Quantifying the Relationship between SARS-CoV-2 Wastewater Concentrations and Building-Level COVID-19 Prevalence at an Isolation Residence: A Passive Sampling Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Passive Samples and Wastewater Processing
2.3. Protocol 1: SARS-CoV-2 Concentration and Quantification
2.4. Protocol 2: SARS-CoV-2 Concentration and Quantification
2.5. Isolation Residence Case Data and Clinical Surveillance
2.6. Data Analysis
3. Results and Discussion
3.1. Student Characteristics & Performance of Passive Samples
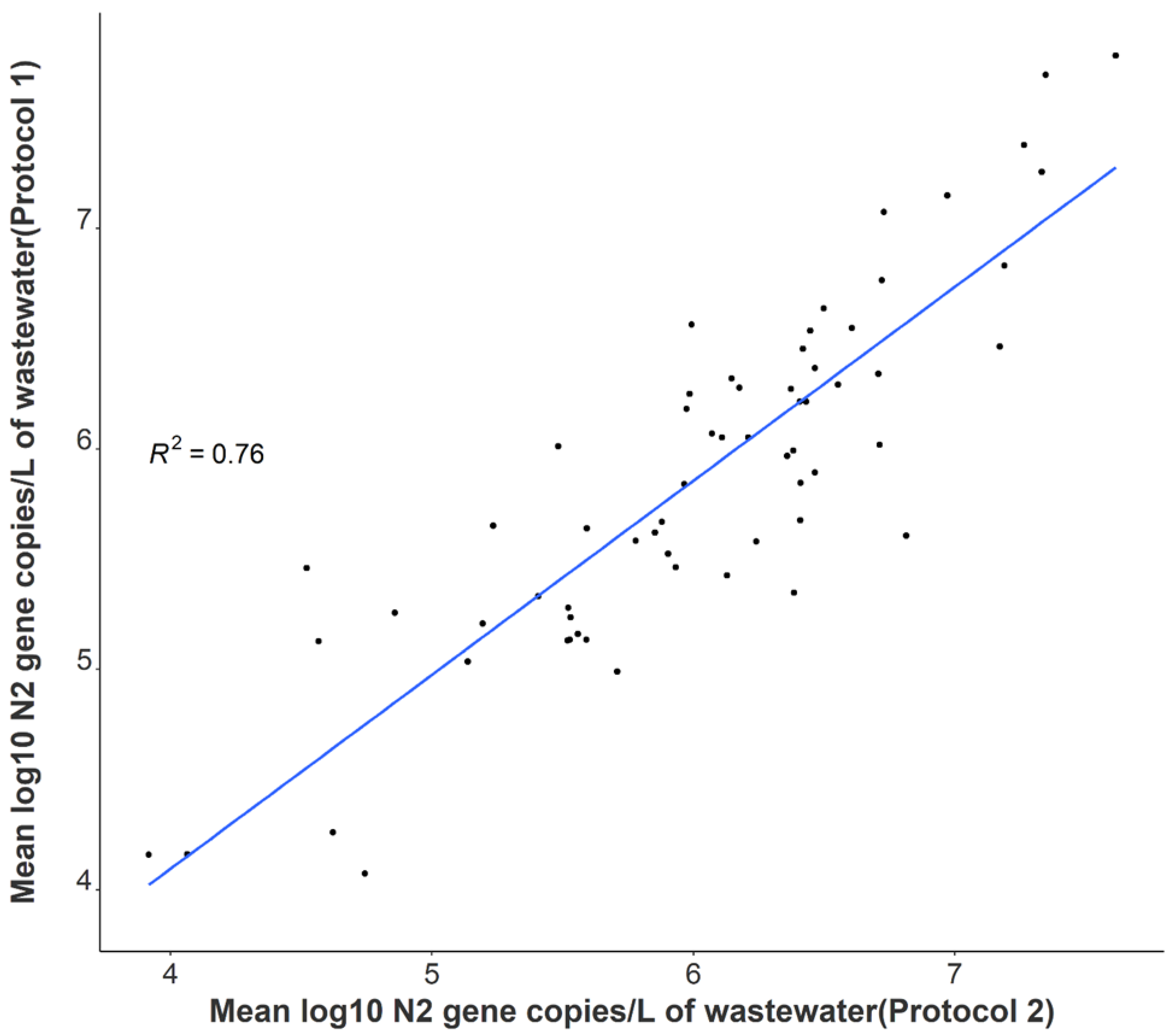
3.2. Comparison of SARS-CoV-2 Detection
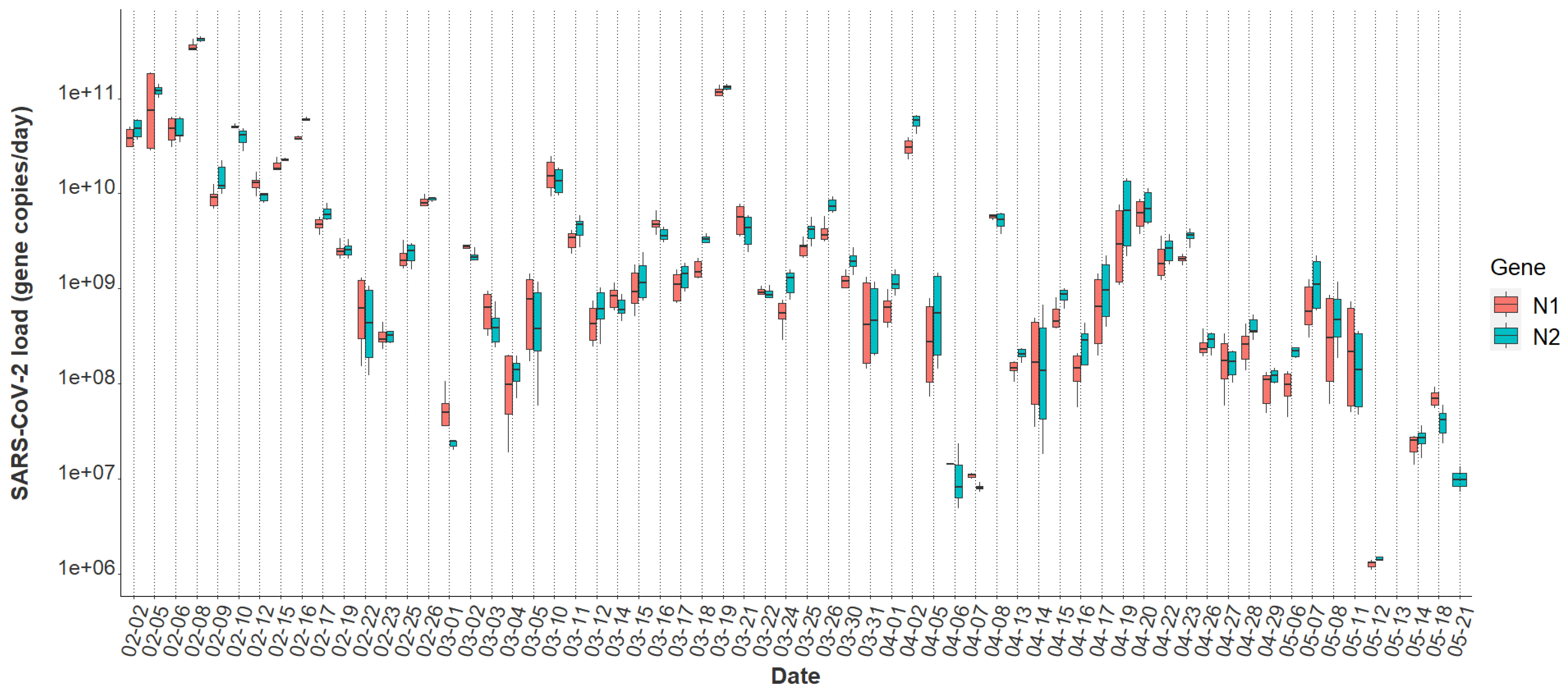
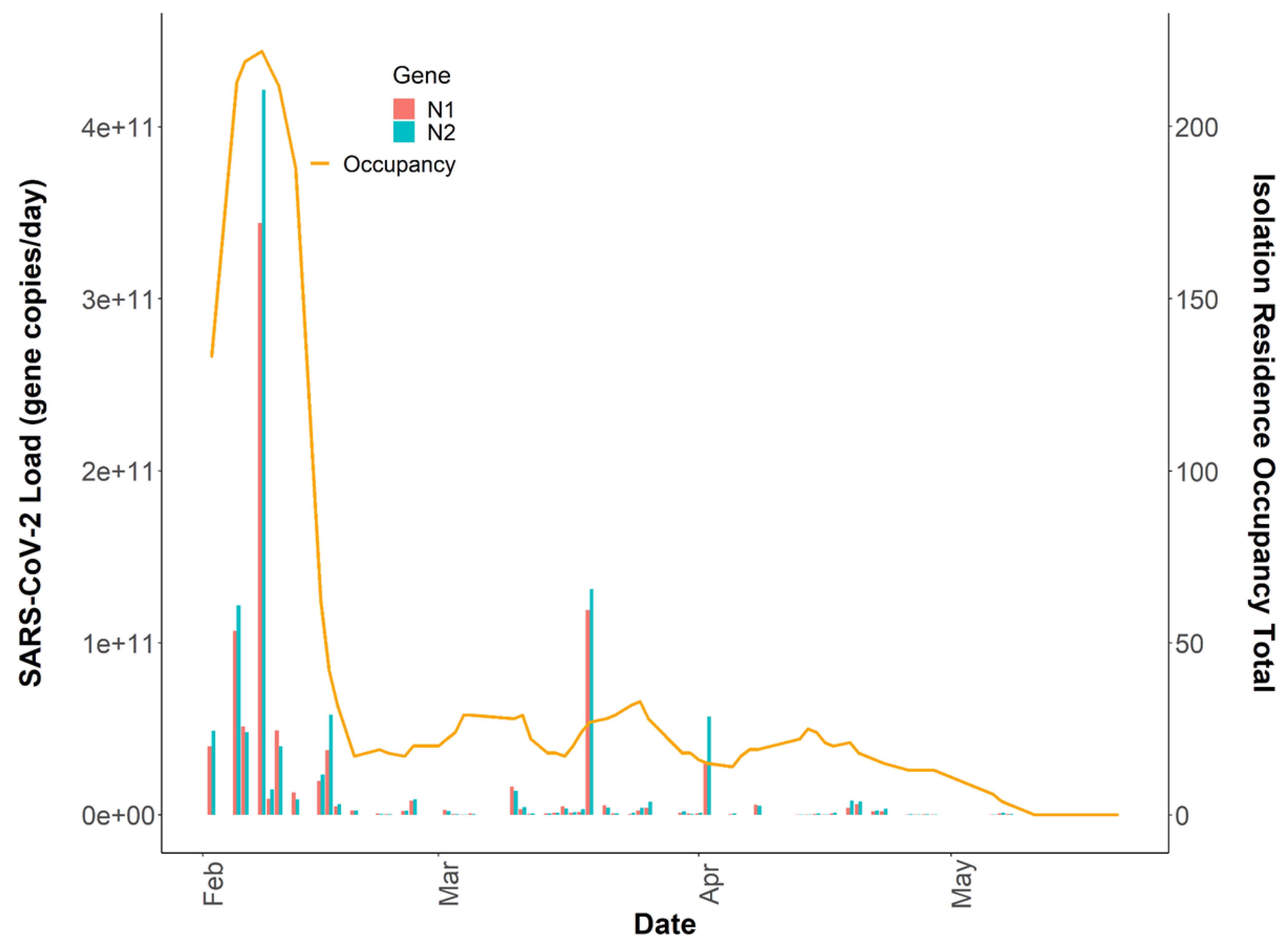
3.3. Variability in SARS-CoV-2 Signal
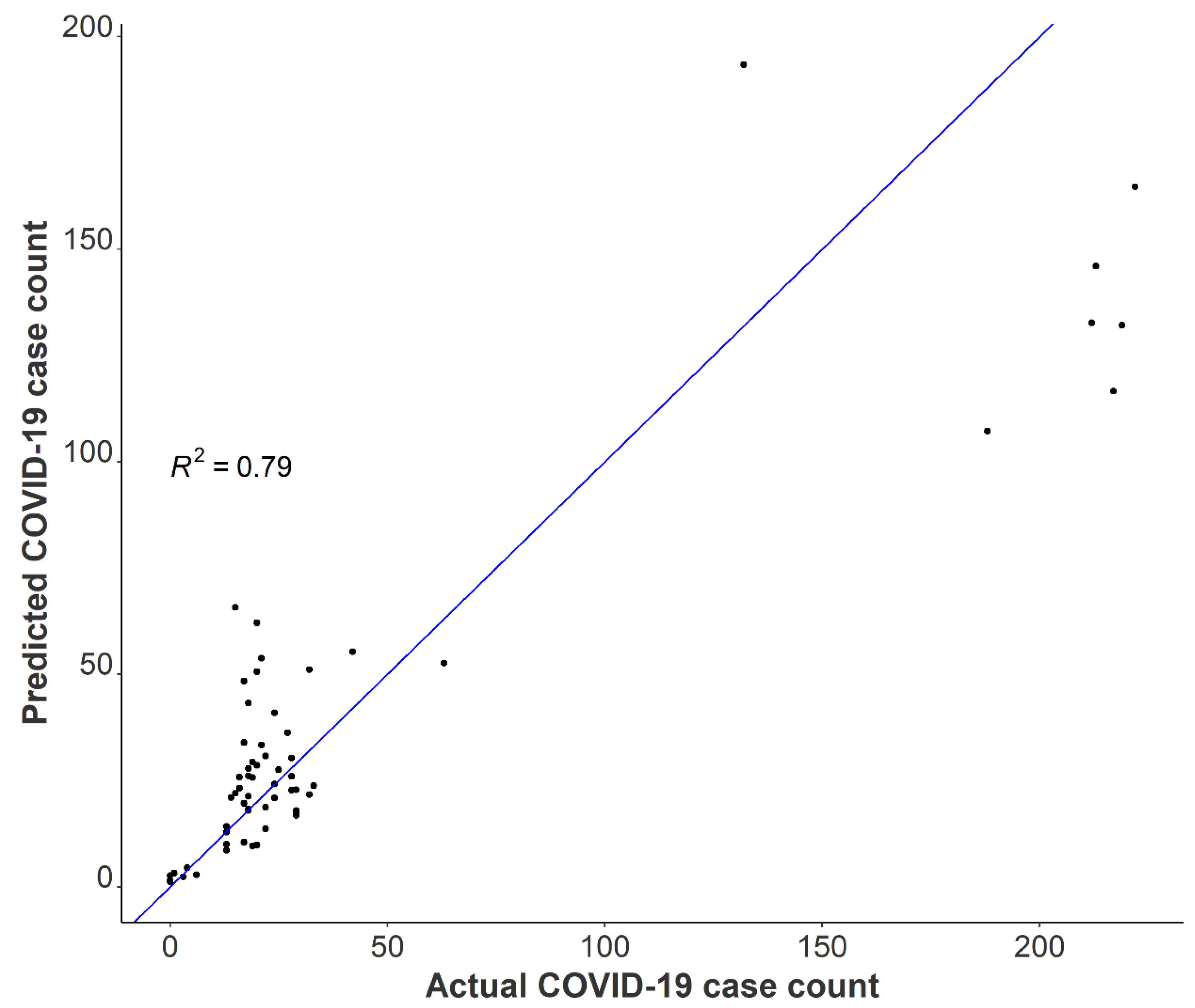
3.4. Relationships between Passive Samplers and Epidemiological Reporting
3.5. Limitations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersen, K.G.; Rambaut, A.; Lipkin, I.W.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: A systematic review and meta-analysis. Lancet Microbe 2020, 2, e13–e22. [Google Scholar] [CrossRef]
- Cheung, K.S.; Hung, I.F.N.; Chan, P.P.Y.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples from a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology 2020, 159, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Sun, J.; Xu, Y.; Li, F.; Huang, X.; Li, H.; Zhao, J.; Huang, J.; Zhao, J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 2020, 26, 1920–1922. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Xu, S.-B.; Lin, Y.-X.; Tian, D.; Zhu, Z.-Q.; Dai, F.-H.; Wu, F.; Song, Z.-G.; Huang, W.; Chen, J.; et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020, 133, 1039–1043. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Lo, I.L.; Lio, C.F.; Cheong, H.H.; Lei, C.I.; Cheong, T.H.; Zhong, X.; Tian, Y.; Sin, N.N. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020, 16, 1698–1707. [Google Scholar] [CrossRef]
- Chan, V.W.-S.; Chiu, P.K.-F.; Yee, C.-H.; Yuan, Y.; Ng, C.-F.; Teoh, J.Y.-C. A systematic review on COVID-19: Urological manifestations, viral RNA detection and special considerations in urological conditions. World J. Urol. 2020, 39, 3127–3138. [Google Scholar] [CrossRef]
- Wu, B.; Lei, Z.Y.; Wu, K.L.; He, J.R.; Cao, H.J.; Fu, J.; Chen, F.; Chen, Y.; Chen, B.; Zhou, X.L.; et al. Epidemiological and Clinical Features of Imported and Local Patients with Coronavirus Disease 2019 (COVID-19) in Hainan, China. SSRN 2020. [Google Scholar] [CrossRef]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e3. [Google Scholar] [CrossRef]
- Han, M.S.; Seong, M.-W.; Kim, N.; Shin, S.; Cho, S.I.; Park, H.; Kim, T.S.; Park, S.S.; Choi, E.H. Viral RNA Load in Mildly Symptomatic and Asymptomatic Children with COVID-19, Seoul. Emerg. Infect. Dis. 2020, 26, 2497–2499. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Gong, Y.; Meng, F.; Shi, Y.; Wang, J.; Mao, P.; Chuai, X.; Bi, Y.; Yang, P.; Wang, F. Comparative study on virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. Sci. China Life Sci. 2020, 64, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Li, F.; Chen, F.; Liu, H.; Zheng, J.; Lei, C.; Wu, X. Viral dynamics in asymptomatic patients with COVID-19. Int. J. Infect. Dis. 2020, 96, 288–290. [Google Scholar] [CrossRef]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef]
- McMahan, C.S.; Self, S.; Rennert, L.; Kalbaugh, C.; Kriebel, D.; Graves, D.; Colby, C.; Deaver, J.A.; Popat, S.C.; Karanfil, T.; et al. COVID-19 wastewater epidemiology: A model to estimate infected populations. Lancet Planet. Health 2021, 5, e874–e881. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Nguyen, A.; McDonald, D.; Zong, Y.; Ronquillo, N.; Ren, J.; Zou, J.; Farmer, S.; Humphrey, G.; Henderson, D.; et al. Rapid, Large-Scale Wastewater Surveillance and Automated Reporting System Enable Early Detection of Nearly 85% of COVID-19 Cases on a University Campus. mSystems 2021, 6, e00793-21. [Google Scholar] [CrossRef]
- Miura, F.; Kitajima, M.; Omori, R. Duration of SARS-CoV-2 viral shedding in faeces as a parameter for wastewater-based epidemiology: Re-analysis of patient data using a shedding dynamics model. Sci. Total Environ. 2021, 769, 144549. [Google Scholar] [CrossRef]
- Proverbio, D.; Kemp, F.; Magni, S.; Ogorzaly, L.; Cauchie, H.M.; Gonçalves, J.; Skupin, A.; Aalto, A. CoWWAn: Model-based assessment of COVID-19 epidemic dynamics by wastewater analysis. medRxiv 2021. [Google Scholar] [CrossRef]
- Peccia, J.; Zulli, A.; Brackney, D.E.; Grubaugh, N.D.; Kaplan, E.H.; Casanovas-Massana, A.; Ko, A.I.; Malik, A.A.; Wang, D.; Wang, M.; et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020, 38, 1164–1167. [Google Scholar] [CrossRef] [PubMed]
- D’Aoust, P.M.; Mercier, E.; Montpetit, D.; Jia, J.-J.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T.; et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2020, 188, 116560. [Google Scholar] [CrossRef]
- Feng, S.; Roguet, A.; McClary-Gutierrez, J.S.; Newton, R.J.; Kloczko, N.; Meiman, J.G.; McLellan, S.L. Evaluation of sampling frequency and normalization of SARS-CoV-2 wastewater concentrations for capturing COVID-19 burdens in the com-munity. Environ. Sci. Technol. 2021, 8, 1955–1965. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Ronquillo, N.; Belda-Ferre, P.; Alvarado, D.; Javidi, T.; Longhurst, C.A.; Knight, R. High-Throughput Wastewater SARS-CoV-2 Detection Enables Forecasting of Community Infection Dynamics in San Diego County. mSystems 2021, 6, e0045-21. [Google Scholar] [CrossRef]
- Barich, D.; Slonczewski, J.L. Wastewater Virus Detection Complements Clinical COVID-19 Testing to Limit Spread of Infection at Kenyon College. medRxiv 2021. [Google Scholar] [CrossRef]
- Betancourt, W.Q.; Schmitz, B.W.; Innes, G.K.; Prasek, S.M.; Brown, K.M.P.; Stark, E.R.; Foster, A.R.; Sprissler, R.S.; Harris, D.T.; Sherchan, S.P.; et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021, 779, 146408. [Google Scholar] [CrossRef]
- Gibas, C.; Lambirth, K.; Mittal, N.; Juel, A.I.; Barua, V.B.; Brazell, L.R.; Hinton, K.; Lontai, J.; Stark, N.; Young, I.; et al. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021, 782, 146749. [Google Scholar] [CrossRef] [PubMed]
- Reeves, K.; Liebig, J.; Feula, A.; Saldi, T.; Lasda, E.; Johnson, W.; Lilienfeld, J.; Maggi, J.; Pulley, K.; Wilkerson, P.J.; et al. High-resolution within-sewer SARS-CoV-2 surveillance facilitates informed intervention. Water Res. 2021, 204, 117613. [Google Scholar] [CrossRef]
- Travis, S.A.; Best, A.A.; Bochniak, K.S.; Dunteman, N.D.; Fellinger, J.; Folkert, P.D.; Koberna, T.; Kopek, B.G.; Krueger, B.P.; Pestun, J.; et al. Providing a Safe, In-Person, Residential College Experience During the COVID-19 Pandemic. Front. Public Health 2021, 9, 672344. [Google Scholar] [CrossRef]
- Bivins, A.; Lott, M.; Shaffer, M.; Wu, Z.; North, D.; Lipp, E.K.; Bibby, K. Building-Level Wastewater Monitoring for COVID-19 Using Tampon Swabs and RT-LAMP for Rapid SARS-Cov-2 RNA Detection. Environ. Sci. Water Res. Technol. 2021. [Google Scholar] [CrossRef]
- Rafiee, M.; Isazadeh, S.; Mohseni-Bandpei, A.; Mohebbi, S.R.; Jahangiri-Rad, M.; Eslami, A.; Dabiri, H.; Roostaei, K.; Tanhaei, M.; Amereh, F. Moore swab performs equal to composite and outperforms grab sampling for SARS-CoV-2 monitoring in wastewater. Sci. Total Environ. 2021, 790, 148205. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; Kaya, D.; Ahmed, W.; Brown, J.; Butler, C.; Greaves, J.; Leal, R.; Maas, K.; Rao, G.; Sherchan, S.; et al. Passive sampling to scale wastewater surveillance of infectious disease: Lessons learned from COVID-19. Sci. Total Environ. 2022, 835, 155347. [Google Scholar] [CrossRef] [PubMed]
- Brizee, A. How ASU, NAU and UA Are Testing Wastewater for COVID-19, Azcentral. 2021. Available online: https://www.azcentral.com/story/news/local/arizona-education/2021/01/13/how-asu-nau-and-ua-testing-wastewater-covid-19/6340627002/ (accessed on 8 March 2022).
- Corchis-Scott, R.; Geng, Q.; Seth, R.; Ray, R.; Beg, M.; Biswas, N.; Charron, L.; Drouillard, K.D.; D’Souza, R.; Heath, D.D.; et al. Averting an Outbreak of SARS-CoV-2 in a University Residence Hall through Wastewater Surveillance. Microbiol. Spectr. 2021, 9, e00792-21. [Google Scholar] [CrossRef]
- Severance, J. COVID Wastewater Testing Continuing this Spring–with an Absorbent New Twist. UConn Today. 2021. Available online: https://today.uconn.edu/2021/01/covid-wastewater-testing-continuing-spring-absorbent-new-twist/ (accessed on 8 March 2022).
- Schang, C.; Crosbie, N.D.; Nolan, M.; Poon, R.; Wang, M.; Jex, A.; John, N.; Baker, L.; Scales, P.; Schmidt, J.; et al. Passive Sampling of SARS-CoV-2 for Wastewater Surveillance. Environ. Sci. Technol. 2021, 55, 10432–10441. [Google Scholar] [CrossRef]
- Habtewold, J.; McCarthy, D.; McBean, E.; Law, I.; Goodridge, L.; Habash, M.; Murphy, H.M. Passive sampling, a practical method for wastewater-based surveillance of SARS-CoV-2. Environ. Res. 2021, 204, 112058. [Google Scholar] [CrossRef]
- Bivins, A.; Ahmed, W.; North, D.; Bibby, K. Wastewater Concentration by Adsorption and Direct Extraction for SARS-CoV-2 RNA Detection and Quantification Using RT-ddPCR V.2; protocols.io: Berkeley, CA, USA, 2020. [Google Scholar] [CrossRef]
- Forootan, A.; Sjöback, R.; Björkman, J.; Sjögreen, B.; Linz, L.; Kubista, M. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomol. Detect. Quantif. 2017, 12, 1–6. [Google Scholar] [CrossRef]
- Ma, Q.; Liu, J.; Liu, Q.; Kang, L.; Liu, R.; Jing, W.; Wu, Y.; Liu, M. Global Percentage of Asymptomatic SARS-CoV-2 Infections among the Tested Population and Individuals with Confirmed COVID-19 Diagnosis. JAMA Netw. Open 2021, 4, e2137257. [Google Scholar] [CrossRef]
- Graham, K.E.; Loeb, S.K.; Wolfe, M.K.; Catoe, D.; Sinnott-Armstrong, N.; Kim, S.; Yamahara, K.M.; Sassoubre, L.M.; Grijalva, L.M.M.; Roldan-Hernandez, L.; et al. SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed. Environ. Sci. Technol. 2020, 55, 488–498. [Google Scholar] [CrossRef]
- Pecson, B.M.; Darby, E.; Haas, C.N.; Amha, Y.M.; Bartolo, M.; Danielson, R.; Dearborn, Y.; Di Giovanni, G.; Ferguson, C.; Fevig, S.; et al. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: Findings from an interlaboratory methods evaluation in the U.S. Environ. Sci. Water Res. Technol. 2021, 7, 504–520. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Castro-Gutierrez, V.; Hassard, F.; Vu, M.; Leitao, R.; Burczynska, B.; Wildeboer, D.; Stanton, I.; Rahimzadeh, S.; Baio, G.; Garelick, H.; et al. Monitoring occurrence of SARS-CoV-2 in school populations: A wastewater-based approach. PLoS ONE 2022, 17, e0270168. [Google Scholar]
- McFadden, D. Quantitative Methods for Analyzing Travel Behaviour of Individuals: Some Recent Developments. Cowles Foundation for Research in Economics, Yale University. 1977. Available online: https://ideas.repec.org/p/cwl/cwldpp/474.html (accessed on 8 March 2022).
- Zhang, Y.; Cen, M.; Hu, M.; Du, L.; Hu, W.; Kim, J.J.; Dai, N. Prevalence and Persistent Shedding of Fecal SARS-CoV-2 RNA in Patients With COVID-19 Infection: A Systematic Review and Meta-analysis. Clin. Transl. Gastroenterol. 2021, 12, e00343. [Google Scholar] [CrossRef] [PubMed]
- Sierpiński, R.; Pinkas, J.; Jankowski, M.; Zgliczyński, W.S.; Wierzba, W.; Gujski, M.; Szumowski, Ł. Sex differences in the frequency of gastrointestinal symptoms and olfactory or taste disorders in 1942 nonhospitalized patients with coronavirus disease 2019 (COVID-19). Pol. Arch. Intern. Med. 2020, 130, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Morales Medina, W.R.M.; D’Elia, S.; Fahrenfeld, N.L. Accumulation of SARS-CoV-2 RNA in Sewer Biofilms. ACS ES&T Water 2022. [Google Scholar] [CrossRef]
| Characteristic | Value |
|---|---|
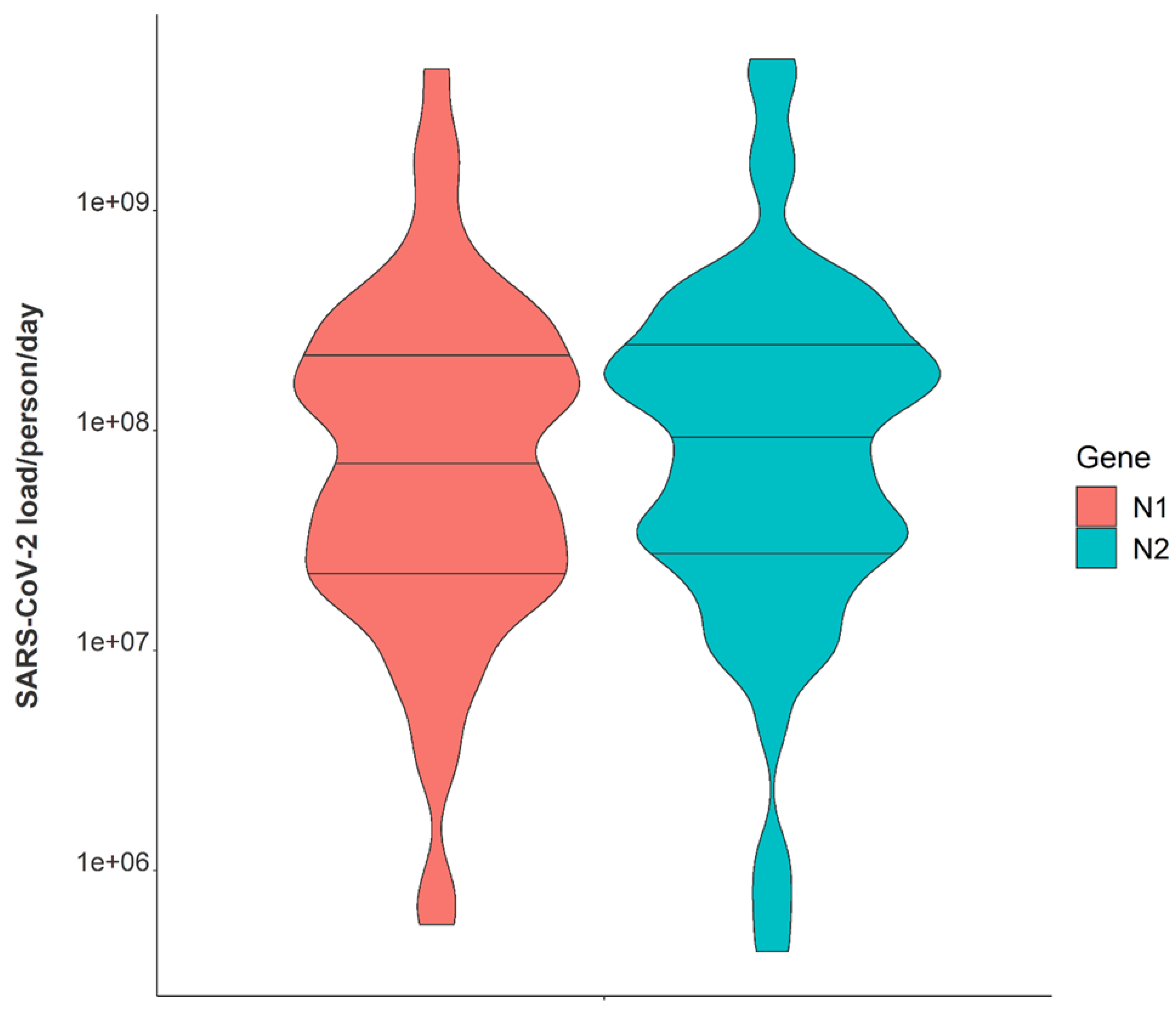
| Viral gene copies per day | |
| Median, N2 | 1.29 × 109 |
| Range (min/max), N2 | 1.38 × 106, 4.53 × 1011 |
| Median, N1 | 1.04 × 109 |
| Range (min/max), N1 | 1.11 × 106, 4.27 × 1011 |
| Viral gene copies per person per day | |
| Median, N2 | 1.01 × 108 |
| Range (min/max), N2 | 4.29 × 105, 4.86 × 109 |
| Median, N1 | 6.87 × 107 |
| Range (min/max), N1 | 5.70 × 105, 4.41 × 109 |
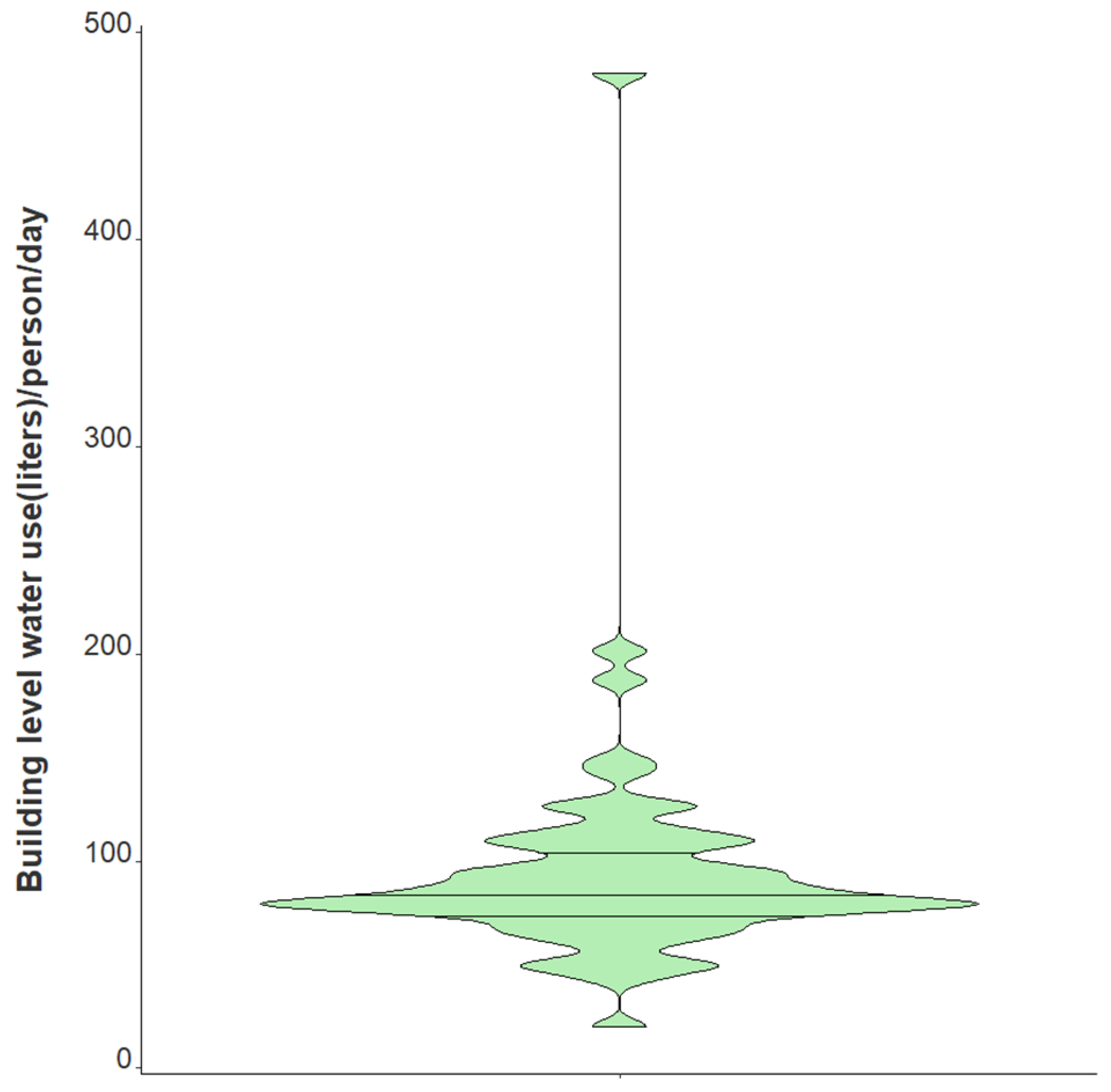
| Water usage per person per day (L) | |
| Median | 83.4 |
| Range (min/max) | 20.3, 479.7 |
| Factor | Univariate IRR | Univariate p-Value | Adjusted IRR | Adjusted p-Value |
|---|---|---|---|---|
| BRSV Recovery | 0.99 | 0.40 | 0.99 | 0.02 |
| % Female Occupants | 1.05 | <0.001 | 1.05 | <0.001 |
| Log Viral Gene Copies | 2.08 | <0.001 | 1.47 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acer, P.T.; Kelly, L.M.; Lover, A.A.; Butler, C.S. Quantifying the Relationship between SARS-CoV-2 Wastewater Concentrations and Building-Level COVID-19 Prevalence at an Isolation Residence: A Passive Sampling Approach. Int. J. Environ. Res. Public Health 2022, 19, 11245. https://doi.org/10.3390/ijerph191811245
Acer PT, Kelly LM, Lover AA, Butler CS. Quantifying the Relationship between SARS-CoV-2 Wastewater Concentrations and Building-Level COVID-19 Prevalence at an Isolation Residence: A Passive Sampling Approach. International Journal of Environmental Research and Public Health. 2022; 19(18):11245. https://doi.org/10.3390/ijerph191811245
Chicago/Turabian StyleAcer, Patrick T., Lauren M. Kelly, Andrew A. Lover, and Caitlyn S. Butler. 2022. "Quantifying the Relationship between SARS-CoV-2 Wastewater Concentrations and Building-Level COVID-19 Prevalence at an Isolation Residence: A Passive Sampling Approach" International Journal of Environmental Research and Public Health 19, no. 18: 11245. https://doi.org/10.3390/ijerph191811245
APA StyleAcer, P. T., Kelly, L. M., Lover, A. A., & Butler, C. S. (2022). Quantifying the Relationship between SARS-CoV-2 Wastewater Concentrations and Building-Level COVID-19 Prevalence at an Isolation Residence: A Passive Sampling Approach. International Journal of Environmental Research and Public Health, 19(18), 11245. https://doi.org/10.3390/ijerph191811245








