Methylation in the Promoter Region of the Dopamine Transporter DAT1 Gene in People Addicted to Nicotine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Methylation Status Assessment of Dopamine Gene Transporter (DAT1) Promoter
2.3. Statistical Analysis
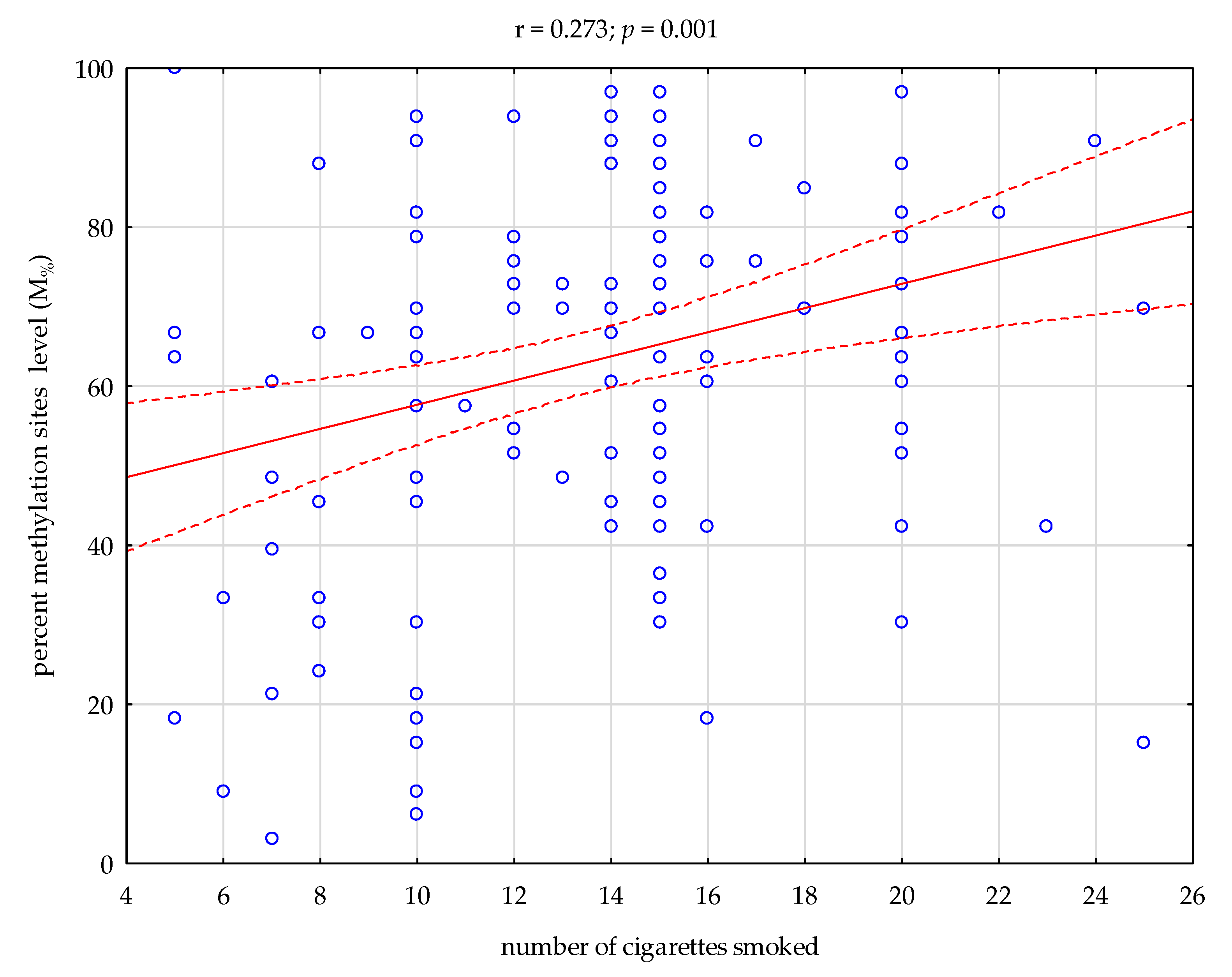
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- National Center for Chronic Disease Prevention and Health Promotion (US) Office on Smoking and Health. Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General; Centers for Disease Control and Prevention (US): Atlanta, GA, USA, 2014.
- Jha, P.; Ramasundarahettige, C.; Landsman, V.; Rostron, B.; Thun, M.; Anderson, R.N.; Mcafee, T.; Peto, R. 21st-Century Hazards of Smoking and Benefits of Cessation in the United States. N. Engl. J. Med. 2013, 368, 341–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Who Report on the Global Tobacco Epidemic: 2021 Addressing New and Emerging Products; World Health Organization: Geneva, Switzerland, 2021.
- World Health Organization. Who Global Report on Trends in Prevalence of Tobacco Use 2000–2025, 3rd ed.; World Health Organization: Geneva, Switzerland, 2019; ISBN 978-92-4-000003-2.
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [PubMed] [Green Version]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An Operational Definition of Epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaenisch, R.; Bird, A. Epigenetic Regulation of Gene Expression: How the Genome Integrates Intrinsic and Environmental Signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef]
- Speybroeck, L. From Epigenesis to Epigenetics. Ann. N. Y. Acad. Sci. 2006, 981, 61–81. [Google Scholar] [CrossRef]
- Nagre, N.N.; Subbanna, S.; Shivakumar, M.; Psychoyos, D.; Basavarajappa, B.S. CB1-Receptor Knockout Neonatal Mice Are Protected against Ethanol-Induced Impairments of DNMT1, DNMT3A, and DNA Methylation. J. Neurochem. 2015, 132, 429–442. [Google Scholar] [CrossRef] [Green Version]
- Blankman, J.L.; Simon, G.M.; Cravatt, B.F. A Comprehensive Profile of Brain Enzymes That Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem. Biol. 2007, 14, 1347–1356. [Google Scholar] [CrossRef] [Green Version]
- Maze, I.; Nestler, E.J. The Epigenetic Landscape of Addiction. Ann. N. Y. Acad. Sci. 2011, 1216, 99–113. [Google Scholar] [CrossRef]
- Tong, M.; Gao, S.; Qi, W.; Shi, C.; Qiu, M.; Yang, F.; Bai, S.; Li, H.; Wang, Z.; Sun, Z.; et al. 5-Hydroxymethylcytosine as a Potential Epigenetic Biomarker in Papillary Thyroid Carcinoma. Oncol. Lett. 2019, 18, 2304–2309. [Google Scholar] [CrossRef] [Green Version]
- Renthal, W.; Nestler, E.J. Epigenetic Mechanisms in Drug Addiction. Trends Mol. Med. 2008, 14, 341–350. [Google Scholar] [CrossRef] [Green Version]
- Baedke, J. The Epigenetic Landscape in the Course of Time: Conrad Hal Waddington’s Methodological Impact on the Life Sciences. Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci. 2013, 44, 756–773. [Google Scholar] [CrossRef] [PubMed]
- Szutorisz, H.; Hurd, Y.L. Epigenetic Effects of Cannabis Exposure. Biol. Psychiatry 2016, 79, 586–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, C.T.; Szutorisz, H.; Garg, P.; Martin, Q.; Landry, J.A.; Sharp, A.J.; Hurd, Y.L. Genome-Wide DNA Methylation Profiling Reveals Epigenetic Changes in the Rat Nucleus Accumbens Associated With Cross-Generational Effects of Adolescent THC Exposure. Neuropsychopharmacology 2015, 40, 2993–3005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiers, H.; Hannon, E.; Schalkwyk, L.C.; Smith, R.; Wong, C.C.Y.; O’Donovan, M.C.; Bray, N.J.; Mill, J. Methylomic Trajectories across Human Fetal Brain Development. Genome Res. 2015, 25, 338–352. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, A.; Malchow, B.; Hasan, A.; Falkai, P. The Impact of Environmental Factors in Severe Psychiatric Disorders. Front. Neurosci. 2014, 8, 19. [Google Scholar] [CrossRef]
- Tuesta, L.M.; Zhang, Y. Mechanisms of Epigenetic Memory and Addiction. EMBO J. 2014, 33, 1091–1103. [Google Scholar] [CrossRef] [Green Version]
- Joehanes, R.; Just, A.C.; Marioni, R.E.; Pilling, L.C.; Reynolds, L.M.; Mandaviya, P.R.; Guan, W.; Xu, T.; Elks, C.E.; Aslibekyan, S.; et al. Epigenetic Signatures of Cigarette Smoking. Circ. Cardiovasc. Genet. 2016, 9, 436–447. [Google Scholar] [CrossRef] [Green Version]
- Grzywacz, A.; Suchanecka, A.; Chmielowiec, J.; Chmielowiec, K.; Szumilas, K.; Masiak, J.; Balwicki, Ł.; Michałowska-Sawczyn, M.; Trybek, G. Personality Traits or Genetic Determinants—Which Strongly Influences E-Cigarette Users? Int. J. Environ. Res. Public Health 2020, 17, 365. [Google Scholar] [CrossRef] [Green Version]
- Grzywacz, A.; Chmielowiec, K.; Boroń, A.; Michałowska-Sawczyn, M.; Chmielowiec, J.; Trybek, G.; Mroczek, B.; Leźnicka, K.; Cieszczyk, P.; Masiak, J. Influence of DAT1 Promotor Methylation on Sports Performance. Genes 2021, 12, 1425. [Google Scholar] [CrossRef]
- Mehler-Wex, C.; Riederer, P.; Gerlach, M. Dopaminergic Dysbalance in Distinct Basal Ganglia Neurocircuits: Implications for the Pathophysiology of Parkinson’s Disease, Schizophrenia and Attention Deficit Hyperactivity Disorder. Neurotox. Res. 2006, 10, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, T.A.; Schork, N.J.; Eskin, E.; Kelsoe, J.R. Identification of Additional Variants within the Human Dopamine Transporter Gene Provides Further Evidence for an Association with Bipolar Disorder in Two Independent Samples. Mol. Psychiatry 2006, 11, 125–133. [Google Scholar] [CrossRef] [Green Version]
- Camí, J.; Farré, M. Drug Addiction. N. Engl. J. Med. 2003, 349, 975–986. [Google Scholar] [CrossRef] [Green Version]
- Vassoler, F.M.; Sadri-Vakili, G. Mechanisms of Transgenerational Inheritance of Addictive-like Behaviors. Neuroscience 2014, 264, 198–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkow, N.D.; Fowler, J.S.; Wang, G.J.; Swanson, J.M. Dopamine in Drug Abuse and Addiction: Results from Imaging Studies and Treatment Implications. Mol. Psychiatry 2004, 9, 557–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Justinova, Z.; Panlilio, L.V.; Goldberg, S.R. Drug Addiction. Curr. Top Behav. Neurosci. 2009, 1, 309–346. [Google Scholar] [CrossRef]
- Kreek, M.J.; Nielsen, D.A.; LaForge, K.S. Genes Associated with Addiction: Alcoholism, Opiate, and Cocaine Addiction. Neuromolecular Med. 2004, 5, 85–108. [Google Scholar] [CrossRef]
- Robison, A.J.; Nestler, E.J. Transcriptional and Epigenetic Mechanisms of Addiction. Nat. Rev. Neurosci. 2011, 12, 623–637. [Google Scholar] [CrossRef] [Green Version]
- Jasiewicz, A.; Rubiś, B.; Samochowiec, J.; Małecka, I.; Suchanecka, A.; Jabłoński, M.; Grzywacz, A. DAT1 Methylation Changes in Alcohol-Dependent Individuals vs. Controls. J. Psychiatr. Res. 2015, 64, 130–133. [Google Scholar] [CrossRef]
- Parrish, R.R.; Day, J.J.; Lubin, F.D. Direct Bisulfite Sequencing for Examination of DNA Methylation with Gene and Nucleotide Resolution from Brain Tissues. Curr. Protoc. Neurosci. 2012, 60, 7–24. [Google Scholar] [CrossRef] [Green Version]
- de Nardi, L.; Carpentieri, V.; Pascale, E.; Pucci, M.; D’addario, C.; Cerniglia, L.; Adriani, W.; Cimino, S. Involvement of DAT1 Gene on Internet Addiction: Cross-Correlations of Methylation Levels in 5′-UTR and 3’-UTR Genotypes, Interact with Impulsivity and Attachment-Driven Quality of Relationships. Int. J. Environ. Res. Public Health 2020, 17, 7956. [Google Scholar] [CrossRef]
- Sherman, C.B. Health Effects of Cigarette Smoking. Clin. Chest Med. 1991, 12, 643–658. [Google Scholar] [CrossRef]
- Kannel, W.B.; D’Agostino, R.B.; Belanger, A.J. Fibrinogen, Cigarette Smoking, and Risk of Cardiovascular Disease: Insights from the Framingham Study. Am. Heart J. 1987, 113, 1006–1010. [Google Scholar] [CrossRef]
- Bartecchi, C.E.; MacKenzie, T.D.; Schrier, R.W. The Human Costs of Tobacco Use. N. Engl. J. Med. 1994, 330, 907–912. [Google Scholar] [CrossRef]
- Doll, R.; Peto, R.; Wheatley, K.; Gray, R.; Sutherland, I. Mortality in Relation to Smoking: 40 Years’ Observations on Male British Doctors. BMJ 1994, 309, 901. [Google Scholar] [CrossRef] [Green Version]
- Power, C.; Atherton, K.; Thomas, C. Maternal Smoking in Pregnancy, Adult Adiposity and Other Risk Factors for Cardiovascular Disease. Atherosclerosis 2010, 211, 643–648. [Google Scholar] [CrossRef]
- Syme, C.; Abrahamowicz, M.; Mahboubi, A.; Leonard, G.T.; Perron, M.; Richer, L.; Veillette, S.; Gaudet, D.; Paus, T.; Pausova, Z. Prenatal Exposure to Maternal Cigarette Smoking and Accumulation of Intra-Abdominal Fat During Adolescence. Obesity 2010, 18, 1021–1025. [Google Scholar] [CrossRef]
- Winans, B.; Humble, M.C.; Lawrence, B.P. Environmental Toxicants and the Developing Immune System: A Missing Link in the Global Battle against Infectious Disease? Reprod. Toxicol. 2011, 31, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Haghighi, A.; Schwartz, D.H.; Abrahamowicz, M.; Leonard, G.T.; Perron, M.; Richer, L.; Veillette, S.; Gaudet, D.; Paus, T.; Pausova, Z. Prenatal Exposure to Maternal Cigarette Smoking, Amygdala Volume, and Fat Intake in Adolescence. JAMA Psychiatry 2013, 70, 98–105. [Google Scholar] [CrossRef] [Green Version]
- Breitling, L.P.; Yang, R.; Korn, B.; Burwinkel, B.; Brenner, H. Tobacco-Smoking-Related Differential DNA Methylation: 27K Discovery and Replication. Am. J. Hum. Genet. 2011, 88, 450–457. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.J.; Hansch, C. The Relative Toxicity of Compounds in Mainstream Cigarette Smoke Condensate. Food Chem. Toxicol. 2000, 38, 637–646. [Google Scholar] [CrossRef]
- Suter, M.; Abramovici, A.; Showalter, L.; Hu, M.; do Shope, C.; Varner, M.; Aagaard-Tillery, K. In Utero Tobacco Exposure Epigenetically Modifies Placental CYP1A1 Expression. Metabolism 2010, 59, 1481–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Okuka, M.; Lu, W.; Tsibris, J.C.M.; McLean, M.P.; Keefe, D.L.; Liu, L. Telomere Shortening and DNA Damage of Embryonic Stem Cells Induced by Cigarette Smoke. Reprod. Toxicol. 2013, 35, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Mortusewicz, O.; Schermelleh, L.; Walter, J.; Cardoso, M.C.; Leonhardt, H. Recruitment of DNA Methyltransferase I to DNA Repair Sites. Proc. Natl. Acad. Sci. USA 2005, 102, 8905–8909. [Google Scholar] [CrossRef] [Green Version]
- Cuozzo, C.; Porcellini, A.; Angrisano, T.; Morano, A.; Lee, B.; di Pardo, A.; Messina, S.; Iuliano, R.; Fusco, A.; Santillo, M.R.; et al. DNA Damage, Homology-Directed Repair, and DNA Methylation. PLoS Genet. 2007, 3, e110. [Google Scholar] [CrossRef]
- Lee, E.W.; D’alonzo, G.E. Cigarette Smoking, Nicotine Addiction, and Its Pharmacologic Treatment. Arch. Intern. Med. 1993, 153, 34–48. [Google Scholar] [CrossRef]
- Mercer, B.A.; Wallace, A.M.; Brinckerhoff, C.E.; D’Armiento, J.M. Identification of a Cigarette Smoke-Responsive Region in the Distal MMP-1 Promoter. Am. J. Respir. Cell Mol. Biol. 2009, 40, 4–12. [Google Scholar] [CrossRef]
- Di, Y.P.; Zhao, J.; Harper, R. Cigarette Smoke Induces MUC5AC Protein Expression through the Activation of Sp1. J. Biol. Chem. 2012, 287, 27948. [Google Scholar] [CrossRef] [Green Version]
- Kadonaga, J.T.; Carner, K.R.; Masiarz, F.R.; Tjian, R. Isolation of CDNA Encoding Transcription Factor Sp1 and Functional Analysis of the DNA Binding Domain. Cell 1987, 51, 1079–1090. [Google Scholar] [CrossRef]
- Han, L.; Lin, I.G.; Hsieh, C.-L. Protein Binding Protects Sites on Stable Episomes and in the Chromosome from De Novo Methylation. Mol. Cell. Biol. 2001, 21, 3416. [Google Scholar] [CrossRef] [Green Version]
- Olson, K.R. Carbon Monoxide Poisoning: Mechanisms, Presentation, and Controversies in Management. J. Emerg. Med. 1984, 1, 233–243. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, L.; Zhao, Y.; Zhang, J.; Wang, D.; Chen, J.; He, Y.; Wu, J.; Zhang, Z.; Liu, Z. Hypoxia Induces Genomic DNA Demethylation through the Activation of HIF-1α and Transcriptional Upregulation of MAT2A in Hepatoma Cells. Mol. Cancer Ther. 2011, 10, 1113–1123. [Google Scholar] [CrossRef] [Green Version]
- Grzywacz, A.; Barczak, W.; Chmielowiec, J.; Chmielowiec, K.; Suchanecka, A.; Trybek, G.; Masiak, J.; Jagielski, P.; Grocholewicz, K.; Rubiś, B. Contribution of Dopamine Transporter Gene Methylation Status to Cannabis Dependency. Brain Sci. 2020, 10, 400. [Google Scholar] [CrossRef]
- Madalena, K.M.; Lerch, J.K. The Effect of Glucocorticoid and Glucocorticoid Receptor Interactions on Brain, Spinal Cord, and Glial Cell Plasticity. Neural Plast. 2017, 2017, 8640970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deroche-Gamonet, V.; Sillaber, I.; Aouizerate, B.; Izawa, R.; Jaber, M.; Ghozland, S.; Kellendonk, C.; le Moal, M.; Spanagel, R.; Schütz, G.; et al. The Glucocorticoid Receptor as a Potential Target to Reduce Cocaine Abuse. J. Neurosci. 2003, 23, 4785. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ding, K.; Liu, R.; Zhang, J.; Yang, R.; Zhou, H.; Yang, C.; Liu, L.; Kang, C. Association between Methylation Status of CpG Island in DAT1 and DRD4 Genes and Clinical Symptoms of ADHD. Chin. J. Nerv. Ment. Dis. 2017, 12, 93–97. [Google Scholar]
- Marzilli, E.; Cerniglia, L.; Tambelli, R.; Cimino, S. Children’s ADHD and Dysregulation Problems, DAT1 Genotype and Methylation, and Their Interplay with Family Environment. Child Youth Care Forum 2022, 2022, 1–29. [Google Scholar] [CrossRef]
- Cimino, S.; Cerniglia, L.; Ballarotto, G.; Marzilli, E.; Pascale, E.; D’Addario, C.; Adriani, W.; Icro Maremmani, A.G.; Tambelli, R. Children’s DAT1 Polymorphism Moderates the Relationship Between Parents’ Psychological Profiles, Children’s DAT Methylation, and Their Emotional/Behavioral Functioning in a Normative Sample. Int. J. Environ. Res. Public Health 2019, 16, 2567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiers, C.E.; Lohoff, F.W.; Lee, J.; Muench, C.; Freeman, C.; Zehra, A.; Marenco, S.; Lipska, B.K.; Auluck, P.K.; Feng, N.; et al. Methylation of the Dopamine Transporter Gene in Blood Is Associated with Striatal Dopamine Transporter Availability in ADHD: A Preliminary Study. Eur. J. Neurosci. 2018, 48, 1884. [Google Scholar] [CrossRef] [PubMed]
- Rubino, A.; D’Addario, C.; di Bartolomeo, M.; Michele Salamone, E.; Locuratolo, N.; Fattapposta, F.; Vanacore, N.; Pascale, E. DNA Methylation of the 5’-UTR DAT 1 Gene in Parkinson’s Disease Patients. Acta. Neurol. Scand. 2020, 142, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Tafani, X.; Pascale, E.; Fattapposta, F.; Pucci, M.; D’Addario, C.; Adriani, W. Cross-Correlations between Motifs in the 5’-UTR of DAT1 Gene: Findings from Parkinson’s Disease. Adv. Biol. Regul. 2020, 78, 100753. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, E.; Pascale, E.; Troianiello, M.; D’Addario, C.; Adriani, W. DAT1 Gene Methylation as an Epigenetic Biomarker in Attention Deficit Hyperactivity Disorder: A Commentary. Front. Genet. 2020, 11, 444. [Google Scholar] [CrossRef] [PubMed]
- Lambacher, G.; Pascale, E.; Pucci, M.; Mangiapelo, S.; D’Addario, C.; Adriani, W. Search for an Epigenetic Biomarker in ADHD Diagnosis, Based on the DAT1 Gene 5’-UTR Methylation: A New Possible Approach. Psychiatry Res. 2020, 291, 113154. [Google Scholar] [CrossRef] [PubMed]
- McLean, K.C.; Syed, M.; Arnett, J.J. Identity Development from Adolescence to Emerging Adulthood. In The Oxford Handbook of Identity Development; Oxford University Press: Oxford, UK, 2015; pp. 53–64. [Google Scholar]
- Steensma, T.D.; Kreukels, B.P.C.; de Vries, A.L.C.; Cohen-Kettenis, P.T. Gender Identity Development in Adolescence. Horm. Behav. 2013, 64, 288–297. [Google Scholar] [CrossRef]
- Offer, D.; Boxer, A. Normal Adolescent Development: Empirical Research Findings. In Child and Adolescent Psychiatry; Lewis, M., Ed.; Williams & Wilkins: Baltimore, MD, USA, 1991. [Google Scholar]
- Shumay, E.; Fowler, J.S.; Volkow, N.D. Genomic Features of the Human Dopamine Transporter Gene and Its Potential Epigenetic States: Implications for Phenotypic Diversity. PLoS ONE 2010, 5, e11067. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.V.; Turner, S.T.; Smith, J.A.; Hammond, P.I.; Lazarus, A.; van de Rostyne, J.L.; Cunningham, J.M.; Kardia, S.L.R. Comparison of the DNA Methylation Profiles of Human Peripheral Blood Cells and Transformed B-Lymphocytes. Hum. Genet. 2010, 127, 651. [Google Scholar] [CrossRef] [Green Version]
| ND | Control | Mann–Whitney U-Test (p) | |
| n | 142 | 238 | |
| Age M (SD) | 27.57 (10.01) | 21.86 (3.55) | −1.726 (0.08432) |
| Number of Cycles | PCR Step | Temperature | Time |
|---|---|---|---|
| 1 | Initial denaturation | 94 °C | 5:00 |
| 35 | Denaturation | 94 °C | 0:25 |
| Annealing | 61 °C | 0:25 | |
| Elongation | 72 °C | 0:25 | |
| 1 | Final elongation | 72 °C | 5:00 |
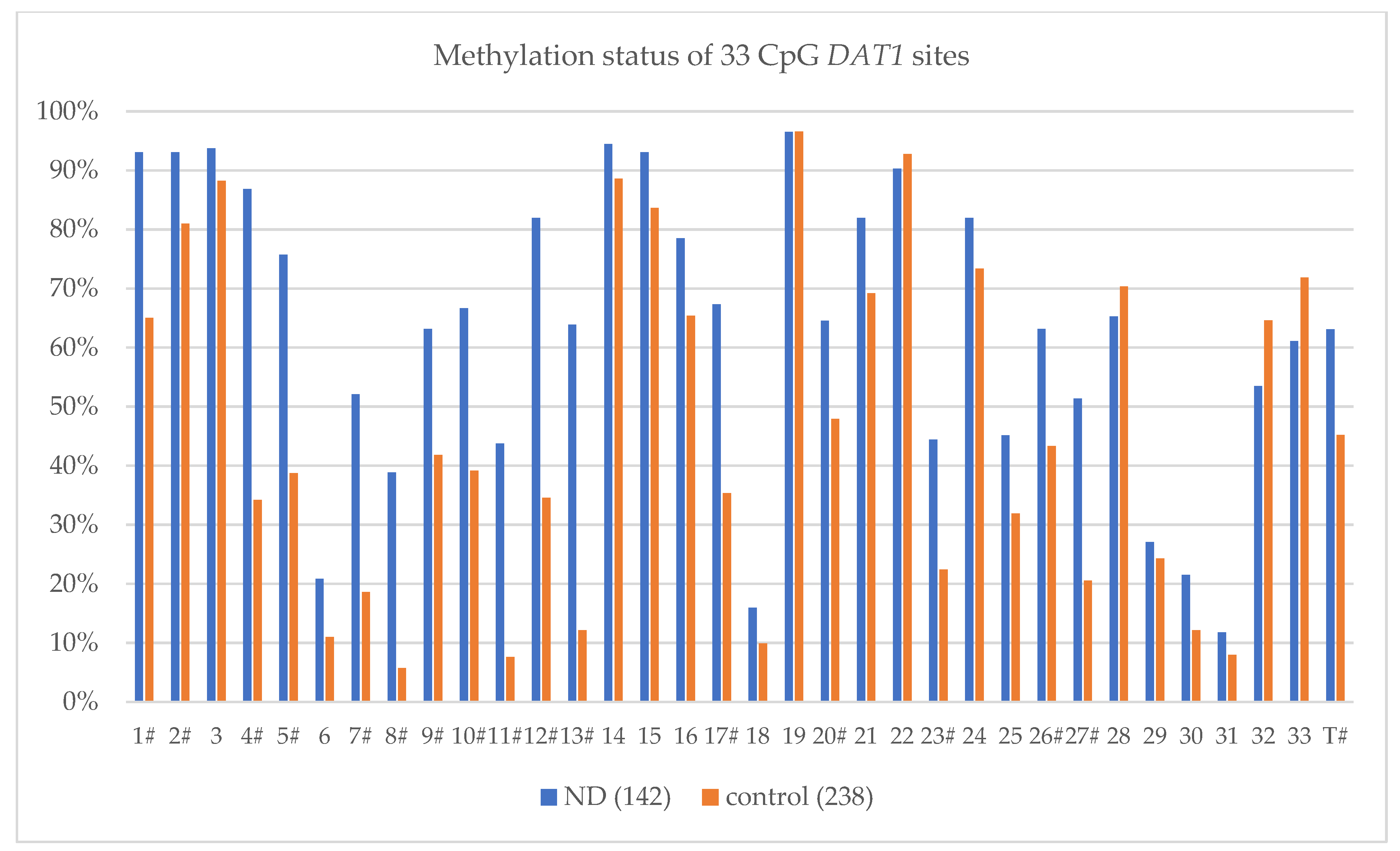
| CpG Site | Studied Group | Methylation Level (%) | χ2(p) | OR | 95% CI (−95%, +95%) |
|---|---|---|---|---|---|
| 1 # | ND (142) | 93.06% | 38.945 (0.000001) | 7.209 | (3.613, 14.383) |
| control (238) | 65.02% | ||||
| 2 # | ND (142) | 93.06% | 10.780 (0.00103) | 3.145 | (1.542, 6.414) |
| control (238) | 80.99% | ||||
| 3 | ND (142) | 93.75% | 3.219 (0.07278) | 2.004 | (0.926, 4.337) |
| control (238) | 88.21% | ||||
| 4 # | ND (142) | 86.81% | 103.251 (0.000001) | 12.646 | (7.327, 21.825) |
| control (238) | 34.22% | ||||
| 5 # | ND (142) | 75.69% | 50.779 (0.000001) | 4.915 | (3.120, 7.744) |
| control (238) | 38.78% | ||||
| 6 | ND (142) | 20.83% | 7.220 (0.00721) | 2.123 | (1.216, 3.707) |
| control (238) | 11.03% | ||||
| 7 # | ND (142) | 52.08% | 49.153 (0.000001) | 4.747 | (3.024, 7.451) |
| control (238) | 18.63% | ||||
| 8 # | ND (142) | 38.89% | 70.829 (0.000001) | 10.479 | (5.639, 19.471) |
| control (238) | 5.73% | ||||
| 9 # | ND (142) | 63.19% | 16.999 (0.00004) | 2.388 | (1.572, 3.627) |
| control (238) | 41.83% | ||||
| 10 # | ND (142) | 66.67% | 28.168 (0.000001) | 3.107 | (2.029, 4.756) |
| control (238) | 39.16% | ||||
| 11 # | ND (142) | 43.75% | 74.885 (0.000001) | 9.450 | (5.385, 16.583) |
| control (238) | 7.60% | ||||
| 12 # | ND (142) | 81.94% | 83.488 (0.000001) | 8.578 | (5.229, 14.070) |
| control (238) | 34.60% | ||||
| 13 # | ND (142) | 63.89% | 117.503 (0.000001) | 12.772 | (7.728, 21.105) |
| control (238) | 12.17% | ||||
| 14 | ND (142) | 94.44% | 3.764 (0.05238) | 2.188 | (0.975, 4.911) |
| control (238) | 88.59% | ||||
| 15 | ND (142) | 93.06% | 7.267 (0.00702) | 2.619 | (1.273, 5.385) |
| control (238) | 83.65% | ||||
| 16 | ND (142) | 78.47% | 7.576 (0.00591) | 1.928 | (1.203, 3.091) |
| control (238) | 65.40% | ||||
| 17 # | ND (142) | 67.36% | 38.282 (0.00000) | 3.772 | (2.453, 5.801) |
| control (238) | 35.36% | ||||
| 18 | ND (142) | 15.97% | 3.255 (0.07121) | 1.733 | (0.949, 3.164) |
| control (238) | 9.89% | ||||
| 19 | ND (142) | 96.53% | 0.0007 (0.97882) | 0.985 | (0.323, 2.996) |
| control (238) | 96.58% | ||||
| 20 # | ND (142) | 64.58% | 10.409 (0.00125) | 1.982 | (1.304, 3.013) |
| control (238) | 47.91% | ||||
| 21 | ND (142) | 81.94% | 7.797 (0.00523) | 2.019 | (1.226, 3.326) |
| control (238) | 69.20% | ||||
| 22 | ND (142) | 90.28% | 0.779 (0.37737) | 0.723 | (0.351, 1.489) |
| control (238) | 92.78% | ||||
| 23 # | ND (142) | 44.44% | 21.378 (0.00000) | 2.766 | (1.785, 4.287) |
| control (238) | 22.43% | ||||
| 24 | ND (142) | 81.94% | 3.783 (0.05177) | 1.646 | (0.993, 2.727) |
| control (238) | 73.38% | ||||
| 25 | ND (142) | 45.14% | 6.986 (0.00821) | 1.753 | (1.154, 2.663) |
| control (238) | 31.94% | ||||
| 26 # | ND (142) | 63.19% | 14.664 (0.00013) | 2.244 | (1.478, 3.406) |
| control (238) | 43.35% | ||||
| 27 # | ND (142) | 51.39% | 41.095 (0.00000) | 4.091 | (2.627, 6.372) |
| control (238) | 20.53% | ||||
| 28 | ND (142) | 65.28% | 1.107 (0.2927) | 0.792 | (0.513, 1.222) |
| control (238) | 70.34% | ||||
| 29 | ND (142) | 27.08% | 0.371 (0.54195) | 1.154 | (0.726, 1.835) |
| control (238) | 24.33% | ||||
| 30 | ND (142) | 21.53% | 6.231 (0.01255) | 1.980 | (1.150, 3.407) |
| control (238) | 12.17% | ||||
| 31 | ND (142) | 11.81% | 1.605 (0.20523) | 1.542 | (0.786, 3.028) |
| control (238) | 7.98% | ||||
| 32 | ND (142) | 53.47% | 4.863 (0.02743) | 0.628 | (0.416, 0.951) |
| control (238) | 64.64% | ||||
| 33 | ND (142) | 61.11% | 4.948 (0.02611) | 0.615 | (0.400, 0.945) |
| control (238) | 71.86% | ||||
| Total Methylation Sites MT ± SD; (MT% ± SD) | |||||
| ND (142) | 20.82 ± 8.01; (63.09% ± 24.28%) | ||||
| control (238) | 14.92 ± 5.81; (45.20% ± 17.61%) | ||||
| Mann–Whitney U-Test Z (p) | 7.881 (0.000001) # | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chmielowiec, J.; Chmielowiec, K.; Strońska-Pluta, A.; Suchanecka, A.; Humińska-Lisowska, K.; Lachowicz, M.; Niewczas, M.; Białecka, M.; Śmiarowska, M.; Grzywacz, A. Methylation in the Promoter Region of the Dopamine Transporter DAT1 Gene in People Addicted to Nicotine. Int. J. Environ. Res. Public Health 2022, 19, 8602. https://doi.org/10.3390/ijerph19148602
Chmielowiec J, Chmielowiec K, Strońska-Pluta A, Suchanecka A, Humińska-Lisowska K, Lachowicz M, Niewczas M, Białecka M, Śmiarowska M, Grzywacz A. Methylation in the Promoter Region of the Dopamine Transporter DAT1 Gene in People Addicted to Nicotine. International Journal of Environmental Research and Public Health. 2022; 19(14):8602. https://doi.org/10.3390/ijerph19148602
Chicago/Turabian StyleChmielowiec, Jolanta, Krzysztof Chmielowiec, Aleksandra Strońska-Pluta, Aleksandra Suchanecka, Kinga Humińska-Lisowska, Milena Lachowicz, Marta Niewczas, Monika Białecka, Małgorzata Śmiarowska, and Anna Grzywacz. 2022. "Methylation in the Promoter Region of the Dopamine Transporter DAT1 Gene in People Addicted to Nicotine" International Journal of Environmental Research and Public Health 19, no. 14: 8602. https://doi.org/10.3390/ijerph19148602
APA StyleChmielowiec, J., Chmielowiec, K., Strońska-Pluta, A., Suchanecka, A., Humińska-Lisowska, K., Lachowicz, M., Niewczas, M., Białecka, M., Śmiarowska, M., & Grzywacz, A. (2022). Methylation in the Promoter Region of the Dopamine Transporter DAT1 Gene in People Addicted to Nicotine. International Journal of Environmental Research and Public Health, 19(14), 8602. https://doi.org/10.3390/ijerph19148602






