Profile of Bacterial Community and Antibiotic Resistance Genes in Typical Vegetable Greenhouse Soil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Soil Sampling
2.2. DNA Extraction
2.3. Illumina High-Throughput Sequencing
2.4. qPCR of ARGs and intI Genes
2.5. Determination of Physicochemical Properties and Heavy Metal Concentration
2.6. Determination of Antibiotics
2.7. Statistical Analysis
3. Results
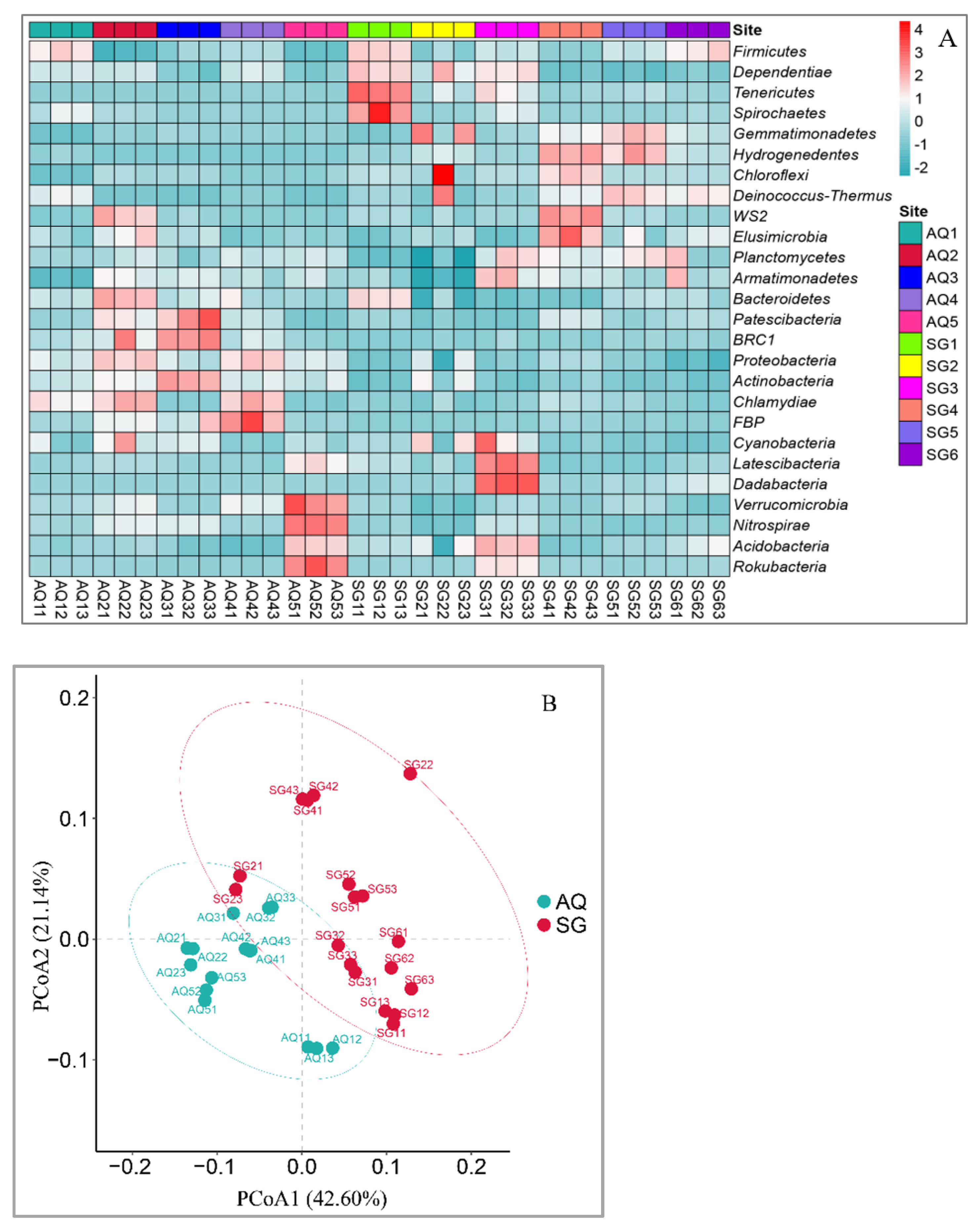
3.1. Characterization of Bacterial Community
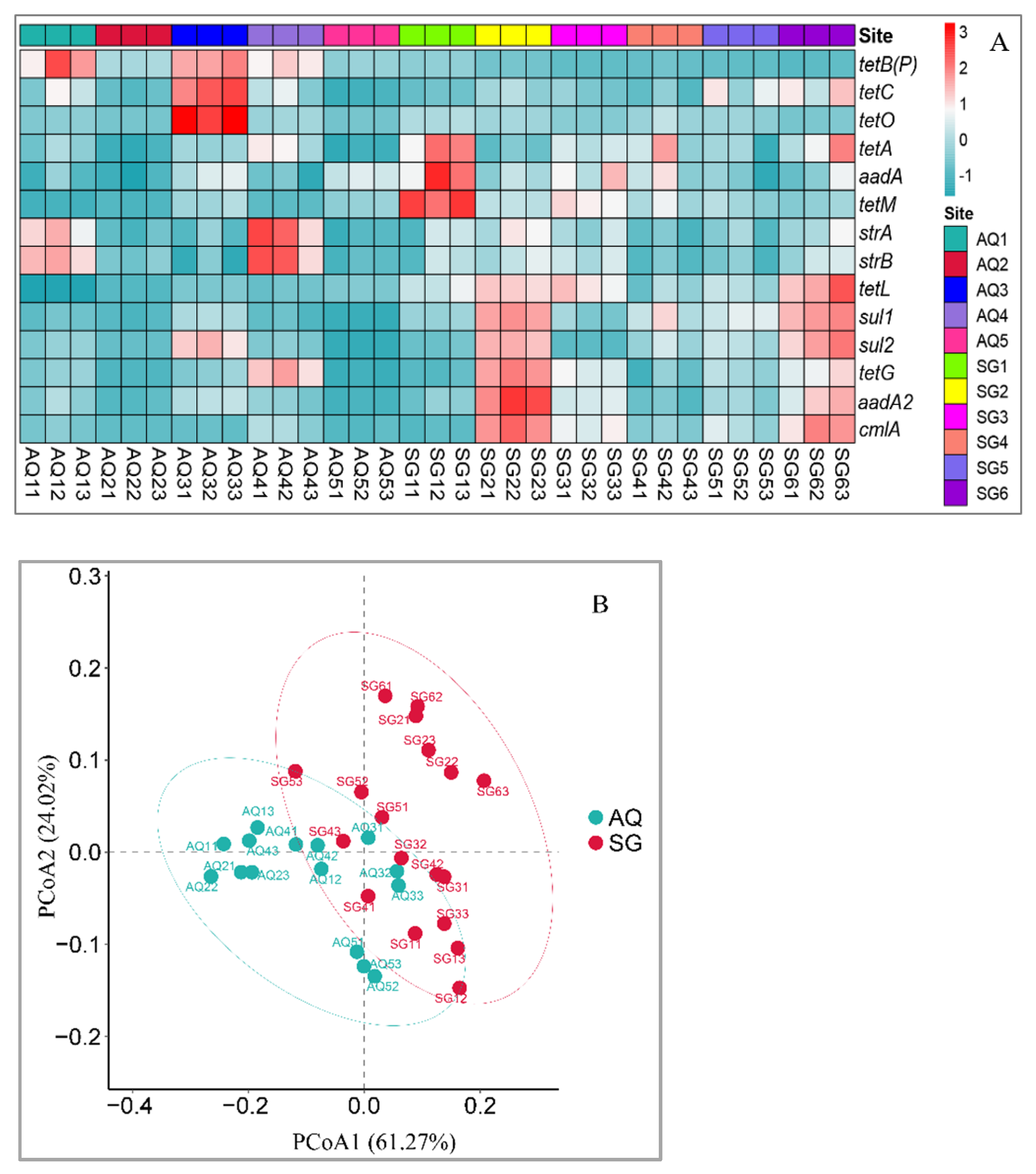
3.2. Abundances of ARGs and intI Gene
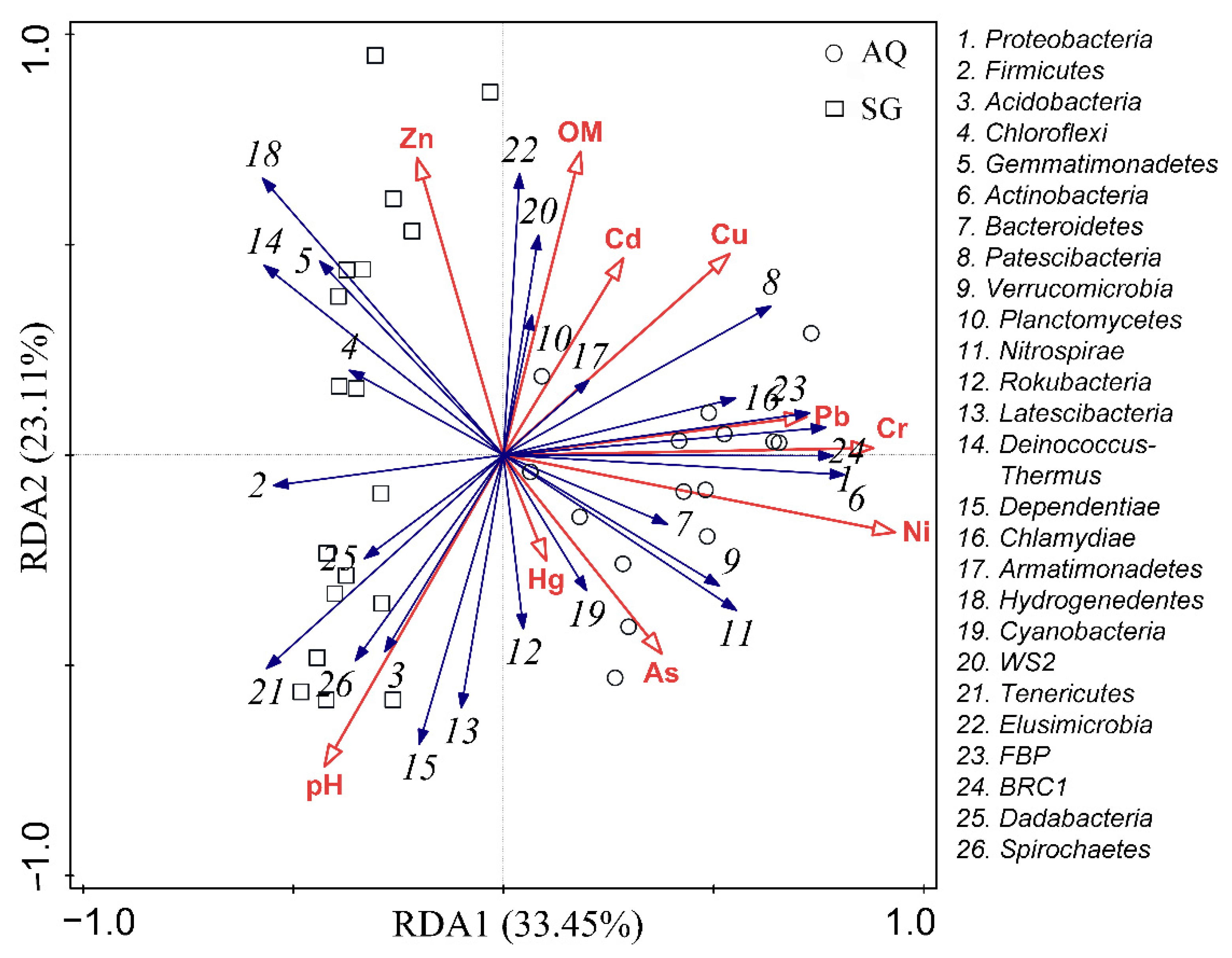
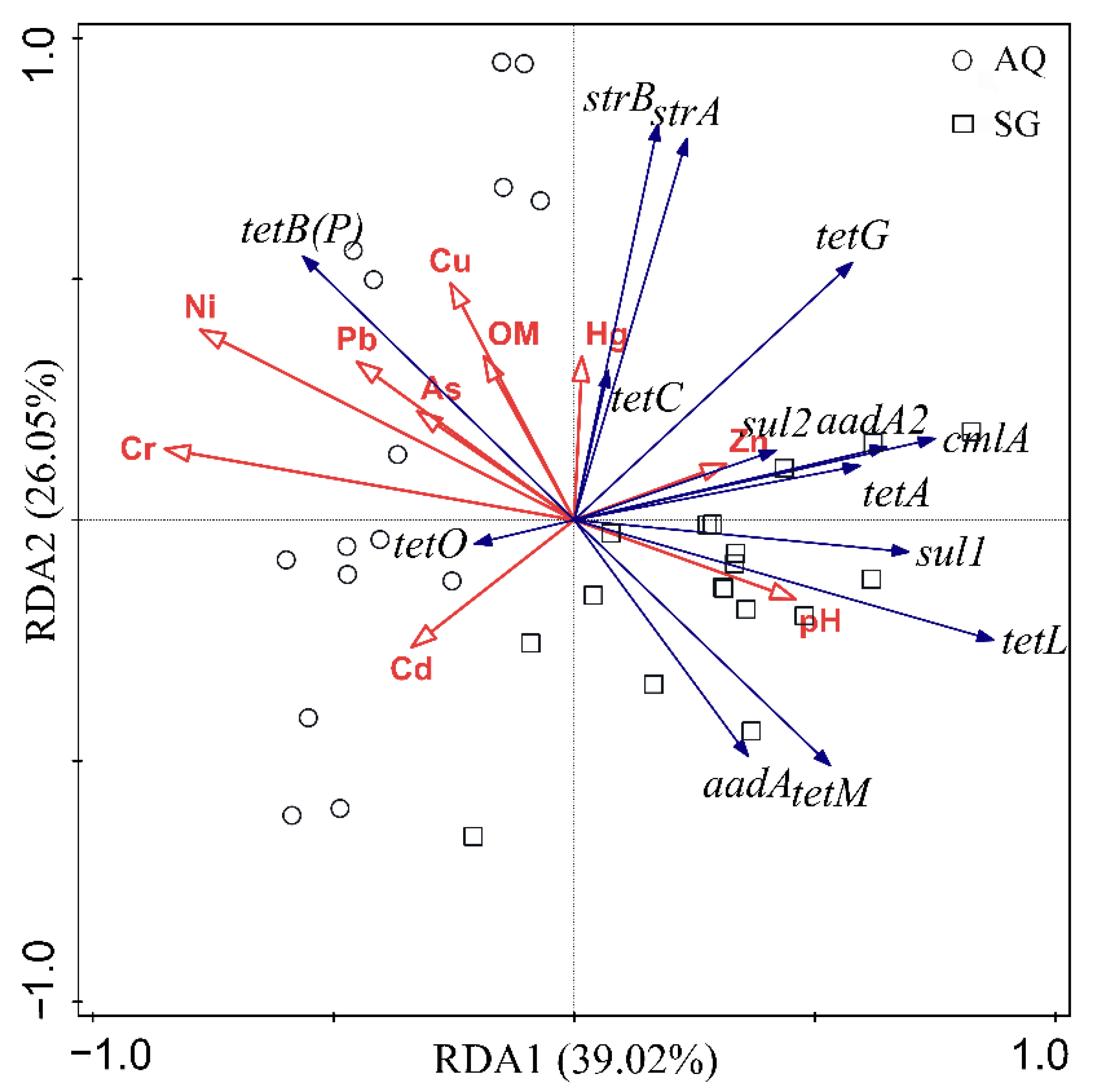
3.3. Factors Influencing ARGs and Bacterial Community
4. Discussion
4.1. Bacterial Community and Correlation with ARGs
4.2. Abundances of ARGs and intI Gene
4.3. Potential Hosts of ARGs and intI Gene
4.4. Contributions of Factors to Bacterial Community and ARGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, L.; Li, T.; Meng, H.; Xie, Y.; Zhang, J.; Hong, J. Effects of seven-year fertilization reclamation on bacterial community in a coal mining subsidence area in Shanxi, China. Int. J. Environ. Res. Public Health 2021, 18, 12504. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.N.; Wu, R.L.; Li, Y.Y.; Qin, Y.F.; Li, Y.L.; Meng, F.Q.; Wang, L.G.; Xu, F.L. Effects of pesticide residues on bacterial community diversity and structure in typical greenhouse soils with increasing cultivation years in Northern China. Sci. Total Environ. 2020, 710, 136321. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Feng, G.; Huang, H.; Chen, Y. Effects of emerging pollutants on the occurrence and transfer of antibiotic resistance genes: A review. J. Hazard. Mater. 2021, 420, 126602. [Google Scholar] [CrossRef] [PubMed]
- Koch, N.; Islam, N.F.; Sonowal, S.; Prasad, R.; Sarma, H. Environmental antibiotics and resistance genes as emerging contaminants: Methods of detection and bioremediation. Curr. Res. Microb. Sci. 2021, 2, 100027. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Song, W.; Lin, H.; Wang, W.; Du, L.; Xing, W. Antibiotics and antibiotic resistance genes in global lakes: A review and meta-analysis. Environ. Int. 2018, 116, 60–73. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Gu, J.; Gao, H.; Qian, X.; Li, H. Abundances of clinically relevant antibiotic resistance genes and bacterial community diversity in the Weihe river, China. Int. J. Environ. Res. Public Health 2018, 15, 708. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Baena, A.M.; Caicedo-Bejarano, L.D.; Chávez-Vivas, M. Structure of Bacterial Community with Resistance to Antibiotics in Aquatic Environments. A Systematic Review. Int. J. Environ. Res. Public Health 2021, 18, 2348. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, W.; Yang, L.; Stedtfeld, R.D.; Peng, A.; Gu, C.; Boyd, S.A.; Li, H. Antibiotic resistance genes and bacterial communities in cornfield and pasture soils receiving swine and dairy manures. Environ. Pollut. 2019, 248, 947–957. [Google Scholar] [CrossRef]
- Kaviani Rad, A.; Astaykina, A.; Streletskii, R.; Afsharyzad, Y.; Etesami, H.; Zarei, M.; Balasundram, S.K. An overview of antibiotic resistance and abiotic stresses affecting antimicrobial resistance in agricultural soils. Int. J. Environ. Res. Public Health 2022, 19, 4666. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, R.; Chen, B.; Zhang, T.; Hu, L.; Zou, S. Characterization of airborne antibiotic resistance genes from typical bioaerosol emission sources in the urban environment using metagenomic approach. Chemosphere 2018, 213, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Li, Y.; Liu, Y.; Sun, Y.; Xia, S.; Zhao, J. Effects of coexistence of tetracycline, copper and microplastics on the fate of antibiotic resistance genes in manured soil. Sci. Total Environ. 2021, 790, 148087. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Sun, R.; Hu, H.; Duan, G.; Meng, L.; Qiao, M. The overlap of soil and vegetable microbes drives the transfer of antibiotic resistance genes from manure-amended soil to vegetables. Sci. Total Environ. 2022, 828, 154463. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qiu, T.; Gao, M.; Sun, Y.; Cheng, S.; Gao, H.; Wang, X. Diversity and abundance of antibiotic resistance genes in rhizosphere soil and endophytes of leafy vegetables: Focusing on the effect of the vegetable species. J. Hazard. Mater. 2021, 415, 125595. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gu, A.Z.; Cen, T.; Li, X.; He, M.; Li, D.; Chen, J. Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ. Pollut. 2018, 237, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lan, B.; Fei, H.; Wang, S.; Zhu, G. Heavy metal could drive co-selection of antibiotic resistance in terrestrial subsurface soils. J. Hazard. Mater. 2021, 411, 124848. [Google Scholar] [CrossRef]
- Deng, W.; Zhang, A.; Chen, S.; He, X.; Jin, L.; Yu, X.; Yang, S.; Li, B.; Fan, L.; Ji, L.; et al. Heavy metals, antibiotics and nutrients affect the bacterial community and resistance genes in chicken manure composting and fertilized soil. J. Environ. Manag. 2020, 257, 109980. [Google Scholar] [CrossRef]
- Peng, S.; Feng, Y.; Wang, Y.; Guo, X.; Chu, H.; Lin, X. Prevalence of antibiotic resistance genes in soils after continually applied with different manure for 30 years. J. Hazard. Mater. 2017, 340, 16–25. [Google Scholar] [CrossRef]
- Zhao, X.; Shen, J.-P.; Zhang, L.-M.; Du, S.; Hu, H.-W.; He, J.-Z. Arsenic and cadmium as predominant factors shaping the distribution patterns of antibiotic resistance genes in polluted paddy soils. J. Hazard. Mater. 2020, 389, 121838. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.; Wang, J.; Zhu, L.; Conkle, J.L.; Yang, R. Soil types influence the characteristic of antibiotic resistance genes in greenhouse soil with long-term manure application. J. Hazard. Mater. 2020, 392, 122334. [Google Scholar] [CrossRef]
- Kianpoor Kalkhajeh, Y.; Huang, B.; Hu, W.; Ma, C.; Gao, H.; Thompson, M.L.; Bruun Hansen, H.C. Environmental soil quality and vegetable safety under current greenhouse vegetable production management in China. Agric. Ecosyst. Environ. 2021, 307, 107230. [Google Scholar] [CrossRef]
- Sun, J.; Pan, L.; Li, Z.; Zeng, Q.; Wang, L.; Zhu, L. Comparison of greenhouse and open field cultivations across China: Soil characteristics, contamination and microbial diversity. Environ. Pollut. 2018, 243, 1509–1516. [Google Scholar] [CrossRef]
- Pan, Z.; Yang, S.; Zhao, L.; Li, X.; Weng, L.; Sun, Y.; Li, Y. Temporal and spatial variability of antibiotics in agricultural soils from Huang-Huai-Hai Plain, northern China. Chemosphere 2021, 272, 129803. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, J.; Zhu, L.; Wang, J. Field-based evidence for enrichment of antibiotic resistance genes and mobile genetic elements in manure-amended vegetable soils. Sci. Total Environ. 2019, 654, 906–913. [Google Scholar] [CrossRef] [PubMed]
- Pu, Q.; Zhao, L.X.; Li, Y.T.; Su, J.Q. Manure fertilization increase antibiotic resistance in soils from typical greenhouse vegetable production bases, China. J. Hazard. Mater. 2020, 391, 122267. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Bing, H.; Fang, L.; Wu, Y.; Yu, J.; Shen, G.; Jiang, M.; Wang, X.; Zhang, X. Diversity patterns of the rhizosphere and bulk soil microbial communities along an altitudinal gradient in an alpine ecosystem of the eastern Tibetan Plateau. Geoderma 2019, 338, 118–127. [Google Scholar] [CrossRef]
- Xu, S.; Tian, L.; Chang, C.; Li, X.; Tian, C. Cultivated rice rhizomicrobiome is more sensitive to environmental shifts than that of wild rice in natural environments. Appl. Soil Ecol. 2019, 140, 68–77. [Google Scholar] [CrossRef]
- Yuan, X.; Gan, Y.; Zhang, Y.; Dong, B. Level, source, and risk assessment of toxic elements in traditional agricultural soils and coping strategies. Environ. Monit. Assess. 2021, 193, 568. [Google Scholar] [CrossRef]
- Zhi, S.; Zhou, J.; Liu, H.; Wu, H.; Zhang, Z.; Ding, Y.; Zhang, K. Simultaneous extraction and determination of 45 veterinary antibiotics in swine manure by liquid chromatography-tandem mass spectrometry. J. Chromatogr. B 2020, 1154, 122286. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Q.; Liang, B.; Li, J. Changes in the abundance and structure of bacterial communities in the greenhouse tomato cultivation system under long-term fertilization treatments. Appl. Soil Ecol. 2017, 121, 82–89. [Google Scholar] [CrossRef]
- Newton, R.J.; Mcmahon, K.D. Seasonal differences in bacterial community composition following nutrient additions in a eutrophic lake. Environ. Microbiol. 2011, 13, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Jia, S.; He, X.; Zhang, X.; Ye, L. Different impacts of manure and chemical fertilizers on bacterial community structure and antibiotic resistance genes in arable soils. Chemosphere 2017, 188, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Guo, Z.; Gu, X.; Gao, H.; Weng, S.; Zhou, J.; Gu, D.; Lu, H.; Zhou, X. Rare rather than abundant microbial communities drive the effects of long-term greenhouse cultivation on ecosystem functions in subtropical agricultural soils. Sci. Total Environ. 2020, 706, 136004. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, P.; Tian, H.; Xiao, Q.; Jiang, H. Pyrosequencing-based assessment of soil microbial community structure and analysis of soil properties with vegetable planted at different years under greenhouse conditions. Soil Tillage Res. 2019, 187, 1–10. [Google Scholar] [CrossRef]
- Pornsukarom, S.; Thakur, S. Assessing the impact of manure application in commercial swine farms on the transmission of antimicrobial resistant Salmonella in the environment. PLoS ONE 2016, 11, e0164621. [Google Scholar] [CrossRef]
- Pornsukarom, S.; Thakur, S. Horizontal dissemination of antimicrobial resistance determinants in multiple Salmonella serotypes following isolation from the commercial swine operation environment after manure application. Appl. Environ. Microbiol. 2017, 83, e01503–e01517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, S.; Wang, Y.; Zhou, B.; Lin, X. Long-term application of fresh and composted manure increase tetracycline resistance in the arable soil of eastern China. Sci. Total Environ. 2015, 506–507, 279–286. [Google Scholar] [CrossRef]
- Peng, S.; Dolfing, J.; Feng, Y.; Wang, Y.; Lin, X. Enrichment of the antibiotic resistance gene tet(L) in an alkaline soil fertilized with plant derived organic manure. Front. Microbiol. 2018, 9, 1140. [Google Scholar] [CrossRef]
- You, Y.; Hilpert, M.; Ward, M.J. Detection of a common and persistent tet(L)-carrying plasmid in chicken-waste-impacted farm soil. Appl. Environ. Microbiol. 2012, 78, 3203–3213. [Google Scholar] [CrossRef] [Green Version]
- Obayiuwana, A.; Ogunjobi, A.; Yang, M.; Ibekwe, M. Characterization of bacterial communities and their antibiotic resistance profiles in wastewaters obtained from pharmaceutical facilities in Lagos and Ogun States, Nigeria. Int. J. Environ. Res. Public Health 2018, 15, 1365. [Google Scholar] [CrossRef] [Green Version]
- Duan, M.; Gu, J.; Wang, X.; Li, Y.; Zhang, R.; Hu, T.; Zhou, B. Factors that affect the occurrence and distribution of antibiotic resistance genes in soils from livestock and poultry farms. Ecotoxicol. Environ. Saf. 2019, 180, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Radu, E.; Woegerbauer, M.; Rab, G.; Oismuller, M.; Strauss, P.; Hufnagl, P.; Gottsberger, R.A.; Krampe, J.; Weyermair, K.; Kreuzinger, N. Resilience of agricultural soils to antibiotic resistance genes introduced by agricultural management practices. Sci. Total Environ. 2021, 756, 143699. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; An, X.; Li, H.; Zhu, Y.; Su, J.; Cui, L. Do manure-borne or indigenous soil microorganisms influence the spread of antibiotic resistance genes in manured soil? Soil Biol. Biochem. 2017, 114, 229–237. [Google Scholar] [CrossRef]
- Dong, P.; Wang, H.; Fang, T.; Wang, Y.; Ye, Q. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 2019, 125, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Han, X.M.; Hu, H.W.; Chen, Q.L.; Yang, L.Y.; Li, H.L.; Zhu, Y.G.; Li, X.Z.; Ma, Y.B. Antibiotic resistance genes and associated bacterial communities in agricultural soils amended with different sources of animal manures. Soil Biol. Biochem. 2018, 126, 91–102. [Google Scholar] [CrossRef]
- Canal, N.; Meneghetti, K.L.; de Almeida, C.P.; da Rosa Bastos, M.; Otton, L.M.; Corcao, G. Characterization of the variable region in the class 1 integron of antimicrobial-resistant Escherichia coli isolated from surface water. Braz. J. Microbiol. 2016, 47, 337–344. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Qiu, X.; Wu, X.; Lu, S. Horizontal gene transfer is a key determinant of antibiotic resistance genes profiles during chicken manure composting with the addition of biochar and zeolite. J. Hazard. Mater. 2020, 408, 124883. [Google Scholar] [CrossRef]
- Li, J.; Xin, Z.; Zhang, Y.; Chen, J.; Yan, J.; Li, H.; Hu, H. Long-term manure application increased the levels of antibiotics and antibiotic resistance genes in a greenhouse soil. Appl. Soil Ecol. 2017, 121, 193–200. [Google Scholar] [CrossRef]
- Du, B.; Yang, Q.; Wang, R.; Wang, R.; Wang, Q.; Xin, Y. Evolution of antibiotic resistance and the relationship between the antibiotic resistance genes and microbial compositions under long-term exposure to tetracycline and sulfamethoxazole. Int. J. Environ. Res. Public Health 2019, 16, 4681. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Chen, Y.; Li, X.; Zhang, Y.; Ye, J.; Huang, H.; Zhu, C. Temporal effects of repeated application of biogas slurry on soil antibiotic resistance genes and their potential bacterial hosts. Environ. Pollut. 2020, 258, 113652. [Google Scholar] [CrossRef]
- Rahman, M.M.; Shan, J.; Yang, P.; Shang, X.; Xia, Y.; Yan, X. Effects of long-term pig manure application on antibiotics, abundance of antibiotic resistance genes (ARGs), anammox and denitrification rates in paddy soils. Environ. Pollut. 2018, 240, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Tseng, S.; Liang, C.; Chia, T.; Ton, S. Changes in the Composition of the Soil Bacterial Community in Heavy Metal-Contaminated Farmland. Int. J. Environ. Res. Public Health 2021, 18, 8661. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Shen, Q.; Wang, L.; Qiu, G.; Shi, J.; Xu, J.; Brookes, P.C.; Liu, X. Effects of Cd, Cu, Zn and their combined action on microbial biomass and bacterial community structure. Environ. Pollut. 2018, 243, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; LIang, C.M.; Yang, T.I.; Cheng, J.L.; Wang, W.K. Shift of bacterial communities in heavy metal-contaminated agricultural land during a remediation process. PLoS ONE 2021, 16, e0255137. [Google Scholar] [CrossRef]
- Tang, X.; Lou, C.; Wang, S.; Lu, Y.; Liu, M.; Hashmi, M.Z.; Liang, X.; Li, Z.; Liao, Y.; Qin, W.; et al. Effects of long-term manure applications on the occurrence of antibiotics and antibiotic resistance genes (ARGs) in paddy soils: Evidence from four field experiments in south of China. Soil Biol. Biochem. 2015, 90, 179–187. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y. Effects of clay combined with moisture management on Cd immobilization and fertility index of polluted rice field. Ecotoxicol. Environ. Saf. 2018, 158, 182–186. [Google Scholar] [CrossRef]
- He, L.; Ying, G.; Liu, Y.; Su, H.; Chen, J.; Liu, S.; Zhao, J. Discharge of swine wastes risks water quality and food safety: Antibiotics and antibiotic resistance genes from swine sources to the receiving environments. Environ. Int. 2016, 92–93, 210–219. [Google Scholar] [CrossRef]



| Factors | ARGs | Bacterial Community | ||
|---|---|---|---|---|
| Contribution % | p | Contribution % | p | |
| Cr | 29.2 | 0.002 | 2.1 | 0.46 |
| Cu | 12.2 | 0.006 | 7.9 | 0.004 |
| Cd | 12.4 | 0.006 | 13.5 | 0.002 |
| OM | 11.7 | 0.002 | 12.1 | 0.002 |
| pH | 10.3 | 0.004 | 17.7 | 0.002 |
| Ni | 8.0 | 0.01 | 30.6 | 0.002 |
| Zn | 7.6 | 0.016 | 6.5 | 0.008 |
| As | 3.9 | 0.092 | 4.9 | 0.032 |
| Pb | 2.4 | 0.238 | 0.9 | 0.896 |
| Hg | 2.3 | 0.226 | 3.9 | 0.092 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, X.; Zhang, Y.; Sun, C.; Wang, W.; Wu, Y.; Fan, L.; Liu, B. Profile of Bacterial Community and Antibiotic Resistance Genes in Typical Vegetable Greenhouse Soil. Int. J. Environ. Res. Public Health 2022, 19, 7742. https://doi.org/10.3390/ijerph19137742
Yuan X, Zhang Y, Sun C, Wang W, Wu Y, Fan L, Liu B. Profile of Bacterial Community and Antibiotic Resistance Genes in Typical Vegetable Greenhouse Soil. International Journal of Environmental Research and Public Health. 2022; 19(13):7742. https://doi.org/10.3390/ijerph19137742
Chicago/Turabian StyleYuan, Xuexia, Yong Zhang, Chenxi Sun, Wenbo Wang, Yuanjuan Wu, Lixia Fan, and Bing Liu. 2022. "Profile of Bacterial Community and Antibiotic Resistance Genes in Typical Vegetable Greenhouse Soil" International Journal of Environmental Research and Public Health 19, no. 13: 7742. https://doi.org/10.3390/ijerph19137742
APA StyleYuan, X., Zhang, Y., Sun, C., Wang, W., Wu, Y., Fan, L., & Liu, B. (2022). Profile of Bacterial Community and Antibiotic Resistance Genes in Typical Vegetable Greenhouse Soil. International Journal of Environmental Research and Public Health, 19(13), 7742. https://doi.org/10.3390/ijerph19137742