CTX-M-Producing Bacteria Isolated from a Highly Polluted River System in Portugal
Abstract
1. Introduction
2. Materials and Methods
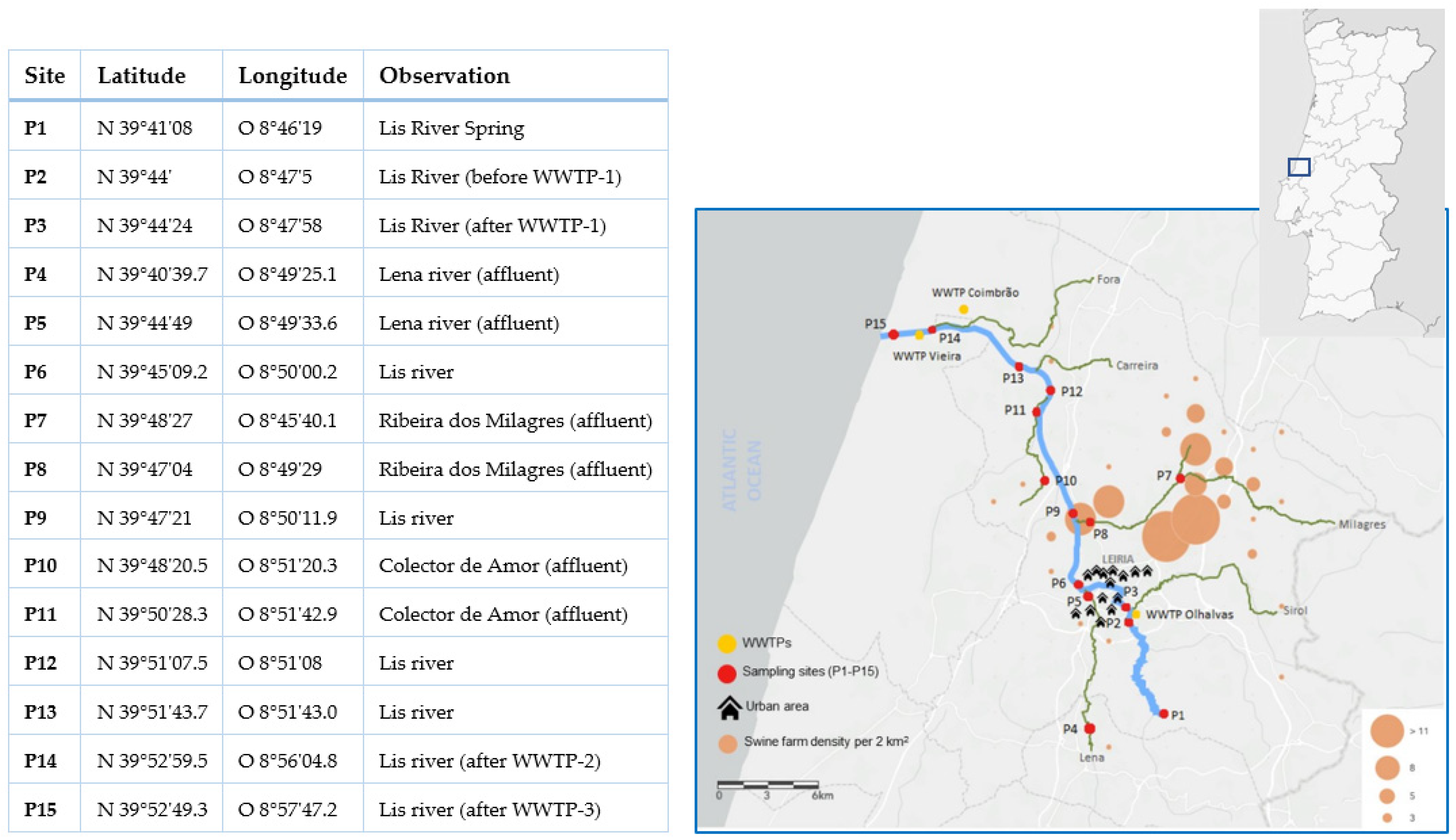
2.1. Study Area
2.2. Sampling, Water Quality Evaluation, and Cefotaxime-Resistant Bacteria Isolation
2.3. Phylogenetic Affiliation
2.4. blaCTX-M Screening, Genetic Environment, and Molecular Typing
2.5. Antibiotic Susceptibility Testing of blaCTX-M Positives
2.6. Conjugation Assays
3. Results
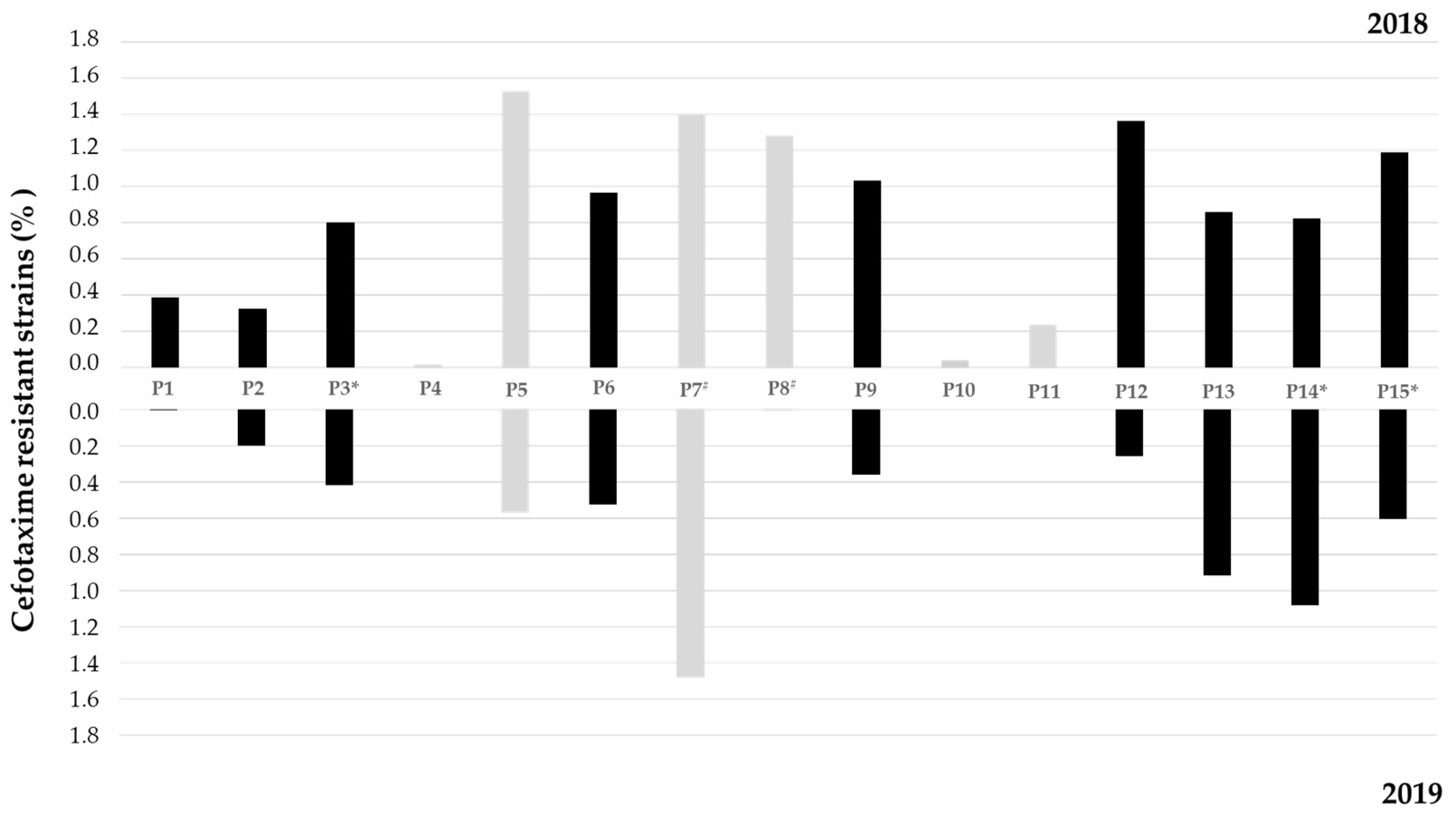
3.1. Water Quality Analysis and Cefotaxime-Resistant Bacteria Incidence
3.2. Phylogenetic Affiliation of Cefotaxime-Resistant Isolates
3.3. Prevalence and Diversity of the blaCTX-M Gene among the Cefotaxime-Resistant Isolates
3.4. Antibiotic Susceptibility Testing of blaCTX-M Gene Carriers
3.5. Conjugation Assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saravanan, A.; Kumar, P.S.; Jeevanantham, S.; Karishma, S.; Tajsabreen, B.; Yaashikaa, P.R.; Reshma, B. Effective water/wastewater treatment methodologies for toxic pollutants removal: Processes and applications towards sustainable development. Chemosphere 2021, 280, 130595. [Google Scholar] [CrossRef] [PubMed]
- United Nations. The United Nations World Water Development Report 2021: Valuing Water; UNESCO: Paris, France, 2021. [Google Scholar]
- Schwarzenbach, R.P.; Escher, B.I.; Fenner, K.; Hofstetter, T.B.; Johnson, C.A.; Von Gunten, U.; Wehrli, B. The challenge of micropollutants in aquatic systems. Science 2006, 313, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.R.; Caraco, N.F.; Correll, D.L.; Howarth, R.W.; Sharpley, A.N.; Smith, V.H. Nonpoint pollution of surface waters with phosphorus and nitrogen. Ecol. Appl. 1998, 8, 559–568. [Google Scholar] [CrossRef]
- Loos, R.; Gawlik, B.M.; Locoro, G.; Rimaviciute, E.; Contini, S.; Bidoglio, G. EU-wide survey of polar organic persistent pollutants in European river waters. Environ. Pollut. 2009, 157, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Tejerina-Garro, F.L.; Maldonado, M.; Ibañez, C.; Pont, D.; Roset, N.; Oberdorff, T. Effects of natural and anthropogenic environmental changes on riverine fish assemblages: A framework for ecological assessment of rivers. Braz. Arch. Biol. Technol. 2005, 48, 91–108. [Google Scholar] [CrossRef]
- Gad, M.; Elsayed, S.; Moghanm, F.S.; Almarshadi, M.H.; Alshammari, A.S.; Khedher, K.M.; Eid, E.M.; Hussein, H. Combining Water Quality Indices and Multivariate Modeling to Assess Surface Water Quality in the Northern Nile Delta, Egypt. Water 2020, 12, 2142. [Google Scholar] [CrossRef]
- Gad, M.; Saleh, A.H.; Hussein, H.; Farouk, M.; Elsayed, S. Appraisal of Surface Water Quality of Nile River Using Water Quality Indices, Spectral Signature and Multivariate Modeling. Water 2022, 14, 1131. [Google Scholar] [CrossRef]
- Yan, T.; Shen, S.-L.; Zhou, A. Indices and models of surface water quality assessment: Review and perspectives. Environ. Pollut. 2022, 308, 119611. [Google Scholar] [CrossRef]
- Marti, E.; Variatza, E.; Balcazar, J.L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef]
- Taylor, N.G.H.; Verner-Jeffreys, D.W.; Baker-Austin, C. Aquatic systems: Maintaining, mixing and mobilising antimicrobial resistance? Trends Ecol. Evol. 2011, 26, 278–284. [Google Scholar] [CrossRef]
- Wellington, E.M.H.; Boxall, A.B.A.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bactéria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Silva, I.; Tacão, M.; Tavares, R.D.S.; Miranda, R.; Araújo, S.; Manaia, C.M.; Henriques, I. Fate of cefotaxime-resistant Enterobacteriaceae and ESBL-producers over a full-scale wastewater treatment process with UV disinfection. Sci. Total Environ. 2018, 639, 1028–1037. [Google Scholar] [CrossRef] [PubMed]
- Tavares, R.D.S.; Tacão, M.; Figueiredo, A.S.; Duarte, A.S.; Esposito, F.; Lincopan, N.; Manaia, C.M.; Henriques, I. Genotypic and phenotypic traits of blaCTX-M-carrying Escherichia coli strains from an UV-C-treated wastewater effluent. Water Res. 2020, 184, 116079. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Coyne, S.; Berendonk, T.U. Origin and evolution of antibiotic resistance: The common mechanisms of emergence and spread in water bodies. Front. Microbiol. 2012, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Adler, A.; Katz, D.E.; Marchaim, D. The continuing plague of Extended-Spectrum β-lactamase-producing Enterobacteriaceae infections. Infect. Dis. Clin. N. Am. 2020, 30, 347–375. [Google Scholar] [CrossRef]
- Chong, Y.; Shimoda, S.; Shimono, N. Current epidemiology, genetic evolution and clinical impact of Extended-Spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumonia. Infect. Genet. Evol. 2018, 61, 185–188. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC); European Food Safety Authority (EFSA); European Medicines Agency (EMA). ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals: Joint Interagency Antimicrobial Consumption and Resistance Analysis (JIACRA) Report. EFSA J. 2017, 15, e04872. [Google Scholar]
- Bush, K. Past and present perspectives on β-lactamases. Antimicrob. Agents Chemother. 2018, 62, e01076-18. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef] [PubMed]
- Knothe, H.; Shah, P.; Krcmery, V.; Antal, M.; Mitsuhashi, S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 1983, 11, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Ojer-Usoz, E.; González, D.; García-Jalón, I.; Vitas, A.I. High dissemination of Extended-Spectrum β-lactamase-producing Enterobacteriaceae in effluents from wastewater treatment plants. Water Res. 2014, 56, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M enzymes: Origin and diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef]
- Hussain, H.I.; Aqib, A.I.; Seleem, M.N.; Shabbir, M.A.; Hao, H.; Iqbal, Z.; Kulyar, M.F.; Zaheer, T.; Li, K. Genetic basis of molecular mechanisms in β-lactam resistant gram-negative bacteria. Microb. Pathog. 2021, 158, 105040. [Google Scholar] [CrossRef]
- Tacão, M.; Moura, A.; Correia, A.; Henriques, I. Co-resistance to different classes of antibiotics among ESBL-producers from aquatic systems. Water Res. 2014, 48, 100–107. [Google Scholar] [CrossRef]
- Toleman, M.A.; Bennett, P.M.; Walsh, T.R. ISCR elements: Novel gene-capturing systems of the 21st Century? Microbiol. Mol. Biol. Rev. 2006, 70, 296–316. [Google Scholar] [CrossRef]
- Decousser, J.W.; Poirel, L.; Nordmann, P. Characterization of a chromosomally encoded Extended-Spectrum class A beta-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 2001, 45, 3595–3598. [Google Scholar] [CrossRef]
- Humeniuk, C.; Arlet, G.; Gautier, V.; Grimont, P.; Labia, R.; Philippon, A. Beta-lactamases of Kluyvera ascorbata; probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 2002, 46, 3045–3049. [Google Scholar] [CrossRef]
- Poirel, L.; Kampfer, P.; Nordmann, P. Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana; a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 2002, 46, 4038–4040. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.M.; Power, P.; Radice, M.; Vay, C.; Famiglietti, A.; Galleni, M.; Ayala, J.A.; Gutkind, G. Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: A possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob. Agents Chemother. 2004, 48, 4895–4897. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vieira, J.; Fonseca, A.; Vilar, V.J.P.; Boaventura, R.A.R.; Botelho, C.M.S. Water quality in Lis river; Portugal. Environ. Monit. Assess. 2012, 184, 7125–7140. [Google Scholar] [CrossRef] [PubMed]
- Vieira, J.; Fonseca, A.; Vilar, V.J.P.; Boaventura, R.A.R.; Botelho, C.M.S. Water quality modelling of Lis River; Portugal. Environ. Sci. Pollut. Res. 2013, 20, 508–524. [Google Scholar] [CrossRef] [PubMed]
- Vieira, J.S.; Botelho, C.M.S.; Boaventura, R.A.R. Trace metal fractionation by the sequential extraction method in sediments from the Lis River (Portugal). Soil Sediment Contam. Int. J. 2009, 18, 102–119. [Google Scholar] [CrossRef]
- Paíga, P.; Santos, L.H.M.L.M.; Ramos, S.; Jorge, S.; Silva, J.G.; Delerue-Matos, C. Presence of pharmaceuticals in the Lis river (Portugal): Sources; fate and seasonal variation. Sci. Total Environ. 2016, 573, 164–177. [Google Scholar] [CrossRef]
- Teixeira, P.; Tacão, M.; Pureza, L.; Gonçalves, J.; Silva, A.; Cruz-Schneider, M.P.; Henriques, I. Occurrence of carbapenemase-producing Enterobacteriaceae in a Portuguese river: blaNDM; blaKPC and blaGES among the detected genes. Environ. Pollut. 2020, 260, 113913. [Google Scholar] [CrossRef]
- Teixeira, P.; Tacão, M.; Henriques, I. Occurrence and distribution of Carbapenem-resistant Enterobacterales and carbapenemase genes along a highly polluted hydrographic basin. Environ. Pollut. 2022, 300, 118958. [Google Scholar] [CrossRef]
- Tacão, M.; Correia, A.; Henriques, I. Resistance to broad-spectrum antibiotics in aquatic systems: Anthropogenic activities modulate the dissemination of blaCTX-M-like genes. Appl. Environ. Microbiol. 2012, 78, 4134–4140. [Google Scholar] [CrossRef]
- Araújo, S.; Silva, I.; Tacão, M.; Patinha, C.; Alves, A.; Henriques, I. Characterization of antibiotic resistant and pathogenic Escherichia coli in irrigation water and vegetables in household farms. Int. J. Food Microbiol. 2017, 257, 192–200. [Google Scholar] [CrossRef]
- Eckert, C.; Gautier, V.; Arlet, G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 2006, 57, 14–23. [Google Scholar] [CrossRef]
- Lartigue, M.F.; Poirel, L.; Nordmann, P. Diversity of genetic environment of blaCTX-M genes. FEMS Microbiol. Lett. 2004, 234, 201–207. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant; extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Fonseca, A.; Botelho, C.; Boaventura, R.A.R.; Vilar, V.J.P. Integrated hydrological and water quality model for river management: A case study on Lena River. Sci. Total Environ. 2014, 485–486, 474–489. [Google Scholar] [CrossRef]
- Girard, M.; Nikiema, J.; Brzezinski, R.; Buelna, G.; Heitz, M. A review of the environmental pollution originating from the piggery industry and of the available mitigation technologies: Towards the simultaneous biofiltration of swine slurry and methane. Can. J. Civ. Eng. 2009, 36, 1946–1957. [Google Scholar] [CrossRef]
- Han, B.; Yang, F.; Tian, X.; Mu, M.; Zhang, K. Tracking antibiotic resistance gene transfer at all seasons from swine waste to receiving environments. Ecotoxicol. Environ. Saf. 2021, 219, 112335. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Z.; Xing, S.; Liao, X. The correlation between antibiotic resistance gene abundance and microbial community resistance in pig farm wastewater and surrounding rivers. Ecotoxicol. Environ. Saf. 2019, 182, 109452. [Google Scholar] [CrossRef]
- Gonçalves, J.M.; Ferreira, S.; Nunes, M.; Eugénio, R.; Amador, P.; Filipe, O.; Duarte, I.M.; Teixeira, M.; Vasconcelos, T.; Oliveira, F.; et al. Developing irrigation management at district scale based on water monitoring: Study on Lis valley, Portugal. AgriEngineering 2020, 2, 78–95. [Google Scholar] [CrossRef]
- Leonard, A.F.C.; Zhang, L.; Balfour, A.J.; Garside, R.; Hawkey, P.M.; Murray, A.K.; Ukoumunne, O.C.; Gaze, W.H. Exposure to and colonisation by antibiotic-resistant E coli in UK coastal water users: Environmental surveillance; exposure assessment; and epidemiological study (Beach Bum Survey). Environ. Int. 2018, 114, 326–333. [Google Scholar] [CrossRef]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research; Discovery; and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Carvalho, I.; Alonso, C.A.; Silva, V.; Pimenta, P.; Cunha, R.; Martins, C.; Igrejas, G.; Torres, C.; Poeta, P. Extended-Spectrum Beta-Lactamase-producing Klebsiella pneumoniae isolated from healthy and sick dogs in Portugal (2020). Microb. Drug Resist. 2020, 26, 709–715. [Google Scholar] [CrossRef]
- Carvalho, I.; Safia Chenouf, N.; Cunha, R.; Martins, C.; Pimenta, P.; Pereira, A.R.; Martínez-Álvarez, S.; Ramos, S.; Silva, V.; Igrejas, G.; et al. Antimicrobial resistance genes and diversity of clones among ESBL- and acquired AmpC-producing Escherichia coli isolated from fecal samples of healthy and sick cats in Portugal. Antibiotics 2021, 10, 262. [Google Scholar] [CrossRef]
- Stedt, J.; Bonnedahl, J.; Hernandez, J.; Waldenström, J.; McMahon, B.J.; Tolf, C.; Olsen, B.; Drobni, M. Carriage of CTX-M type extended spectrum β-lactamases (ESBLs) in gulls across Europe. Acta Vet. Scand. 2015, 57, 74. [Google Scholar] [CrossRef]
- Leão, C.; Clemente, L.; Moura, L.; Seyfarth, A.M.; Hansen, I.M.; Hendriksen, R.S.; Amaro, A. Emergence and clonal spread of CTX-M-65-producing Escherichia coli from retail meat in Portugal. Front. Microbiol. 2021, 12, 653595. [Google Scholar] [CrossRef]
- Calhau, V.; Mendes, C.; Pena, A.; Mendonça, N.; Silva, G.J. Virulence and plasmidic resistance determinants of Escherichia coli isolated from municipal and hospital wastewater treatment plants. J. Water Health 2015, 13, 311–318. [Google Scholar] [CrossRef]
- Mesquita, E.; Ribeiro, R.; Silva, C.J.C.; Alves, R.; Baptista, R.; Condinho, S.; Rosa, M.J.; Perdigão, J.; Caneiras, C.; Duarte, A. An update on wastewater multi-resistant bacteria: Identification of clinical pathogens such as Escherichia coli O25b:H4-B2-ST131-producing CTX-M-15 ESBL and KPC-3 carbapenemase-producing Klebsiella oxytoca. Microorganisms 2021, 9, 576. [Google Scholar] [CrossRef]
- Furlan, J.P.; Lopes, R.; Gonzalez, I.H.L.; Ramos, P.L.; Stehling, E.G. Comparative analysis of multidrug resistance plasmids and genetic background of CTX-M-producing Escherichia coli recovered from captive wild animals. Appl. Microbiol. Biotechnol. 2020, 104, 6707–6717. [Google Scholar] [CrossRef]
- Palmeira, J.D.; Cunha, M.V.; Carvalho, J.; Ferreira, H.; Fonseca, C.; Torres, R.T. Emergence and spread of cephalosporinases in Wildlife: A review. Animals 2021, 11, 1765. [Google Scholar] [CrossRef]
- Sun, Y.; Zeng, Z.; Chen, S.; Ma, J.; He, L.; Liu, Y.; Deng, Y.; Lei, T.; Zhao, J.; Liu, J.-H. High prevalence of blaCTX-M extended-spectrum B-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 2010, 16, 1475–1481. [Google Scholar] [CrossRef]
- Girlich, D.; Bonnin, R.A.; Naas, T. Occurrence and diversity of CTX-M-producing Escherichia coli from the Seine River. Front. Microbiol. 2020, 11, 603578. [Google Scholar] [CrossRef]
- Hassen, B.; Abbassi, M.S.; Benlabidi, S.; Ruiz-Ripa, L.; Mama, O.M.; Ibrahim, C.; Hassen, A.; Hammami, S.; Torres, C. Genetic characterization of ESBL-producing Escherichia coli and Klebsiella pneumoniae isolated from wastewater and river water in Tunisia: Predominance of CTX-M-15 and high genetic diversity. Environ. Sci. Pollut. Res. 2020, 27, 44368–44377. [Google Scholar] [CrossRef]
- Kittinger, C.; Lipp, M.; Folli, B.; Kirschner, A.; Baumert, R.; Galler, H.; Grisold, A.J.; Luxner, J.; Weissenbacher, M.; Farnleitner, A.; et al. Enterobacteriaceae isolated from the River Danube: Antibiotic resistances; with a focus on the presence of ESBL and carbapenemases. PLoS ONE 2016, 11, 1–17. [Google Scholar] [CrossRef]
- Lopes, R.; Fuentes-Castillo, D.; Fontana, H.; Rodrigues, L.; Dantas, K.; Cerdeira, L.; Henriques, I.; Lincopan, N. Endophytic lifestyle of global clones of extended-spectrum b-lactamase producing priority pathogens in fresh vegetables: A Trojan horse strategy favoring human colonization? mSystems 2021, 6, e01125-20. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, BY, USA, 1991; pp. 115–175. [Google Scholar]
- Henriques, I.S.; Fonseca, F.; Alves, A.; Saavedra, M.J.; Correia, A. Occurrence and diversity of integrons and β-lactamase genes among ampicillin-resistant isolates from estuarine waters. Res. Microbiol. 2006, 157, 938–947. [Google Scholar] [CrossRef]
- Versalovic, J.; Koeuth, T.; Lupski, R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef]
- Saladin, M.; Cao, V.T.; Lambert, T.; Donay, J.L.; Herrmann, J.L.; Ould-Hocine, Z.; Verdet, C.; Delisle, F.; Philippon, A.; Arlet, G. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 2002, 209, 161–168. [Google Scholar]


| Affiliation | Campaign Year | Site | Strain a | Antibiotic Susceptibility b | blaCTX-M variantc | Genetic Environment of blaCTX-M | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AML | AMC | PRL | TZP | TIC | TIM | ATM | CAZ | CTX | FEP | IPM | CN | CIP | TE | C | SXT | ISEcp1 | IS26 | orf477 | IS903 | In Figure 3 | |||||
| Escherichia | 2018 | 1 | E1 | nd | + | + | nd | ||||||||||||||||||
| 1 | E2 * | −1 | + | + | A | ||||||||||||||||||||
| 5 | E3 * | −32 | + | + | H | ||||||||||||||||||||
| 5 | E4 | −65 | + | + | J | ||||||||||||||||||||
| 7 | E5 * | −1 | + | + | A | ||||||||||||||||||||
| 8 | E6 | −1 | + | + | A | ||||||||||||||||||||
| 8 | E7 * | −15 | + | + | D | ||||||||||||||||||||
| 9 | E8 * | −65 | + | + | J | ||||||||||||||||||||
| 9 | E9 | nd | + | + | nd | ||||||||||||||||||||
| 11 | E10 * | −14 | + | + | C | ||||||||||||||||||||
| 13 | E11 | nd | + | + | nd | ||||||||||||||||||||
| 13 | E12 | −15 | + | + | E | ||||||||||||||||||||
| 15 | E13 * | −15 | + | + | D | ||||||||||||||||||||
| 1 | E14 | nd | + | nd | |||||||||||||||||||||
| 1 | E15 | −55 | + | + | I | ||||||||||||||||||||
| 2 | E16 | −65 | + | + | J | ||||||||||||||||||||
| 3 | E17 | −15 | + | + | D | ||||||||||||||||||||
| 3 | E18 | −15 | + | + | D | ||||||||||||||||||||
| 5 | E19 * | −32 | + | + | H | ||||||||||||||||||||
| 6 | E20 | −15 | + | + | + | F | |||||||||||||||||||
| 2019 | 2 | E21 | −15 | + | + | D | |||||||||||||||||||
| 5 | E22 * | −27 | + | + | G | ||||||||||||||||||||
| 6 | E23 * | −27 | + | + | G | ||||||||||||||||||||
| 6 | E24 * | −55 | + | + | I | ||||||||||||||||||||
| 7 | E25 | nd | + | nd | |||||||||||||||||||||
| 7 | E26 | −32 | + | + | H | ||||||||||||||||||||
| 7 | E27 | −15 | + | + | D | ||||||||||||||||||||
| 8 | E28 | −15 | + | + | D | ||||||||||||||||||||
| 9 | E29 | −27 | + | + | G | ||||||||||||||||||||
| 15 | E30 | −15 | + | + | D | ||||||||||||||||||||
| 15 | E31 * | −65 | + | + | J | ||||||||||||||||||||
| 15 | E32 | −1 | + | + | A | ||||||||||||||||||||
| Klebsiella | 2018 | 5 | K1 * | −15 | + | + | D | ||||||||||||||||||
| 8 | K2 | nd | + | + | nd | ||||||||||||||||||||
| 8 | K3 | −15 | + | + | D | ||||||||||||||||||||
| 9 | K4 | −15 | + | + | D | ||||||||||||||||||||
| 14 | K5 | −15 | + | + | D | ||||||||||||||||||||
| 14 | K6 * | −15 | + | + | D | ||||||||||||||||||||
| 2019 | 3 | K7 | −15 | + | + | D | |||||||||||||||||||
| 3 | K8 * | −15 | + | + | D | ||||||||||||||||||||
| 3 | K9 | nd | + | + | nd | ||||||||||||||||||||
| 3 | K10 | −15 | + | + | D | ||||||||||||||||||||
| 5 | K11 | −15 | + | + | D | ||||||||||||||||||||
| 6 | K12 | nd | + | + | nd | ||||||||||||||||||||
| 9 | K13 | nd | + | + | nd | ||||||||||||||||||||
| 9 | K14 | nd | + | + | nd | ||||||||||||||||||||
| 12 | K15 | −15 | + | + | + | F | |||||||||||||||||||
| 13 | K16 | nd | + | + | nd | ||||||||||||||||||||
| 14 | K17 | nd | + | + | nd | ||||||||||||||||||||
| 14 | K18 | −15 | + | + | D | ||||||||||||||||||||
| Citrobacter | 2018 | 12 | C1 | nd | + | nd | |||||||||||||||||||
| 2019 | 12 | C2 * | −3 | + | + | B | |||||||||||||||||||
| 12 | C3 * | nd | + | + | nd | ||||||||||||||||||||
| Enterobacter | 13 | C4 * | −32 | + | + | H | |||||||||||||||||||
| Antibiotic Susceptibility | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor/Transconjugant Strains and blaCTX-M Gene | AML | AMC | PRL | TZP | TIC | TIM | ATM | CAZ | CTX | FEP | IPM | CN | CIP | TE | C | SXT | |
| Escherichia E3:: blaCTX-M-32 | E3D | ||||||||||||||||
| E3T | |||||||||||||||||
| Escherichia E5:: blaCTX-M-1 | E5 | ||||||||||||||||
| E5T | |||||||||||||||||
| Escherichia E19:: blaCTX-M-32 | E19 | ||||||||||||||||
| E19T | |||||||||||||||||
| Escherichia E23:: blaCTX-M-27 | E23 | ||||||||||||||||
| E23T | |||||||||||||||||
| Escherichia E24:: blaCTX-M-55 | E24 | ||||||||||||||||
| E24T | |||||||||||||||||
| Klebsiella K1:: blaCTX-M-15 | K1 | ||||||||||||||||
| K1T | |||||||||||||||||
| Klebsiella K6:: blaCTX-M-15 | K6 | ||||||||||||||||
| K6T | |||||||||||||||||
| Citrobacter C3:: blaCTX-M-nd | C3 | ||||||||||||||||
| C3T | |||||||||||||||||
| Citrobacter C4:: blaCTX-M-32 | C4 | ||||||||||||||||
| C4T | |||||||||||||||||
| Recipient strain E. coli CV601 | - | ||||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tacão, M.; Laço, J.; Teixeira, P.; Henriques, I. CTX-M-Producing Bacteria Isolated from a Highly Polluted River System in Portugal. Int. J. Environ. Res. Public Health 2022, 19, 11858. https://doi.org/10.3390/ijerph191911858
Tacão M, Laço J, Teixeira P, Henriques I. CTX-M-Producing Bacteria Isolated from a Highly Polluted River System in Portugal. International Journal of Environmental Research and Public Health. 2022; 19(19):11858. https://doi.org/10.3390/ijerph191911858
Chicago/Turabian StyleTacão, Marta, José Laço, Pedro Teixeira, and Isabel Henriques. 2022. "CTX-M-Producing Bacteria Isolated from a Highly Polluted River System in Portugal" International Journal of Environmental Research and Public Health 19, no. 19: 11858. https://doi.org/10.3390/ijerph191911858
APA StyleTacão, M., Laço, J., Teixeira, P., & Henriques, I. (2022). CTX-M-Producing Bacteria Isolated from a Highly Polluted River System in Portugal. International Journal of Environmental Research and Public Health, 19(19), 11858. https://doi.org/10.3390/ijerph191911858







