Reliability of Early Estimates of the Basic Reproduction Number of COVID-19: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Methods
2.1. Search Strategy and Selection Criteria
2.1.1. Database Search
2.1.2. Search Strategy
2.1.3. Study Selection
2.1.4. Data Extraction and Quality Assessment
2.2. Data Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 21 November 2021).
- Nishiura, H. Correcting the Actual Reproduction Number: A Simple Method to Estimate R0 from Early Epidemic Growth Data. Int. J. Environ. Res. Public Health. 2010, 7, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Mindermann, S.; Brauner, J.; Leech, G.; Stephenson, A.; Gavenčiak, T.; Kulveit, J.; The, Y.W.; Chindelevitch, L.; Gal, Y. How Robust are the Estimated Effects of Nonpharmaceutical Interventions against COVID-19? In Proceedings of the 34th Conference on Neural Information Processing Systems (NeurIPS 2020), Online, 6–12 December 2020; Available online: https://github.com/epidemics/COVIDNPIs/tree/neurips (accessed on 2 February 2022).
- Adam, D. A guide to R—The pandemic’s misunderstood metric. Nature 2020, 583, 346–348. [Google Scholar] [CrossRef]
- Gurdasani, D.; Drury, J.; Greenhalgh, T.; Griffin, S.; Haque, Z.; Hyde, Z.; Katzourakis, A.; McKee, M.; Michie, S.; Pagel, C.; et al. Mass infection is not an option: We must do more to protect our young. Lancet 2021, 398, 297–298. [Google Scholar] [CrossRef]
- UK Health Security Agency. The R Value and Growth Rate. Available online: https://www.gov.uk/guidance/the-r-value-and-growth-rate (accessed on 26 January 2022).
- Reproduction Live. Is COVID-19 Multiplying or Dying Out in Your Region? Available online: https://reproduction.live/ (accessed on 12 March 2022).
- Arroyo-Marioli, F.; Bullano, F.; Kucinskas, S.; Rondón-Moreno, C. Tracking R of COVID-19: A new real-time estimation using the Kalman filter. PLoS ONE 2021, 16, e0244474. [Google Scholar] [CrossRef]
- Yu, C.J.; Wang, Z.X.; Xu, Y.; Hu, M.X.; Chen, K.; Qin, G. Assessment of basic reproductive number for COVID-19 at global level: A meta-analysis. Medicine 2021, 100, e25837. [Google Scholar] [CrossRef]
- Billah, M.A.; Miah, M.M.; Khan, M.N. Reproductive number of coronavirus: A systematic review and meta-analysis based on global level evidence. PLoS ONE 2020, 15, e0242128. [Google Scholar] [CrossRef] [PubMed]
- Dhungel, B.; Rahman, S.; Rahman, M.; Bhandari, A.K.C.; Phuong, L.M.; Biva, N.A.; Gilmour, S. A Systematic Review and Meta-Analysis of the Transmissibility and Severity of COVID-19. PROSPERO. 2021. Available online: https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=279514 (accessed on 26 January 2022).
- NIH. Study Quality Assessment Tools. Available online: https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools (accessed on 26 January 2022).
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Inthout, J.; Ioannidis, J.P.; Borm, G.F. The Hartung-Knapp-Sidik-Jonkman method for random effects meta-analysis is straightforward and considerably outperforms the standard DerSimonian-Laird method. BMC Med. Res. Methodol. 2014, 14, 25. [Google Scholar] [CrossRef]
- Furuya-Kanamori, L.; Barendregt, J.J.; Doi, S.A.R. A new improved graphical and quantitative method for detecting bias in meta-analysis. Int. J. Evid. Based Healthc. 2018, 16, 195–203. [Google Scholar] [CrossRef]
- Shi, L.; Lin, L. The trim-and-fill method for publication bias: Practical guidelines and recommendations based on a large database of meta-analyses. Medicine 2019, 98, e15987. [Google Scholar] [CrossRef]
- Bi, Q.; Wu, Y.; Mei, S.; Ye, C.; Zou, X.; Zhang, Z.; Liu, X.; Wei, L.; Truelove, S.A.; Zhang, T.; et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: A retrospective cohort study. Lancet Infect. Dis. 2020, 20, 911–919. [Google Scholar] [CrossRef]
- Lu, Z.; Yu, Y.; Chen, Y.; Ren, Y.; Xu, C.; Wang, S.; Yin, Z. A fractional-order SEIHDR model for COVID-19 with inter-city networked coupling effects. Nonlinear Dyn. 2020, 101, 1717–1730. [Google Scholar] [CrossRef] [PubMed]
- Rocklöv, J.; Sjödin, H.; Wilder-Smith, A. COVID-19 outbreak on the Diamond Princess cruise ship: Estimating the epidemic potential and effectiveness of public health countermeasures. J. Travel Med. 2020, 27, taaa030. [Google Scholar] [CrossRef]
- Hoque, M.E. Estimation of the number of affected people due to the Covid-19 pandemic using susceptible, infected and recover model. Int. J. Mod. Phys. C 2020, 31, 2050111. [Google Scholar] [CrossRef]
- World Health Organization. Statement on the Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus 2019 (n-CoV) on 23 January 2020. Available online: https://www.who.int/news/item/23-01-2020-statement-on-the-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov) (accessed on 4 February 2022).
- Riley, S.; Fraser, C.; Donnelly, C.A.; Ghani, A.C.; Abu-Raddad, L.J.; Hedley, A.J.; Leung, G.M.; Ho, L.-M.; Lam, T.-H.; Thach, T.Q.; et al. Transmission Dynamics of the Etiological Agent of SARS in Hong Kong: Impact of Public Health Interventions. Science 2003, 300, 1961–1966. [Google Scholar] [CrossRef]
- Cauchemez, S.; Fraser, C.; van Kerkhove, M.D.; Donnelly, C.A.; Riley, S.; Rambaut, A.; Enouf, V.; van der Werf, S.; Ferguson, N.M. Middle East respiratory syndrome coronavirus: Quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect. Dis. 2014, 14, 50–56. [Google Scholar] [CrossRef]
- Kwok, K.O.; Tang, A.; Wei, V.W.I.; Park, W.H.; Yeoh, E.K.; Riley, S. Epidemic Models of Contact Tracing: Systematic Review of Transmission Studies of Severe Acute Respiratory Syndrome and Middle East Respiratory Syndrome. Comput. Struct. Biotechnol. J. 2019, 17, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Chertow, D.S.; Kindrachuk, J. Influenza, Measles, SARS, MERS, and Smallpox. In Highly Infectious Diseases in Critical Care; Hidalgo, J., Woc-Colburn, L., Eds.; Springer: Cham, Switzerland, 2020; pp. 69–96. [Google Scholar] [CrossRef]
- National Emerging Special Pathogens Training & Education Center. Playing the Numbers Game: R0. Available online: https://netec.org/2020/01/30/playing-the-numbers-game-r0/ (accessed on 19 March 2022).
- Gani, R.; Leach, S. Transmission potential of smallpox in contemporary populations. Nature 2001, 414, 748–751. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention; World Health Organization. History and Epidemiology of Global Smallpox Eradication: Day 1 Module 1—Smallpox: Disease, Prevention, and Intervention (Training Course) (Presentation). 2014. Available online: https://stacks.cdc.gov/view/cdc/27929 (accessed on 19 March 2022).
- Health Service Executive of Ireland. Chapter 23 Varicella-Zoster: Hospitalisation Notifiable, Outbreak Notifiable; Health Service Executive of Ireland: Dublin, Ireland, 2022. [Google Scholar]
- Guerra, F.M.; Bolotin, S.; Lim, G.; Heffernan, J.; Deeks, S.L.; Li, Y.; Crowcroft, N.S. The basic reproduction number (R 0) of measles: A systematic review. Lancet Infect. Dis. 2017, 17, e420–e428. [Google Scholar] [CrossRef]
- Secunder Kermani. Coronavirus: Whitty and Vallance Faced “Herd Immunity” Backlash, Emails Show. BBC News. Available online: https://www.bbc.com/news/uk-politics-54252272 (accessed on 26 January 2022).
- Dan Diamond. U.S. to Narrowly Miss Biden’s July 4 Vaccination Goal, White House says. The Washington Post. Available online: https://www.washingtonpost.com/health/2021/06/22/white-house-vaccination-goal/ (accessed on 26 January 2022).
- Delamater, P.L.; Street, E.J.; Leslie, T.F.; Yang, Y.T.; Jacobsen, K.H. Complexity of the Basic Reproduction Number (R0). Emerg. Infect. Dis. 2019, 25, 1. [Google Scholar] [CrossRef]
- Ali, S.T.; Wang, L.; Lau, E.H.Y.; Xu, X.-K.; Du, Z.; Wu, Y.; Leung, G.M.; Cowling, B.J. Serial interval of SARS-CoV-2 was shortened over time by nonpharmaceutical interventions. Science 2020, 369, 1106–1109. [Google Scholar] [CrossRef]
- Reuters. Bosnia Reports Sharp Rise in Coronavirus Cases after Relaxing Lockdown. Available online: https://www.reuters.com/article/us-health-coronavirus-bosnia-cases-idUSKBN22B2DB (accessed on 12 March 2022).
- Lewnard, J.A.; Ndeffo Mbah, M.L.; Alfaro-Murillo, J.A.; Altice, F.L.; Bawo, L.; Nyenswah, T.G.; Galvani, A.P. Dynamics and control of Ebola virus transmission in Montserrado, Liberia: A mathematical modelling analysis. Lancet Infect. Dis. 2014, 14, 1189–1195. [Google Scholar] [CrossRef]
- Gedeon, T.; Bodelón, C.; Kuenzi, A. Hantavirus Transmission in Sylvan and Peridomestic Environments. Bull. Math. Biol. 2010, 72, 541–564. [Google Scholar] [CrossRef] [PubMed]

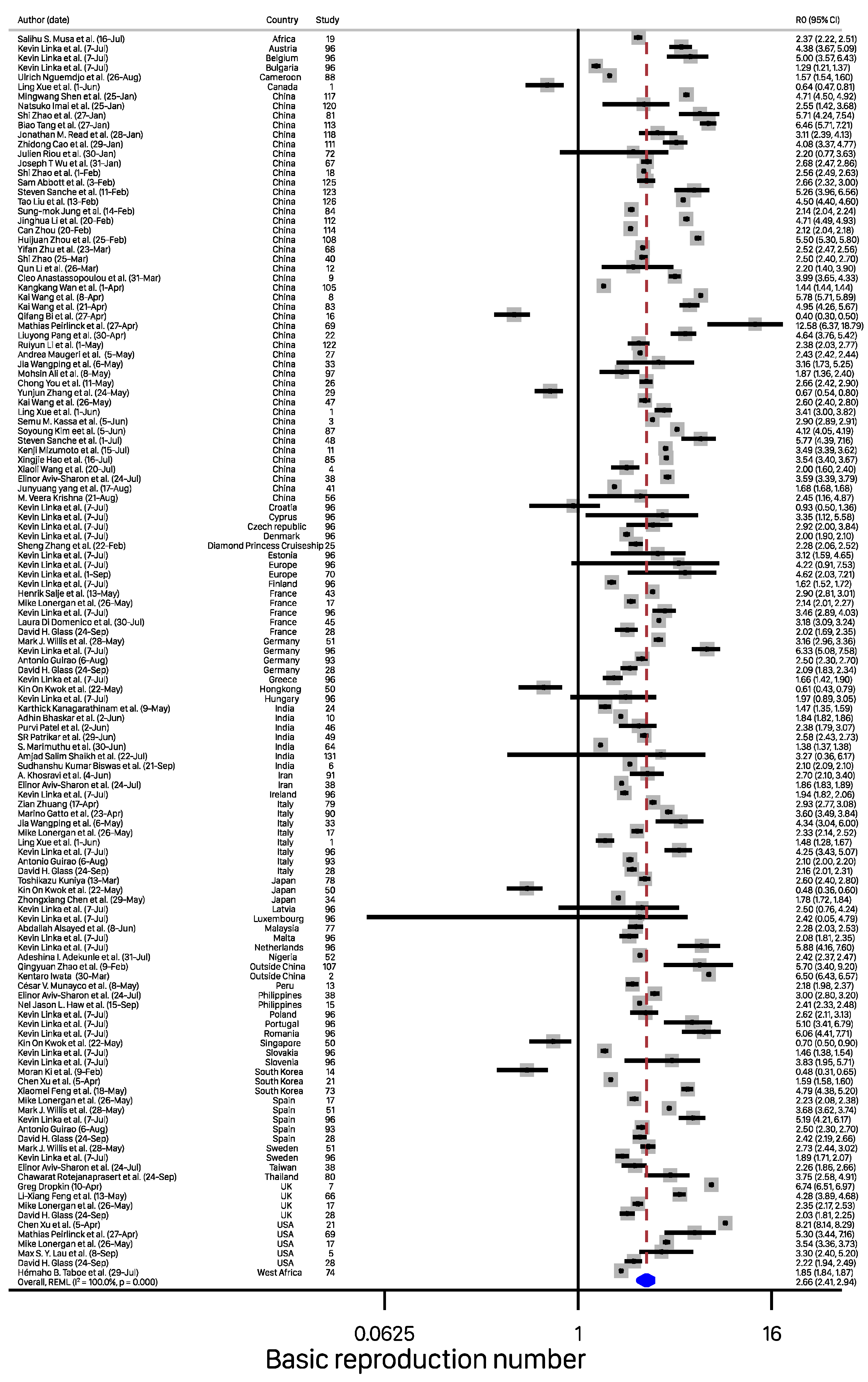


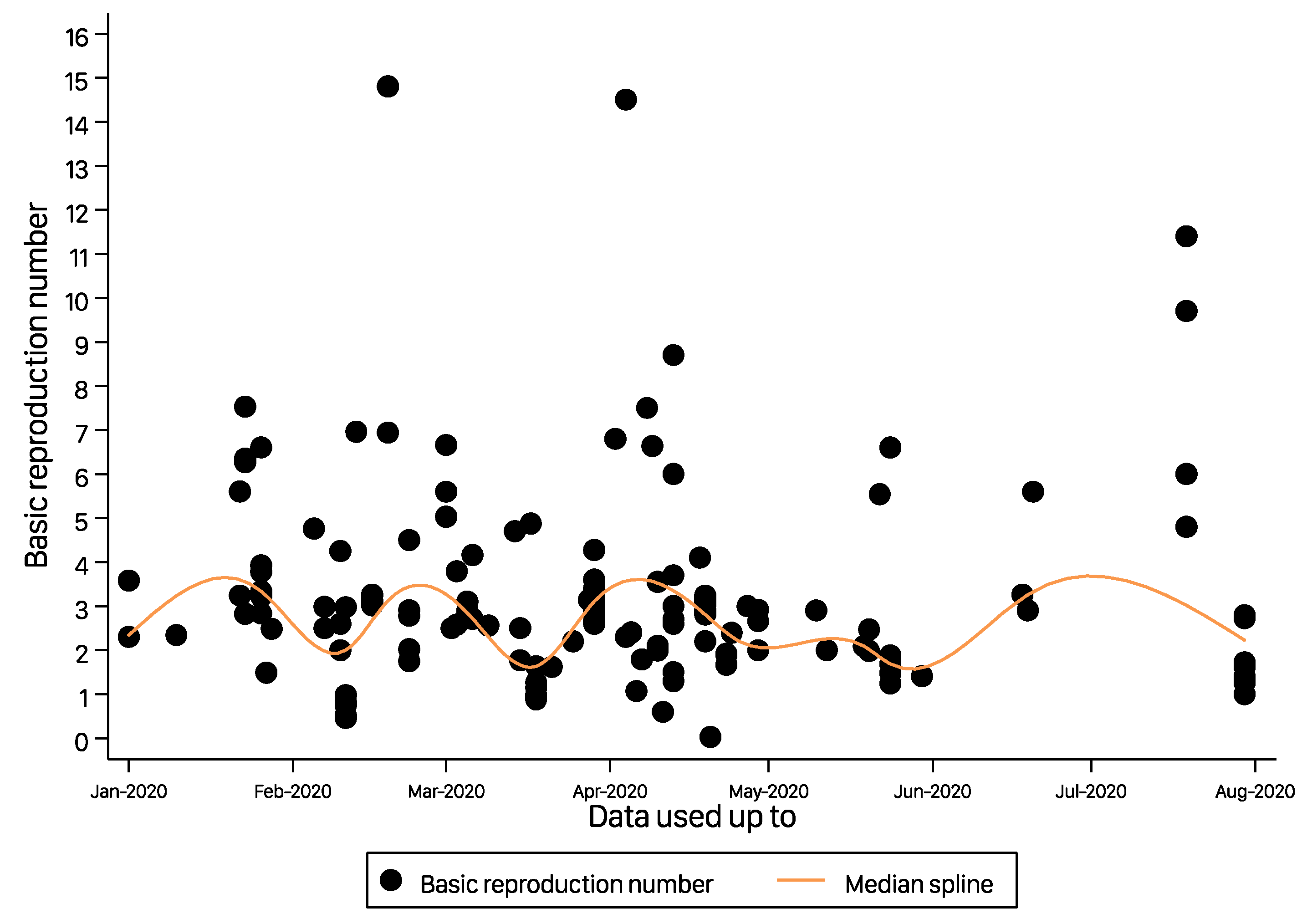
| Characteristics | Number of Reporting | R0 (95% CI) | p Value |
|---|---|---|---|
| Heterogeneity | |||
| Method considered | (n = 161) | ||
| 20 | 3.06 (2.32–4.03) | <0.001 |
| 6 | 2.47 (2.13–2.86) | <0.001 |
| 87 | 2.99 (2.67–3.35) | <0.001 |
| 4 | 2.60 (1.94–3.48) | <0.001 |
| 44 | 2.24 (1.87–2.69) | <0.001 |
| Duration of data | (n = 127) | ||
| 15 | 2.74 (2.26–3.31) | <0.001 |
| 28 | 2.70 (2.11–3.46) | <0.001 |
| 53 | 2.45 (2.06–2.91) | <0.001 |
| 31 | 2.86 (2.47–3.32) | <0.001 |
| Last month of data | (n = 128) | ||
| 25 | 3.34 (2.89–3.87) | <0.001 |
| 14 | 2.23 (1.40–3.56) | <0.001 |
| 30 | 2.18 (1.73–2.76) | <0.001 |
| 13 | 2.72 (1.99–3.71) | <0.001 |
| 12 | 2.69 (2.40–3.01) | <0.001 |
| 30 | 2.80 (2.31–3.39) | <0.001 |
| 4 | 2.60 (1.94–3.48) | <0.001 |
| Month of publication | (n = 130) | ||
| 8 | 3.87 (2.97–5.03) | <0.001 |
| 11 | 2.90 (1.92–4.37) | <0.001 |
| 6 | 3.18 (2.28–4.45) | <0.001 |
| 11 | 3.37 (1.93–5.89) | <0.001 |
| 26 | 2.22 (1.74–2.85) | <0.001 |
| 11 | 2.12 (1.58–2.86) | <0.001 |
| 40 | 2.83 (2.43–3.28) | <0.001 |
| 6 | 2.04 (1.70–2.45) | <0.001 |
| 8 | 2.27 (2.12–2.43) | <0.001 |
| Country | (n = 130) | ||
| 43 | 3.02 (2.55–3.59) | <0.001 |
| 49 | 2.24 (1.87–2.68) | <0.001 |
| 5 | 4.09 (2.60–6.43) | <0.001 |
| 8 | 2.69 (2.08–3.48) | <0.001 |
| 7 | 1.91 (1.56–2.33) | <0.001 |
| 5 | 2.68 (2.18–3.29) | <0.001 |
| 4 | 3.43 (1.99–5.91) | <0.001 |
| 5 | 3.02 (2.22–4.09) | <0.001 |
| 3 | 3.18 (1.99–5.08) | <0.001 |
| Continent | (n = 126) | ||
| 66 | 2.54 (2.18–2.96) | <0.001 |
| 50 | 2.78 (2.46–3.15) | <0.001 |
| 8 | 2.74 (1.62–4.64) | 0.002 |
| 2 | 1.94 (1.27–2.98) | <0.001 |
| Type of central estimate | (n = 130) | ||
| 34 | 2.99 (2.43–3.68) | <0.001 |
| 13 | 2.39 (1.91–2.98) | <0.001 |
| 83 | 2.58 (2.28–2.92) | <0.001 |
| Location in China | (n = 43) | ||
| 8 | 3.40 (2.61–4.44) | <0.001 |
| 2 | 3.39 (2.48–4.64) | <0.001 |
| 6 | 1.50 (0.76–2.96) | <0.001 |
| 27 | 3.39 (2.84–4.04) | <0.001 |
| Reproduction Number Threshold | Number (%) | Cumulative Number (%) | Mean | Range |
|---|---|---|---|---|
| Epidemic containment ( < 1) | 21 (6.2%) | 21 (6.2%) | 0.69 | 0.03–0.99 |
| Influenza (1 ≤ < 1.5) [26] | 19 (5.6%) | 40 (11.8%) | 1.33 | 1.00–1.49 |
| SARS-CoV (1.5 ≤ < 4) [23] | 231 (68.3%) | 271 (80.2%) | 2.61 | 1.50–3.99 |
| HIV (4 ≤ < 5) [27] | 26 (7.7%) | 297 (87.9%) | 4.43 | 4.02–4.95 |
| Smallpox (5 ≤ < 6) [28] | 16 (4.7%) | 313 (92.6%) | 5.47 | 5.00–5.88 |
| Rubella/Polio (6 ≤ < 7) [29] | 16 (4.7%) | 329 (97.3%) | 6.49 | 6–6.96 |
| Chickenpox (7 ≤ < 12) [30] | 6 (1.8%) | 335 (99.1%) | 8.84 | 7.50–11.40 |
| Measles (12 ≤ < 18) [31] | 3 (0.9%) | 338 (100%) | 13.96 | 12.58–14.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhungel, B.; Rahman, M.S.; Rahman, M.M.; Bhandari, A.K.C.; Le, P.M.; Biva, N.A.; Gilmour, S. Reliability of Early Estimates of the Basic Reproduction Number of COVID-19: A Systematic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2022, 19, 11613. https://doi.org/10.3390/ijerph191811613
Dhungel B, Rahman MS, Rahman MM, Bhandari AKC, Le PM, Biva NA, Gilmour S. Reliability of Early Estimates of the Basic Reproduction Number of COVID-19: A Systematic Review and Meta-Analysis. International Journal of Environmental Research and Public Health. 2022; 19(18):11613. https://doi.org/10.3390/ijerph191811613
Chicago/Turabian StyleDhungel, Bibha, Md. Shafiur Rahman, Md. Mahfuzur Rahman, Aliza K. C. Bhandari, Phuong Mai Le, Nushrat Alam Biva, and Stuart Gilmour. 2022. "Reliability of Early Estimates of the Basic Reproduction Number of COVID-19: A Systematic Review and Meta-Analysis" International Journal of Environmental Research and Public Health 19, no. 18: 11613. https://doi.org/10.3390/ijerph191811613
APA StyleDhungel, B., Rahman, M. S., Rahman, M. M., Bhandari, A. K. C., Le, P. M., Biva, N. A., & Gilmour, S. (2022). Reliability of Early Estimates of the Basic Reproduction Number of COVID-19: A Systematic Review and Meta-Analysis. International Journal of Environmental Research and Public Health, 19(18), 11613. https://doi.org/10.3390/ijerph191811613








