Viral Eco-Genomic Tools: Development and Implementation for Aquatic Biomonitoring
Abstract
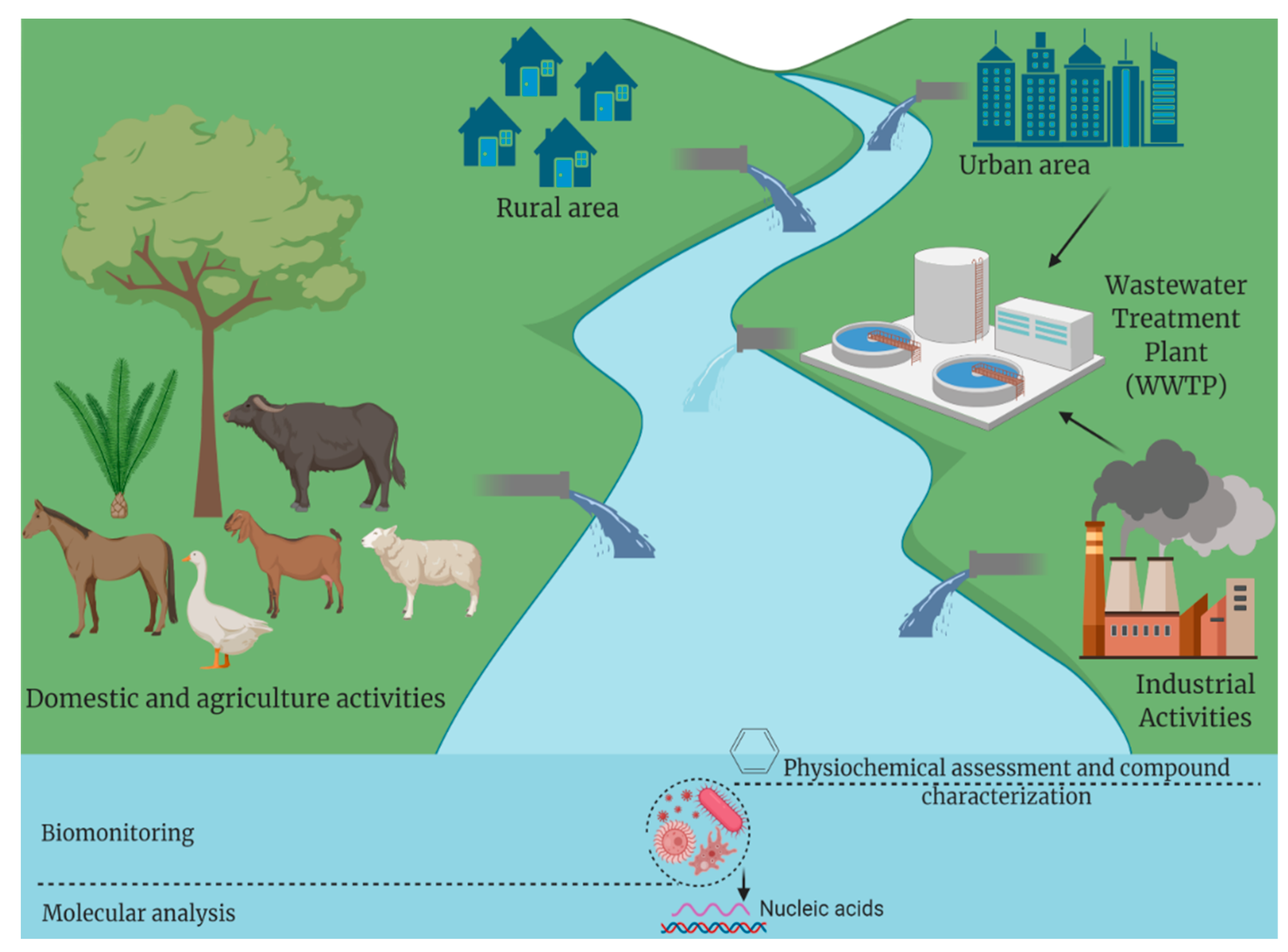
1. Introduction
2. Enteric Viruses in Water and Their Impact on Public Health
| Family | Genus | Virus Name | Genome | Size (Nm) | Types | Disease | Ref. |
|---|---|---|---|---|---|---|---|
| Picorna-viridae | Hepatovirus | Hepatitis virus | +ssRNA | 27–32 | 3 types | Hepatitis | [22] |
| Kobuvirus | Aichivirus | +ssRNA | 30 | 6 types | Gastroenteritis | [23] | |
| Enterovirus | Poliovirus | dsDNA | 40 | 14 species | Paralysis, aseptic meningitis | [24] | |
| Coxsackievirus | +ssRNA | 30 | Cox A1-23 and B1-6 | Myocarditis, aseptic meningitis, Bornholm disease and epidemic pleurodynia | [25] | ||
| Echovirus | +ssRNA | 30 | 28 types | Fever, rash, respiratory and heart disease, aseptic meningitis | [26] | ||
| Enterovirus | +ssRNA | 30 | 12 species | Gastroenteritis | [27] | ||
| Parechovirus | HPeV | +ssRNA | 28 | 19 genotypes | Gastroenteritis, respiratory and CNS diseases, and sepsis | [28] | |
| Cosavirus | HCoSV | +ssRNA | 30 | 5 species | Gastroenteritis non-polio AFP | [29] | |
| Aphthovirus | FMDV | +ssRNA | 25 | 7 serotypes | Respiratory diseases | [23] | |
| Adeno-viridae | Mastadenovirus | Adenovirus (AdV) | dsDNA | 70 | 60 types |
| [30] |
| Parvo-viridae | Erythrovirus | Parvovirus | ssDNA | 22 | 3 genotypes | Gastroenteritis | [31] |
| Bocavirus | Bocavirus | ssDNA | 20 | 4 genotypes | Respiratory diseases | [31,32] | |
| Reoviridae | Rotavirus | Rotaviruses | dsRNA | 80 | 9 species | Gastroenteritis | [33] |
| Hepeviridae | Hepevirus | Hepatitis E virus | +ssRNA | 27–34 | 4 genotypes | Infectious hepatitis | [34] |
| Picobirna-viridae | Picobirnavirus | Picobirnavirus | dsRNA | 35 | Human and Rabbit Picobirna-virus | Respiratory diseases and gastroenteritis | [35] |
| Astroviridae | Mamastrovirus | Astrovirus | +ssRNA | 35 | 8 serotypes | Gastroenteritis | [36] |
| Bunya-viridae | Hantavirus | Hantavirus | −ssRNA | 120 | 4 genera | Hemorrhagic fever and cardiopulmonary syndrome | [37] |
| Flavi-viridae | Flavivirus | TBEV | +ssRNA | 50 | 5 subtypes | Fever, meningitis and encephalitis | [38] |
| Arena-viridae | Arenavirus | Arenavirus | −ssRNA | 40–200 | 4 genera | Aseptic meningitis and hemorrhagic fever | [39] |
| Corona-viridae | Alphacorona-virus | HCoV-229E | +ssRNA | 120–140 | 7 subtypes |
| [11,40] |
| HCoV-NL63 | |||||||
| Betacoronavirus | HCoV-OC43 | ||||||
| HCoV-HKU1 | |||||||
| MERS-CoV | |||||||
| SARS-CoV | |||||||
| SARS-CoV-2 | |||||||
| Orthomyxo-viridae | Influenza A virus | AIVs (HSN1 and H9N2) | −ssRNA | 100 | Many subtypes |
| [41] |
| Paramyxo-viridae | Henipavirus | Nipah virus | −ssRNA | 40 | 2 genotypes (M and B) |
| [42] |
| Calici-viridae | Norovirus | Norovirus | +ssRNA | 27–40 | 9 genotypes | Gastroenteritis | [43,44] |
| Sapovirus | Sapovirus | +ssRNA | 27–40 | 18 genotypes | Gastroenteritis |
3. Waterborne Viruses Concentration Methods
3.1. Adsorption/Elution Method
3.2. Ultrafiltration
3.3. Ultracentrifugation
3.4. Hydro-Extraction Method
3.5. Freeze-Drying Technique
3.6. Antibody-Capture Technique
4. Conventional Viral Detection Tools for Aquatic Biomonitoring
4.1. Electron Microscopy
4.2. Cell Culture Systems
4.3. Immunological Methods
4.4. Biosensors
5. Viral Genetic Tools for Aquatic Biomonitoring
5.1. Polymerase Chain Reaction (PCR), Sequencing and Phylogenetic Analysis
5.2. Isothermal Nucleic Acid Amplification-Based Assays
5.3. DNA Microarray Technology
5.4. Next Generation Sequencing (NGS) and Metagenomic Technology
5.5. ChIP-Seq Analysis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bashir, I.; Lone, F.A.; Bhat, R.A.; Mir, S.A.; Dar, Z.A.; Dar, S.A. Concerns and threats of contamination on aquatic ecosystems. Bioremediat. Biotechnol. 2020, 1–26. [Google Scholar]
- Cordier, T.; Lanzén, A.; Apothéloz-Perret-Gentil, L.; Stoeck, T.; Pawlowski, J. Embracing environmental genomics and machine learning for routine biomonitoring. Trends Microbiol. 2019, 27, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Fong, T.-T.; Lipp, E.K. Enteric viruses of humans and animals in aquatic environments: Health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2005, 69, 357–371. [Google Scholar] [CrossRef] [PubMed]
- Okoh, A.I.; Sibanda, T.; Gusha, S.S. Inadequately treated wastewater as a source of human enteric viruses in the environment. Int. J. Environ. Res. Public Health 2010, 7, 2620–2637. [Google Scholar] [CrossRef]
- Tschope, C.; Ammirati, E.; Bozkurt, B.; Caforio, A.L.P.; Cooper, L.T.; Felix, S.B.; Hare, J.M.; Heidecker, B.; Heymans, S.; Hubner, N.; et al. Myocarditis and inflammatory cardiomyopathy: Current evidence and future directions. Nat. Rev. Cardiol. 2020, 18, 169–193. [Google Scholar] [CrossRef]
- Da Silva Silveira, C.; Maya, L.; Casaux, M.L.; Schild, C.; Caffarena, D.; Aráoz, V.; da Costa, R.A.; Macías-Rioseco, M.; Perdomo, Y.; Castells, M.; et al. Diseases associated with bovine viral diarrhea virus subtypes 1a and 2b in beef and dairy cattle in uruguay. Braz. J. Microbiol. 2020, 51, 357–368. [Google Scholar] [CrossRef]
- Bivins, A.; North, D.; Ahmad, A.; Ahmed, W.; Alm, E.; Been, F.; Bhattacharya, P.; Bijlsma, L.; Boehm, A.B.; Brown, J.; et al. Wastewater-based epidemiology: Global collaborative to maximize contributions in the fight against covid-19. Environ. Sci. Technol. 2020, 54, 7754–7757. [Google Scholar] [CrossRef]
- Haas, C.N.; Rose, J.B.; Gerba, C.P. Quantitative Microbial Risk Assessment; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- Bishop, R.F. Enteric viruses. In Encyclopedia of Virology, 2nd ed.; Granoff, A., Webster, R.G., Eds.; Elsevier: Oxford, UK, 1999; pp. 441–449. [Google Scholar]
- Rasa, S.; Nora-Krukle, Z.; Henning, N.; Eliassen, E.; Shikova, E.; Harrer, T.; Scheibenbogen, C.; Murovska, M.; Prusty, B.K.; The European Network on, M.C. Chronic viral infections in myalgic encephalomyelitis/chronic fatigue syndrome (me/cfs). J. Transl. Med. 2018, 16, 268. [Google Scholar] [CrossRef]
- Bouseettine, R.; Hassou, N.; Bessi, H.; Ennaji, M.M. Waterborne transmission of enteric viruses and their impact on public health. Emerg. Reemerging Viral Pathog. 2020, 1, 907–932. [Google Scholar] [CrossRef]
- Collivignarelli, M.C.; Abbà, A.; Benigna, I.; Sorlini, S.; Torretta, V. Overview of the main disinfection processes for wastewater and drinking water treatment plants. Sustainability 2018, 10, 86. [Google Scholar] [CrossRef]
- Thevenin, T.; Lobert, P.-E.; Hober, D. Inactivation of an enterovirus by airborne disinfectants. BMC Infect. Dis. 2013, 13, 177. [Google Scholar] [CrossRef] [PubMed]
- Hrušková, T.; Sasáková, N.; Gregova, G.; Papajová, I.; Bujdošová, Z. Disinfection of Water Used for Human and Animal Consumption; IntechOpen Limited: London, UK, 2018. [Google Scholar]
- Farkas, K.; Walker, D.I.; Adriaenssens, E.M.; McDonald, J.E.; Hillary, L.S.; Malham, S.K.; Jones, D.L. Viral indicators for tracking domestic wastewater contamination in the aquatic environment. Water Res. 2020, 181, 115926. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.; Salvador, D.; Brandão, J.; Ahmed, W.; Sadowsky, M.J.; Valério, E. Environmental and adaptive changes necessitate a paradigm shift for indicators of fecal contamination. Microbiol. Spectr. 2020, 8, 1–20. [Google Scholar] [CrossRef]
- Teixeira, P.; Costa, S.; Brown, B.; Silva, S.; Rodrigues, R.; Valério, E. Quantitative pcr detection of enteric viruses in wastewater and environmental water sources by the lisbon municipality: A case study. Water 2020, 12, 544. [Google Scholar] [CrossRef]
- Bondaruk, J.; Janson, E.; Wysocka, M.; Chałupnik, S. Identification of hazards for water environment in the upper silesian coal basin caused by the discharge of salt mine water containing particularly harmful substances and radionuclides. J. Sustain. Min. 2015, 14, 179–187. [Google Scholar] [CrossRef]
- Keck, F.; Vasselon, V.; Tapolczai, K.; Rimet, F.; Bouchez, A. Freshwater biomonitoring in the information age. Front. Ecol. Environ. 2017, 15, 266–274. [Google Scholar] [CrossRef]
- Zolkefli, N.; Sharuddin, S.S.; Yusoff, M.Z.M.; Hassan, M.A.; Maeda, T.; Ramli, N. A review of current and emerging approaches for water pollution monitoring. Water 2020, 12, 3417. [Google Scholar] [CrossRef]
- Hrdy, J.; Vasickova, P. Virus detection methods for different kinds of food and water samples—The importance of molecular techniques. Food Control 2022, 134, 108764. [Google Scholar] [CrossRef]
- Shouval, D. Chapter 29—Hepatitis a. In Zakim and Boyer’s Hepatology, 6th ed.; Boyer, T.D., Manns, M.P., Sanyal, A.J., Eds.; W.B. Saunders: Saint Louis, MO, USA, 2012; pp. 531–539. [Google Scholar]
- Rivadulla, E.; Romalde, J.L. A comprehensive review on human aichi virus. Virol. Sin. 2020, 35, 501–516. [Google Scholar] [CrossRef]
- Romero, J.R.; Modlin, J.F. 173—Poliovirus. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 8th ed.; Bennett, J.E., Dolin, R., Blaser, M.J., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2015; pp. 2073–2079.e2072. [Google Scholar]
- Johannessen, I.; Burns, S.M. 48—picornaviruses: Meningitis; paralysis; rashes; intercostal myositis; myocarditis; infectious hepatitis; common cold. In Medical Microbiology, 18th ed.; Greenwood, D., Barer, M., Slack, R., Irving, W., Eds.; Churchill Livingstone: Edinburgh, UK, 2012; pp. 483–496. [Google Scholar]
- Hyypiä, T.; Harvala, H. Echoviruses☆. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Romero, J.R. 164—Human enteroviruses. In Infectious Diseases, 4th ed.; Cohen, J., Powderly, W.G., Opal, S.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1406–1416.e1401. [Google Scholar]
- Olijve, L.; Jennings, L.; Walls, T. Human parechovirus: An increasingly recognized cause of sepsis-like illness in young infants. Clin. Microbiol. Rev. 2018, 31, e00047-17. [Google Scholar] [CrossRef]
- Yang, Y.; Ju, A.; Xu, X.; Cao, X.; Tao, Y. A novel type of cosavirus from children with nonpolio acute flaccid paralysis. Virol. J. 2016, 13, 169. [Google Scholar] [CrossRef] [PubMed]
- Flint, J.; Nemerow, G.R. Human Adenoviruses; World Scientific: Singapore, 2016; p. 416. [Google Scholar]
- Payne, S. Chapter 29—Family parvoviridae. In Viruses; Payne, S., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 237–241. [Google Scholar]
- Ison, M.G.; Lee, N. 173—Noninfluenza respiratory viruses. In Infectious Diseases, 4th ed.; Cohen, J., Powderly, W.G., Opal, S.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1472–1482.e1475. [Google Scholar]
- El-Senousy, W.M.; Abu Senna, A.S.M.; Mohsen, N.A.; Hasan, S.F.; Sidkey, N.M. Clinical and environmental surveillance of rotavirus common genotypes showed high prevalence of common p genotypes in egypt. Food Environ. Virol. 2020, 12, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Debing, Y.; Moradpour, D.; Neyts, J.; Gouttenoire, J. Update on hepatitis e virology: Implications for clinical practice. J. Hepatol. 2016, 65, 200–212. [Google Scholar] [CrossRef]
- Dolin, R.; Treanor, J.J. 179—Astroviruses and picobirnaviruses. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 8th ed.; Bennett, J.E., Dolin, R., Blaser, M.J., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2015; pp. 2128–2130.e2121. [Google Scholar]
- Hargest, V.; Davis, A.; Schultz-Cherry, S. Astroviridae. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Payne, S. Chapter 24—Family bunyaviridae. In Viruses; Payne, S., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 209–214. [Google Scholar]
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Chapter 36—Flaviviruses. In Fenner and White’s Medical Virology, 5th ed.; Burrell, C.J., Howard, C.R., Murphy, F.A., Eds.; Academic Press: London, UK, 2017; pp. 493–518. [Google Scholar]
- Radoshitzky, S.R.; Buchmeier, M.J.; Charrel, R.N.; Clegg, J.C.S.; Gonzalez, J.-P.J.; Günther, S.; Hepojoki, J.; Kuhn, J.H.; Lukashevich, I.S.; Romanowski, V.; et al. Ictv virus taxonomy profile: Arenaviridae. J. Gen. Virol. 2019, 100, 1200–1201. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of sars-cov-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef]
- De Graaf, M.; Beck, R.; Caccio, S.M.; Duim, B.; Fraaij, P.L.A.; Le Guyader, F.S.; Lecuit, M.; Le Pendu, J.; de Wit, E.; Schultsz, C. Sustained fecal-oral human-to-human transmission following a zoonotic event. Curr. Opin. Virol. 2017, 22, 1–6. [Google Scholar] [CrossRef]
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Chapter 26—paramyxoviruses. In Fenner and White’s Medical Virology, 5th ed.; Burrell, C.J., Howard, C.R., Murphy, F.A., Eds.; Academic Press: London, UK, 2017; pp. 367–382. [Google Scholar]
- Gelaw, A.; Pietsch, C.; Mann, P.; Liebert, U.G. Molecular detection and characterisation of sapoviruses and noroviruses in outpatient children with diarrhoea in northwest ethiopia. Epidemiol. Infect. 2019, 147, e218. [Google Scholar] [CrossRef]
- Payne, S. Chapter 12—Family caliciviridae. In Viruses; Payne, S., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 115–119. [Google Scholar]
- Haramoto, E.; Kitajima, M.; Hata, A.; Torrey, J.R.; Masago, Y.; Sano, D.; Katayama, H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018, 135, 168–186. [Google Scholar] [CrossRef]
- Block, J.C. Viruses in Water Systems: Detection and Identification; VCH: New York, NY, USA, 1989. [Google Scholar]
- Jothikumar, N.; Khanna, P.; Paulmurugan, R.; Kamatchiammal, S.; Padmanabhan, P. A simple device for the concentration and detection of enterovirus, hepatitis e virus and rotavirus from water samples by reverse transcription-polymerase chain reaction. J. Virol. Methods 1995, 55, 401–415. [Google Scholar] [CrossRef]
- Gerba, C. Recovering Viruses from Sewage, Effluents, and Water; CRC Press: Boca Raton, FL, USA, 2018; pp. 1–23. [Google Scholar]
- Rotem-Borensztajn, Y.; Belfort, G.; Katzenelson, E. Virus concentration by ultrafiltration. J. Environ. Eng. Div. 1979, 105, 401–407. [Google Scholar] [CrossRef]
- Urase, T.; Yamamoto, K.; Ohgaki, S. Evaluation of virus removal in membrane separation processes using coliphage qβ. Water Sci. Technol. 1993, 28, 9–15. [Google Scholar] [CrossRef]
- Prata, C.; Ribeiro, A.; Cunha, Â.; Gomes, N.C.; Almeida, A. Ultracentrifugation as a direct method to concentrate viruses in environmental waters: Virus-like particle enumeration as a new approach to determine the efficiency of recovery. J. Environ. Monit. JEM 2012, 14, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Fumian, T.M.; Leite, J.P.; Castello, A.A.; Gaggero, A.; Caillou, M.S.; Miagostovich, M.P. Detection of rotavirus a in sewage samples using multiplex qpcr and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J. Virol. Methods 2010, 170, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Schultz, A.C.; Perelle, S.; Di Pasquale, S.; Kovac, K.; De Medici, D.; Fach, P.; Sommer, H.M.; Hoorfar, J. Collaborative validation of a rapid method for efficient virus concentration in bottled water. Int. J. Food Microbiol. 2011, 145, S158–S166. [Google Scholar] [CrossRef]
- Ramia, S.; Sattar, S.A. Second-step concentration of viruses in drinking and surface waters using polyethylene glycol hydroextraction. Can. J. Microbiol. 1979, 25, 587–592. [Google Scholar] [CrossRef]
- Kittigul, L.; Khamoun, P.; Sujirarat, D.; Utrarachkij, F.; Chitpirom, K.; Chaichantanakit, N.; Vathanophas, K. An improved method for concentrating rotavirus from water samples. Mem. Do Inst. Oswaldo Cruz 2001, 96, 815–821. [Google Scholar] [CrossRef]
- Myrmel, M.; Rimstad, E.; Wasteson, Y. Immunomagnetic separation of a norwalk-like virus (genogroup i) in artificially contaminated environmental water samples. Int. J. Food Microbiol. 2000, 62, 17–26. [Google Scholar] [CrossRef]
- Reagan, R.L.; Brueckner, A.L. Morphological observations by electron microscopy of the lansing strain of poliomyelitis virus after propagation in the swiss albino mouse. Tex. Rep. Biol. Med. 1952, 10, 425–428. [Google Scholar]
- Malik, Y.S.; Verma, A.K.; Kumar, N.; Touil, N.; Karthik, K.; Tiwari, R.; Bora, D.P.; Dhama, K.; Ghosh, S.; Hemida, M.G.; et al. Advances in diagnostic approaches for viral etiologies of diarrhea: From the lab to the field. Front. Microbiol. 2019, 10, 1957. [Google Scholar] [CrossRef]
- Goldsmith, C.S.; Miller, S.E. Modern uses of electron microscopy for detection of viruses. Clin. Microbiol. Rev. 2009, 22, 552–563. [Google Scholar] [CrossRef]
- Richert-Pöggeler, K.R.; Franzke, K.; Hipp, K.; Kleespies, R.G. Electron microscopy methods for virus diagnosis and high resolution analysis of viruses. Front. Microbiol. 2019, 9, 3255. [Google Scholar] [CrossRef] [PubMed]
- Spackman, E.; Killian, M.L. Avian influenza virus isolation, propagation, and titration in embryonated chicken eggs. Methods Mol. Biol. 2014, 1161, 125–140. [Google Scholar]
- Jogler, C.; Hoffmann, D.; Theegarten, D.; Grunwald, T.; Uberla, K.; Wildner, O. Replication properties of human adenovirus in vivo and in cultures of primary cells from different animal species. J. Virol. 2006, 80, 3549–3558. [Google Scholar] [CrossRef] [PubMed]
- Kolawole, A.O.; Mirabelli, C.; Hill, D.R.; Svoboda, S.A.; Janowski, A.B.; Passalacqua, K.D.; Rodriguez, B.N.; Dame, M.K.; Freiden, P.; Berger, R.P.; et al. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog. 2019, 15, e1008057. [Google Scholar] [CrossRef] [PubMed]
- Ghietto, L.M.; Toigo D’Angelo, A.P.; Viale, F.A.; Adamo, M.P. Human bocavirus 1 infection of caco-2 cell line cultures. Virology 2017, 510, 273–280. [Google Scholar] [CrossRef]
- Huang, Q.; Deng, X.; Yan, Z.; Cheng, F.; Luo, Y.; Shen, W.; Lei-Butters, D.C.; Chen, A.Y.; Li, Y.; Tang, L.; et al. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog. 2012, 8, e1002899. [Google Scholar] [CrossRef]
- Dijkman, R.; Koekkoek, S.M.; Molenkamp, R.; Schildgen, O.; van der Hoek, L. Human bocavirus can be cultured in differentiated human airway epithelial cells. J. Virol. 2009, 83, 7739. [Google Scholar] [CrossRef] [PubMed]
- Soliman, M.; Kim, D.-S.; Kim, C.; Seo, J.-Y.; Kim, J.-Y.; Park, J.-G.; Alfajaro, M.M.; Baek, Y.-B.; Cho, E.-H.; Park, S.-I.; et al. Porcine sapovirus cowden strain enters llc-pk cells via clathrin- and cholesterol-dependent endocytosis with the requirement of dynamin ii. Vet. Res. 2018, 49, 92. [Google Scholar] [CrossRef] [PubMed]
- Schildgen, O.; Jebbink, M.F.; de Vries, M.; Pyrc, K.; Dijkman, R.; Simon, A.; Müller, A.; Kupfer, B.; van der Hoek, L. Identification of cell lines permissive for human coronavirus nl63. J. Virol. Methods 2006, 138, 207–210. [Google Scholar] [CrossRef]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.J.C.; Kikkert, M.; Barcena, M.; et al. Sars-coronavirus-2 replication in vero e6 cells: Replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020, 101, 925–940. [Google Scholar] [CrossRef]
- Brown, B.A.; Nix, W.A.; Sheth, M.; Frace, M.; Oberste, M.S. Seven strains of enterovirus d68 detected in the united states during the 2014 severe respiratory disease outbreak. Genome Announc. 2014, 2, e01201-14. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, H. Efficient cell culture systems for hepatitis e virus strains in feces and circulating blood. Rev. Med. Virol. 2011, 21, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Sasaki, R.; Masuzaki, R.; Matsumoto, N.; Ogawa, M.; Moriyama, M. Cell culture systems and drug targets for hepatitis a virus infection. Viruses 2020, 12, 533. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.K.; Grau, K.R.; Costantini, V.; Kolawole, A.O.; de Graaf, M.; Freiden, P.; Graves, C.L.; Koopmans, M.; Wallet, S.M.; Tibbetts, S.A.; et al. Human norovirus culture in b cells. Nat. Protoc. 2015, 10, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Hisaie, K.; Kurokawa, S.; Suzuki, A.; Sakon, N.; Uchida, Y.; Yuki, Y.; Kiyono, H. Human norovirus propagation in human induced pluripotent stem cell-derived intestinal epithelial cells. Cell Mol. Gastroenterol. Hepatol. 2019, 7, 686–688.e685. [Google Scholar] [CrossRef]
- Otto, P.H.; Reetz, J.; Eichhorn, W.; Herbst, W.; Elschner, M.C. Isolation and propagation of the animal rotaviruses in ma-104 cells--30 years of practical experience. J. Virol. Methods 2015, 223, 88–95. [Google Scholar] [CrossRef]
- Superti, F.; Tinari, A.; Baldassarri, L.; Donelli, G. Ht-29 cells: A new substrate for rotavirus growth. Arch. Virol. 1991, 116, 159–173. [Google Scholar] [CrossRef]
- Barreto, A.; Rodriguez, L.S.; Rojas, O.L.; Wolf, M.; Greenberg, H.B.; Franco, M.A.; Angel, J. Membrane vesicles released by intestinal epithelial cells infected with rotavirus inhibit t-cell function. Viral Immunol. 2010, 23, 595–608. [Google Scholar] [CrossRef]
- Berry, J.M.; Barnabé, N.; Coombs, K.M.; Butler, M. Production of reovirus type-1 and type-3 from vero cells grown on solid and macroporous microcarriers. Biotechnol. Bioeng. 1999, 62, 12–19. [Google Scholar] [CrossRef]
- Leland, D.S.; Ginocchio, C.C. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 2007, 20, 49–78. [Google Scholar] [CrossRef]
- Lee, H.; Park, Y.; Kim, M.; Jee, Y.; Cheon, D.-S.; Jeong, H.S.; Ko, G. Development of a latex agglutination test for norovirus detection. J. Microbiol. 2010, 48, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Khoris, I.M.; Takemura, K.; Lee, J.; Hara, T.; Abe, F.; Suzuki, T.; Park, E.Y. Enhanced colorimetric detection of norovirus using in-situ growth of ag shell on au nps. Biosens. Bioelectron. 2019, 126, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Henriquez, L.; Brenes-Acuna, M.; Castro-Rojas, A.; Cordero-Salmeron, R.; Lopretti-Correa, M.; Vega-Baudrit, J.R. Biosensors for the detection of bacterial and viral clinical pathogens. Sensors 2020, 20, 6926. [Google Scholar] [CrossRef]
- Khan, A.; Rao, T.S. Chapter 4—Nanobiosensors for virus detection in the environment. In Nanomaterials for Air Remediation; Abdeltif, A., Assadi, A.A., Nguyen-Tri, P., Nguyen, T.A., Rtimi, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 61–87. [Google Scholar]
- Hashemi Goradel, N.; Mirzaei, H.; Sahebkar, A.; Poursadeghiyan, M.; Masoudifar, A.; Malekshahi, Z.V.; Negahdari, B. Biosensors for the detection of environmental and urban pollutions. J. Cell. Biochem. 2018, 119, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Yin, J.; Lv, S.; Wang, B.; Mu, Y. Advanced “lab-on-a-chip” to detect viruses—Current challenges and future perspectives. Biosens. Bioelectron. 2020, 163, 112291. [Google Scholar] [CrossRef]
- Hamza, I.A.; Bibby, K. Critical issues issues in application of molecular methods to environmental virology. J. Virol. Methods 2019, 266, 11–24. [Google Scholar] [CrossRef]
- Borchardt, M.A.; Bertz, P.D.; Spencer, S.K.; Battigelli, D.A. Incidence of enteric viruses in groundwater from household wells in wisconsin. Appl. Environ. Microbiol. 2003, 69, 1172–1180. [Google Scholar] [CrossRef]
- Van Heerden, J.; Ehlers, M.M.; Van Zyl, W.B.; Grabow, W.O.K. Incidence of adenoviruses in raw and treated water. Water Res. 2003, 37, 3704–3708. [Google Scholar] [CrossRef]
- Jiang, S.; Noble, R.; Chu, W. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of southern california. Appl. Environ. Microbiol. 2001, 67, 179–184. [Google Scholar] [CrossRef]
- Katayama, H.; Shimasaki, A.; Ohgaki, S. Development of a virus concentration method and its application to detection of enterovirus and norwalk virus from coastal seawater. Appl. Environ. Microbiol. 2002, 68, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, R.G.; Jones, E.L.; Gerba, C.P. Viruses in recreational water-borne disease outbreaks: A review. J. Appl. Microbiol. 2009, 107, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Hamza, I.A.; Jurzik, L.; Uberla, K.; Wilhelm, M. Methods to detect infectious human enteric viruses in environmental water samples. Int. J. Hyg. Envir. Health 2011, 214, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Ahmed, W.; Oikarinen, S.; Sherchan, S.P.; Heikinheimo, A.; Jiang, G.; Simpson, S.L.; Greaves, J.; Bivins, A. Application of digital pcr for public health-related water quality monitoring. Sci. Total Environ. 2022, 837, 155663. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, M.; Oka, T.; Haramoto, E.; Phanuwan, C.; Takeda, N.; Katayama, K.; Katayama, H. Genetic diversity of genogroup iv noroviruses in wastewater in japan. Lett. Appl. Microbiol. 2011, 52, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Kazama, S.; Miura, T.; Masago, Y.; Konta, Y.; Tohma, K.; Manaka, T.; Liu, X.F.; Nakayama, D.; Tanno, T.; Saito, M.; et al. Environmental surveillance of norovirus genogroups i and ii for sensitive detection of epidemic variants. Appl. Environ. Microb. 2017, 83, e03406-16. [Google Scholar] [CrossRef] [PubMed]
- Wongboot, W.; Okada, K.; Chantaroj, S.; Kamjumphol, W.; Hamada, S. Simultaneous detection and quantification of 19 diarrhea-related pathogens with a quantitative real-time pcr panel assay. J. Microbiol. Methods 2018, 151, 76–82. [Google Scholar] [CrossRef]
- Kowada, K.; Takeuchi, K.; Hirano, E.; Toho, M.; Sada, K. Development of a multiplex real-time pcr assay for detection of human enteric viruses other than norovirus using samples collected from gastroenteritis patients in fukui prefecture, japan. J. Med. Virol. 2018, 90, 67–75. [Google Scholar] [CrossRef]
- Mo, Q.H.; Wang, H.B.; Tan, H.; Wu, B.M.; Feng, Z.L.; Wang, Q.; Lin, J.C.; Yang, Z. Comparative detection of rotavirus rna by conventional rt-pcr, taqman rt-pcr and real-time nucleic acid sequence-based amplification. J. Virol. Methods 2015, 213, 1–4. [Google Scholar] [CrossRef]
- Wang, H.; Cong, F.; Zeng, F.; Lian, Y.; Liu, X.; Luo, M.; Guo, P.; Ma, J. Development of a real time reverse transcription loop-mediated isothermal amplification method (rt-lamp) for detection of a novel swine acute diarrhea syndrome coronavirus (sads-cov). J. Virol. Methods 2018, 260, 45–48. [Google Scholar] [CrossRef]
- Yu, X.; Shi, L.; Lv, X.; Yao, W.; Cao, M.; Yu, H.; Wang, X.; Zheng, S. Development of a real-time reverse transcription loop-mediated isothermal amplification method for the rapid detection of porcine epidemic diarrhea virus. Virol. J. 2015, 12, 76. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, Y.; Yan, H.; Mammel, M.; Chen, H. Sequence-independent amplification coupled with DNA microarray analysis for detection and genotyping of noroviruses. AMB Express 2015, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Jaykus, L.A. Development of a recombinase polymerase amplification assay for detection of epidemic human noroviruses. Sci. Rep. 2017, 7, 40244. [Google Scholar] [CrossRef] [PubMed]
- Aebischer, A.; Wernike, K.; Hoffmann, B.; Beer, M. Rapid genome detection of schmallenberg virus and bovine viral diarrhea virus by use of isothermal amplification methods and high-speed real-time reverse transcriptase pcr. J. Clin. Microbiol. 2014, 52, 1883–1892. [Google Scholar] [CrossRef]
- Tsai, S.K.; Chen, C.C.; Lin, H.J.; Lin, H.Y.; Chen, T.T.; Wang, L.C. Combination of multiplex reverse transcription recombinase polymerase amplification assay and capillary electrophoresis provides high sensitive and high-throughput simultaneous detection of avian influenza virus subtypes. J. Vet. Sci. 2020, 21, e24. [Google Scholar] [CrossRef]
- Gilbride, K. Molecular methods for the detection of waterborne pathogens. In Waterborne Pathogens; Elsevier: Amsterdam, The Netherlands, 2014; pp. 231–290. [Google Scholar]
- Perot, P.; Lecuit, M.; Eloit, M. Astrovirus diagnostics. Viruses 2017, 9, 10. [Google Scholar] [CrossRef]
- Wong, M.V.; Hashsham, S.A.; Gulari, E.; Rouillard, J.-M.; Aw, T.G.; Rose, J.B. Detection and characterization of human pathogenic viruses circulating in community wastewater using multi target microarrays and polymerase chain reaction. J. Water Health 2013, 11, 659–670. [Google Scholar] [CrossRef]
- Kim, J.-M.; Kim, S.Y.; Park, Y.B.; Kim, H.J.; Min, B.S.; Cho, J.-C.; Yang, J.M.; Cho, Y.-H.; Ko, G. Simultaneous detection of major enteric viruses using a combimatrix microarray. J. Microbiol. 2012, 50, 970–977. [Google Scholar] [CrossRef]
- Chin, A.W.H.; Chu, J.T.S.; Perera, M.R.A.; Hui, K.P.Y.; Yen, H.L.; Chan, M.C.W.; Peiris, M.; Poon, L.L.M. Stability of sars-cov-2 in different environmental conditions. Lancet Microbe 2020, 1, e10. [Google Scholar] [CrossRef]
- Angly, F.E.; Felts, B.; Breitbart, M.; Salamon, P.; Edwards, R.A.; Carlson, C.; Chan, A.M.; Haynes, M.; Kelley, S.; Liu, H.; et al. The marine viromes of four oceanic regions. PLoS Biol. 2006, 4, 2121–2131. [Google Scholar] [CrossRef]
- Maljkovic Berry, I.; Melendrez, M.C.; Bishop-Lilly, K.A.; Rutvisuttinunt, W.; Pollett, S.; Talundzic, E.; Morton, L.; Jarman, R.G. Next generation sequencing and bioinformatics methodologies for infectious disease research and public health: Approaches, applications, and considerations for development of laboratory capacity. J. Infect. Dis. 2020, 221, S292–S307. [Google Scholar] [CrossRef] [PubMed]
- Aw, T.G.; Howe, A.; Rose, J.B. Metagenomic approaches for direct and cell culture evaluation of the virological quality of wastewater. J. Virol. Methods 2014, 210, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Hamza, H.; Leifels, M.; Wilhelm, M.; Hamza, I.A. Relative abundance of human bocaviruses in urban sewage in greater cairo, egypt. Food Environ. Virol. 2017, 9, 304–313. [Google Scholar] [CrossRef]
- Bibby, K.; Crank, K.; Greaves, J.; Li, X.; Wu, Z.Y.; Hamza, I.A.; Stachler, E. Metagenomics and the development of viral water quality tools. Npj Clean Water 2019, 2, 9. [Google Scholar] [CrossRef]
- Vazquez-Castellanos, J.F.; Garcia-Lopez, R.; Perez-Brocal, V.; Pignatelli, M.; Moya, A. Comparison of different assembly and annotation tools on analysis of simulated viral metagenomic communities in the gut. BMC Genom. 2014, 15, 37. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.; Oliveira, J.; Sousa, M. Bioinformatics and computational tools for next-generation sequencing analysis in clinical genetics. J. Clin. Med. 2020, 9, 132. [Google Scholar] [CrossRef]
- Hunter, O.V.; Sei, E.; Richardson, R.B.; Conrad, N.K. Chromatin immunoprecipitation and microarray analysis suggest functional cooperation between kaposi’s sarcoma-associated herpesvirus orf57 and k-bzip. J. Virol 2013, 87, 4005–4016. [Google Scholar] [CrossRef]
- Zheng, Y.; Hearing, P. The use of chromatin immunoprecipitation (chip) to study the binding of viral proteins to the adenovirus genome in vivo. Methods Mol. Biol. (Clifton N.J.) 2014, 1089, 79–87. [Google Scholar]
- Yang, J.; Hearing, P. Chromatin immunoprecipitation to study the binding of proteins to the adenovirus genome in vivo. Methods Mol. Med. 2007, 131, 113–121. [Google Scholar]
- Schramlova, J.; Arientova, S.; Hulinska, D. The role of electron microscopy in the rapid diagnosis of viral infections--review. Folia Microbiol. 2010, 55, 88–101. [Google Scholar] [CrossRef]
- Kfir, R.; Genthe, B. Advantages and disadvantages of the use of immunodetection techniques for the enumeration of microorganisms and toxins in water. Wat. Sci. Technol 1993, 27, 243–252. [Google Scholar] [CrossRef]
- Fout, G.S.; Martinson, B.C.; Moyer, M.W.N.; Dahling, D.R. A multiplex reverse transcription-pcr method for detection of human enteric viruses in groundwater. Appl. Environ. Microb. 2003, 69, 3158–3164. [Google Scholar] [CrossRef] [PubMed]
- Greening, G.E.; Hewitt, J.; Lewis, G.D. Evaluation of integrated cell culture-pcr (c-pcr) for virological analysis of environmental samples. J. Appl. Microbiol. 2002, 93, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, Y.; Ding, F.; Chen, D.; Wang, Y.; Jin, X.; Zhu, X. Rational design of electroactive redox enzyme nanocapsules for high-performance biosensors and enzymatic biofuel cell. Biosens. Bioelectron. 2021, 174, 112805. [Google Scholar] [CrossRef]
- Park, P.J. Chip–seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009, 10, 669–680. [Google Scholar] [CrossRef]
| Virus | Cell Line | Origin | Ref. |
|---|---|---|---|
| Avian influenza viruses | SPF-ECE | Specific pathogen-free embryonated chicken egg | [61] |
| MDCK | Madin–Darby Canine Kidney cell | ||
| Vero | African green monkey kidney cell | ||
| Adenovirus | A549 | Human lung carcinoma cell | [62] |
| PK-15 | Porcine kidney epithelial cell | ||
| Astrovirus | HEK | Human embryo kidney | [63] |
| Caco-2 | Human colorectal adenocarcinoma cell | ||
| A549 | Human lung carcinoma cell | ||
| Bocavirus | Caco-2 | Human colorectal adenocarcinoma cell | [64] |
| HEK293 | Human embryonic kidney cell | [65] | |
| HTEpC | Human trachea epithelial primary cell | [66] | |
| Coxsackievirus | HeLa | Human cervical cancer cell | [67] |
| Coronavirus | MRC-5 | Human fetal lung fibroblast cell | [68] |
| Vero-E6 | African green monkey kidney cell | [69] | |
| Enterovirus | RD | Human muscle tissue | [70] |
| HEV | A549 | Human lung carcinoma cell | [71] |
| HAV | Caco-2 | Human colorectal adenocarcinoma cell | Reviewed in [72] |
| HepG2-N | Human hepatoma | ||
| Huh-7 | Hepatocarcinoma cell | ||
| MRC-5 | Human fetal lung fibroblast cell | ||
| Vero | African green monkey kidney cell | ||
| Norovirus | BJAB | Human B cell lines | [73] |
| iPSC–derived IECs | Human induced pluripotent stem cell | [74] | |
| Rotavirus | MA-104 | African green monkey epithelial cell | [75] |
| HT-29 | Human colon carcinoma cell line | [76] | |
| Caco-2 | Human colorectal adenocarcinoma cell | [77] | |
| Reovirus | Vero | African green monkey | [78] |
| Sapovirus | LLC-PK1 | Porcine kidney cell | [67] |
| Technique | Principle | EVs to Detect | Ref. |
|---|---|---|---|
| Nucleic acid sequence-based amplification (NASBA) |
| Human adenovirus and echovirus | [58,99] |
| Loop-mediated isothermal amplification (LAMP) |
| Noroviruses and swine acute diarrhea syndrome-coronavirus | [58,100,101] |
| Single primer iso-thermal amplification (SPIA) |
| Human norovirus | [58,102] |
| Recombinase polymerase amplification (RPA) |
| Human norovirus, avian influenza virus and bovine viral diarrhea virus (BVDV) | [58,103,104,105] |
| Method | Advantages | Disadvantages | Ref. |
|---|---|---|---|
| Electron microscopy |
|
| [121] |
| Cell culture |
|
| [16] |
| Immunological methods (e.g., ELISA and latex agglutination technique) |
|
| [122] |
| Polymerase chain reaction (PCR) |
|
| [16] |
| Nested and semi-nested PCR |
|
| [89] |
| Multiplex PCR |
|
| [123] |
| Real-time PCR (rt-PCR or qPCR) |
|
| [16] |
| ICC-PCR |
|
| [124] |
| Digital PCR |
|
| [16,94] |
| Isothermal nucleic acid amplification-based assays |
|
| [16] |
| Biosensors |
|
| [125] |
| Microarrays |
|
| [16] |
| Metagenomics or next-generation sequencing (NGS) |
|
| [16] |
| ChIP-Seq analysis |
|
| [126] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mostafa-Hedeab, G.; Allayeh, A.K.; Elhady, H.A.; Eledrdery, A.Y.; Mraheil, M.A.; Mostafa, A. Viral Eco-Genomic Tools: Development and Implementation for Aquatic Biomonitoring. Int. J. Environ. Res. Public Health 2022, 19, 7707. https://doi.org/10.3390/ijerph19137707
Mostafa-Hedeab G, Allayeh AK, Elhady HA, Eledrdery AY, Mraheil MA, Mostafa A. Viral Eco-Genomic Tools: Development and Implementation for Aquatic Biomonitoring. International Journal of Environmental Research and Public Health. 2022; 19(13):7707. https://doi.org/10.3390/ijerph19137707
Chicago/Turabian StyleMostafa-Hedeab, Gomaa, Abdou Kamal Allayeh, Hany Abdelfattah Elhady, Abozer Y. Eledrdery, Mobarak Abu Mraheil, and Ahmed Mostafa. 2022. "Viral Eco-Genomic Tools: Development and Implementation for Aquatic Biomonitoring" International Journal of Environmental Research and Public Health 19, no. 13: 7707. https://doi.org/10.3390/ijerph19137707
APA StyleMostafa-Hedeab, G., Allayeh, A. K., Elhady, H. A., Eledrdery, A. Y., Mraheil, M. A., & Mostafa, A. (2022). Viral Eco-Genomic Tools: Development and Implementation for Aquatic Biomonitoring. International Journal of Environmental Research and Public Health, 19(13), 7707. https://doi.org/10.3390/ijerph19137707








