Pandemic. J. Pharm. Res. Int. 2021, 33, 117–127. [Google Scholar] [CrossRef]
- Palanisamy, N.; Vihari, N.; Meena, D.S.; Kumar, D.; Midha, N.; Tak, V.; Sharma, A.; Bohra, G.K.; Kothari, N.; Dutt, N.; et al. Clinical profile of bloodstream infections in COVID-19 patients: A retrospective cohort study. BMC Infect. Dis. 2021, 21, 933. [Google Scholar] [CrossRef]
- Zeshan, B.; Karobari, M.I.; Afzal, N.; Siddiq, A.; Basha, S.; Basheer, S.N.; Peeran, S.W.; Mustafa, M.; Daud, N.H.A.; Ahmed, N.; et al. The Usage of Antibiotics by COVID-19 Patients with Comorbidities: The Risk of Increased Antimicrobial Resistance. Antibiotics 2021, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Boorgula, S.Y.; Yelamanchili, S.; Kottapalli, P.; Naga, M.D. An Update on Secondary Bacterial and Fungal Infections and Their Antimicrobial Resistance Pattern (AMR) in COVID-19 Confirmed Patients at a Tertiary Care Hospital. J. Lab. Physicians 2022, 14, 260–264. [Google Scholar] [CrossRef]
- Tadesse, B.T.; Ashley, E.A.; Ongarello, S.; Havumaki, J.; Wijegoonewardena, M.; González, I.J.; Dittrich, S. Antimicrobial resistance in Africa: A systematic review. BMC Infect. Dis. 2017, 17, 616. [Google Scholar] [CrossRef]
- Azimi, T.; Maham, S.; Fallah, F.; Azimi, L.; Gholinejad, Z. Evaluating the antimicrobial resistance patterns among major bacterial pathogens isolated from clinical specimens taken from patients in mofid children’s hospital, Tehran, Iran: 2013–2018. Infect. Drug Res. 2019, 12, 2089–2102. [Google Scholar] [CrossRef]
- Mogasale, V.V.; Saldanha, P.; Pai, V.; Rekha, P.D.; Mogasale, V. A descriptive analysis of antimicrobial resistance patterns of WHO priority pathogens isolated in children from a tertiary care hospital in India. Sci. Rep. 2021, 11, 5116. [Google Scholar] [CrossRef]
- Alhumaid, S.; Al Mutair, A.; Al Alawi, Z.; Alzahrani, A.J.; Tobaiqy, M.; Alresasi, A.M.; Bu-Shehab, I.; Al-Hadary, I.; Alhmeed, N.; Alismail, M.; et al. Antimicrobial susceptibility of gram-positive and gram-negative bacteria: A 5-year retrospective analysis at a multi-hospital healthcare system in Saudi Arabia. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 43. [Google Scholar] [CrossRef]
- Alshammari, N.; Aly, M.; Al-abdullah, N. Prevalence of Multidrug-Resistant Gram-Negative Bacteria in Saudi Arabia: Meta Review. Biosc. Biotech. Res. Comm. 2021, 14, 12–19. [Google Scholar] [CrossRef]
- Hu, F.; Zhu, D.; Wang, F.; Wang, M. Current Status and Trends of Antibacterial Resistance in China. Clin. Infect. Dis. 2018, 67, S128–S134. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Rabbi, B.; Sultana, S. Antibiotic resistance in Bangladesh: A systematic review. Int. J. Infect. Dis. 2019, 80, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Effah, C.Y.; Sun, T.; Liu, S.; Wu, Y. Klebsiella pneumoniae: An increasing threat to public health. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Subedi, D.; Vijay, A.K.; Willcox, M. Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: An ocular perspective. Clin. Exp. Optom. 2018, 101, 162–171. [Google Scholar] [CrossRef]
- Sato, T.; Shiraishi, T.; Hiyama, Y.; Honda, H.; Shinagawa, M.; Usui, M.; Kuronuma, K.; Masumori, N.; Takahashi, S.; Tamura, Y.; et al. Contribution of novel amino acid alterations in PmrA or PmrB to colistin resistance in mcr-negative Escherichia coli clinical isolates, including major multidrug-resistant lineages O25b:H4-ST131-H30Rx and Non-x. Antimicrob. Agent. Chemother. 2018, 62, e00864-18. [Google Scholar] [CrossRef]
- Abbas, M.; Uçkay, I.; Lipsky, B.A. In diabetic foot infections antibiotics are to treat infection, not to heal wounds. Expert Opin. Pharmacother. 2015, 16, 821–832. [Google Scholar] [CrossRef]
- Chen, Q.; Li, D.; Beiersmann, C.; Neuhann, F.; Moazen, B.; Lu, G.; Müller, O. Risk factors for antibiotic resistance development in healthcare settings in China: A systematic review. Epidemiology Infect. 2021, 149, 1–30. [Google Scholar] [CrossRef]
| Author | Country | Year | Study Design | Duration (months) | Settings | Participants | Age | AR Findings | Secondary Infection | Quality |
|---|---|---|---|---|---|---|---|---|---|---|
| Hassan Mahmoudi | Iran | 2020 | Cross sectional study | 8 | Inpatients and outpatients | 340 patients | NA | Among COVID-19 patients, Enterobacteriaceae had the highest resistance to cotrimoxazol, piperacillin, ceftazidime, and cefepime. | Klebsiella, S. aureus (MSSA), E. coli, S. aureus (MRSA), and Enterobacter species, and P. aeruginosa. | Medium 50% |
| Ehsan Sharifipour | Iran | 2020 | Retrospective observational study | During the COVID-19 era | Inpatients (ICU) | 19 patients | Mean (SD) 67 (± 14.6) | A. baumannii isolates showed high-level resistance to all tested antibiotics. Only colistin showed a 52% resistance rate. | A. baumannii. | Medium 66% |
| Jie Li | China | 2020 | Retrospective electronic medical records reviewed study | 2 | Inpatients (ICU) | 102 patients | Mean (SD) 66.2 (±11.2) | The rate of AR was generally high. Carbapenem-resistant A. baumannii (CRAB) and carbapenem-resistant K. pneumoniae (CRKP) accounted for 91.7% and 76.6% of AR, respectively. Meticillin resistance was present in 100% of S. aureus and coagulase-negative staphylococci. Extended-spectrum beta-lactamase (ESBL) producing E. coli was responsible for 75% of AR. | The top three bacteria causing SBIs were A. baumannii, K. pneumoniae, and S. maltophilia. | High 100% |
| Angela Gomez-Simmonds | United States (New York) | 2020 | Retrospective study | 3 | Inpatients (ICU) | 13 patients | Median age 67 years, IQR (50–72) | Most of (18/20) the isolates showed high-level meropenem resistance. One patient with K. pneumoniae VAP developed ceftazidime/avibactam treatment failure attributable to the development of resistance. | K. pneumoniae and 4 Enterobacter cloacae complex isolates. | High 100% |
| Ling Sang | China | 2020 | Retrospective medical records review study | 3 | Inpatients (ICU) | 190 patients | Mean (SD) 62.68 (±13.3) | The rates of MDR bacteria and CRE were unexpectedly high (K. pneumonia = 94.5%, A. baumannii = 98.3%, and Pseudomonas spp. = 92.5%). | K. pneumoniae, A. baumannii, S. maltophilia, C. albicans, and Pseudomonas spp. | Medium 50% |
| Naveenraj Palanisamy | India | 2021 | Retrospective observational study | 5 | Inpatients (ICU) | 750 patients | Median (IQR) 65 years (54–70) | Out of 64 patients, 57.8% patients had MDRO. The incidence of carbapenem-resistant Gram-negative bacteria was 47.2% (25/53). | A. baumannii, followed by K. pneumonia. | High 87% |
| Aleksa Despotovic | Serbia | 2021 | Retrospective study | 12 | Inpatients (ICU) | 611 patients | Mean (SD) 66.2 (±13.6) | The majority of tested antimicrobials demonstrated high resistant rates, above 80%. Additionally, resistance was significantly higher for imipenem, meropenem, and ciprofloxacin compared with the pre-COVID-19 era. | In COVID-19 patients, Acinetobacter spp. was the dominant cause of HAIs and more frequently isolated than in non-COVID-19 patients. | High 100% |
| Takwa E. Meawed | Egypt | 2021 | Cross-sectional study | 6 | Inpatients (ICU) | 197 patients | Range: from 40 to 83 years | The most frequently isolated bacteria were (PDR) K. pneumoniae, followed by (MDR) A. baumannii. | PDR were K. pneumoniae, followed by MDR A. baumannii. | High 83% |
| Basit Zeshan | Pakistan | 2021 | Retrospective follow-up study | 3 | Inpatients (ICU) | 856 patients | Classified age group. Over 61 was the largest group. | E. coli was mostly resistant to ciprofloxacin and ampicillin. K. pneumoniae was mostly resistant to ampicillin and amoxycillin. | E. coli and K. pneumonia. | Medium 66% |
| Paola Caruso | Italy | 2021 | Retrospective study | 22 | Inpatients and outpatients | 255 patients | Median (IQR), 65.0 (58.0, 74.0) | Compared with the 2019 group, the 2020 group had a significantly higher prevalence of AR. The prevalence of S. aureus resistance to oxacillin and the C. striatum resistance to both vancomycin and linezolid was significantly higher in 2020. Regarding the resistance among Gram-negative bacteria, the 2020 group showed a significantly higher rate of resistance to carbapenems, colistin, and third- and fourth-generation cephalosporins. | The most frequent Gram-positive pathogen isolated in both 2019 and 2020 was S. aureus, whereas, among Gram-negative bacteria, P. aeruginosa was detected more frequently in both cohorts. | High 87% |
| Vikas Saini | India | 2021 | Retrospective review study | Inpatients (ICU) | 7309 samples pre-pandemic and (4968) samples during the pandemic phase in 2020 | Classified age group; however, above 18 Y was significant. | Compared with the pre-COVID-19 era, during COVID-19, bacterial isolates indicated up to 40% of AMR. | Common bacteria during the COVID-19 era included A. baumannii and E. coli. | High 83% | |
| Eustachius Hagni Wardoyo | Indonesia | 2021 | Retrospective study | 13 | Inpatients and outpatients | 148 isolates in group A and 62 isolates in group B | NA | An increase in susceptibility was observed in 10/16 antibiotics, where ofloxacin, aztreonam, and fosfomycin were significant. A significant decrease in susceptibility to piperacillin, amoxicillin, cefadroxil, and ampicillin was seen. | The study focuses on E. coli. | Medium 66% |
| Mustafa Karataş | Turkey | 2021 | Retrospective comparative study | 3 | Inpatients and outpatients | 3532 patients | Median 52 (IQR) (0–99) | The rate of ESBL producing Enterobacterales MDR bacteria pre-COVID-19 was similar to the rate during COVID. | The most common strains pre-COVID-19 and during COVID-19 were the same, as follows: E. coli, K. pneumoniae, and P. aeruginosa. | High 83% |
| Chiara Temperoni | Italy | 2021 | Retrospective observational study | 3 | Inpatients (ICU) | 89 patients | Median 67.1 years | Among Gram-negative and Gram-positive bacteria isolates, MDR was 55.2% and 37.2%, respectively. | The most common Gram-negative bacteria were E. coli, A. baumannii and K. pneumoniae. The most common Gram-positive bacteria were S. aureus and E. faecalis. | Medium 66% |
| Abdulrahman S. Bazaid | Saudi Arabia | 2022 | Retrospective study | 8 | Inpatients and outpatients | 108 patients | Classified age group Half of the study cohort was aged 56 years or over | Overall, the AR rate was higher among ICU patients compared with non-ICU patients. In total, 56% of ICU patients infected by A. baumannii and K. pneumoniae presented with full resistance to all examined antibiotics except colistin. In non-ICU patients, E. coli was highly resistant to piperacillin/tazobactam and trimetho-prim/sulfamethoxazole. | The most prevalent bacteria among ICU patients were A. baumannii and K. pneumoniae. In non-ICU patients, E. coli and P. aeruginosa were predominant organisms. | Medium 66% |
| Samaneh Pourajam | Iran | 2022 | Retrospective study | 6 | Inpatients (ICU) | 553 patients | Median (IQR) 69.4 (21–95) years | Most patients had XDR. The prevalence of carbapenem-resistant Gram-negative bacilli in COVID-19 patients was high. | K. pneumonia and A. baumannii | High 83% |
| Alireza Nikzad Jamnani | Iran | 2022 | Retrospective cohort study | 7 | Inpatients (ICU) | 38 patients | Classified age group. >of 70 years represented the majority. | Acinetobacter spp. had 100% resistance to amikacin, gentamycin, imipenem, and cefxime. Additionally, Klebsiella spp. had 100% resistance to amikacin, cotrimaazol, cefxime, ceftazidime, gentamycin, and ciprofloxacin. The resistance of E. coli to cefxime and cotrimaazol in the corona group was 100%. Among the non-corona group, Acinetobacter spp. and Klebsiella spp. were resistant to almost all tested antibiotics except colistin. | Acinetobacter spp. were the most common bacteria. | Medium 66% |
| Marina Gysin | Switzerland | 2021 | Prospective observational study | 2 | Inpatients (ICU) | 168 isolates | NA | High resistance was found in P. aeruginosa for piperacillin/tazobactam, cefepime, ceftazidime, and meropenem. Low levels of resistance were found in Enterobacterales for piperacillin/tazobactam, ceftriaxone, and ceftazidime. | P. aeruginosa, Enterobacter cloacae, and Klebsiella. | Medium 66% |
| Michalis Polemis | Greece | 2021 | Retrospective observational study | 36 | Inpatients and outpatients | 17,837 isolates | NA | Significant differences were found in the slope of non-susceptibility trends of 1- A. baumannii to amikacin, tigecycline, and colistin; 2- K. pneumoniae to meropenem and tigecycline; 3- P. aeruginosa to imipenem, meropenem, and levofloxacin. Additionally, significant differences were found in the slope of non-susceptibility trends of S. aureus to oxacillin and of E. faecium to glycopeptides. | The most common bacteria were A. baumannii, K. pneumoniae, P. aeruginosa, and E. coli. | Medium 66% |
| Yasemin Genç Bahçe | Turkey | 2022 | Retrospective observational study | 22 | Inpatients (ICU) | 119 isolates before COVID-19; 87 isolates afterwards. | Mean (SD) 71.36 (± 14.93) | AR rates in A. baumannii strains increased following the pandemic, except for tigecycline. High AR was observed after the pandemic for K. pneumoniae; however, these increases were not statistically significant. Except for imipenem, antibiotic resistance rates in P. aeruginosa strains increased following the pandemic. | A. baumannii, K. pneumoniae, P. aeruginosa, and E. faecium were the most common in the pandemic time. | High 83% |
| Khaled Abdulrahman Aldhwaihi | Saudi Arabia | 2021 | Retrospective study | 7 | Inpatients and outpatients | 286 isolates | NA | AR rates were congruent before and during COVID-19 pandemic. | A. baumannii, K. pneumonia, E. Coli. | Medium 50% |
| Sushma Yadav Boorgula | India | 2022 | Retrospective study | 2 | Inpatients | 122 patients | Median (IQR) 58 (51.67) | Bacterial resistance to Carbapenem had an 6% increase among tested isolates. | K. pneumoniae followed by A. baumannii. | High 100% |
| Surbhi Khurana | India | 2021 | Prospective study | 3 | Inpatients (ICU) | 151 patients | Mean (SD) 46.01 ± 19.03 | The hitherto observed resistances were as follows: amoxicillin/clavulanic acid = 84%, levofloxacin = 83%, ciprofloxacin = 79%, piperacillin/tazobactam = 77%, and trimethoprim/sulfamethoxazole = 75%. Generally, resistance to third-generation cephalosporins and carbapenems was (64%– 69%). Notably, all isolates were found to be sensitive to colistin. | K. pneumonia, A. baumannii, E. coli, and P. aeruginosa. | High 100 |
| Antibiotic | A. baumannii Median (IQR) | E. coli Median (IQR) | K. pneumonia Median (IQR) | P. aeruginosa Median (IQR) |
|---|---|---|---|---|
| Amoxicillin clavulanate | - | 85.5 (49–92.75) | 81.8 (79.3–83.75) | - |
| Amikacin | 84.6 (56.3–92.95) | 6 (0–43,35) | 69.85 (58.7–80.12) | 25 (12–28) |
| Ampicillin | - | 87.5 (85.25–93.75) | 100 (90.5–100) | - |
| Aztreonam | - | - | 84.7 (67.27–88.87) | - |
| Cefazolin | - | - | 93 (78–95.5) | - |
| Cefuroxime | - | 65.5 (55.75–77.62) | 88.9 (79.6–91.42) | - |
| Cefepime | 94.4 (93–100) | 0 (0–25) | 81.15 (71.7–87.25) | 14.3 (12.5–47.8) |
| Ceftazidime | 91.2 (50–100) | 18.75 (0–41.87) | 93.5 (83.7–97.9) | 40 (23–41.7) |
| Cefoperazone sulbactam | - | - | 76.2 (73.8–77.9) | - |
| Ceftriaxone | 76.2 (54.75–95.55) | 73 (49.25–93.75) | 84 (77.55–93.4) | 75 (43.75–87.5) |
| Colistin | 2.5 (0–19.62) | 0 (0–7.15) | 21.1 (12.42–69.82) | 4 (0–12.25) |
| Gentamicin | 95.7 (74.2–97.1) | 40 (19–47) | 57.1 (33.45–86.6) | 25 (19.75–58.75) |
| Levofloxacin | 97.05 (91.92–100) | 75 (56.85–87.5) | 80.8 (78.55–90.85) | 43.5 (28.6–80) |
| Meropenem | 92.1 (64.02–95.65) | 2.5 (0–26.17) | 71.25 (55.37–77.37) | 38 (18.37–42.17) |
| Imipenem | 92.1 (80.65–95.72) | 10 (4–26) | 65.7 (19.25–72.87) | 42.9 (19.75–52.9) |
| Ertapenem | - | - | 71.4 (55.55–75.05) | - |
| Ciprofloxacin | 91.2 (65–100) | 71 (28.2–76) | 87.8 (55.1–92.95) | 50 (32.3–62.5) |
| Trimethoprim/sulfamethoxazole | 50 (46.8–84.2) | 50 (40–80) | 73.5 (32–74) | - |
| Tigecycline | 9.5 (8.8–33.3) | 0 (0–22.5) | 31.4 (1.7–44) | - |
| Piperacillin/tazobactam | 93.7 (66.9–100) | 23 (12–37.8) | 77.7 (57.1–79.27) | 11.25 (9.25–13.85) |
| Nitrofurantion | - | - | 51.8 (38.5–60.6) | - |
| S. aureus Median (IQR) | E. faecium Median (IQR) | |
|---|---|---|
| Oxacillin | 48.5 (25.5–63.75) | - |
| Ampicillin | - | 81.8 (52.4–90.9) |
| Erythromycin | - | 90.9 (78.45–95.45) |
| Clindamycin | 33.3 (16.65–50.9) | - |
| Ciprofloxacin | - | 81.8 (77–100) |
| Vancomycin | - | 11 (0–18.1) |
| Tetracycline | - | 66 (60.25–83) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sulayyim, H.J.A.; Ismail, R.; Hamid, A.A.; Ghafar, N.A. Antibiotic Resistance during COVID-19: A Systematic Review. Int. J. Environ. Res. Public Health 2022, 19, 11931. https://doi.org/10.3390/ijerph191911931
Sulayyim HJA, Ismail R, Hamid AA, Ghafar NA. Antibiotic Resistance during COVID-19: A Systematic Review. International Journal of Environmental Research and Public Health. 2022; 19(19):11931. https://doi.org/10.3390/ijerph191911931
Chicago/Turabian StyleSulayyim, Hadi Jaber Al, Rohani Ismail, Abdullah Al Hamid, and Noraini Abdul Ghafar. 2022. "Antibiotic Resistance during COVID-19: A Systematic Review" International Journal of Environmental Research and Public Health 19, no. 19: 11931. https://doi.org/10.3390/ijerph191911931
APA StyleSulayyim, H. J. A., Ismail, R., Hamid, A. A., & Ghafar, N. A. (2022). Antibiotic Resistance during COVID-19: A Systematic Review. International Journal of Environmental Research and Public Health, 19(19), 11931. https://doi.org/10.3390/ijerph191911931







