The Removal of Erythromycin and Its Effects on Anaerobic Fermentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Anaerobic Fermentation Experiment
2.3. Chemical and Microbial Analyses
2.4. Data Analysis
3. Results and Discussion
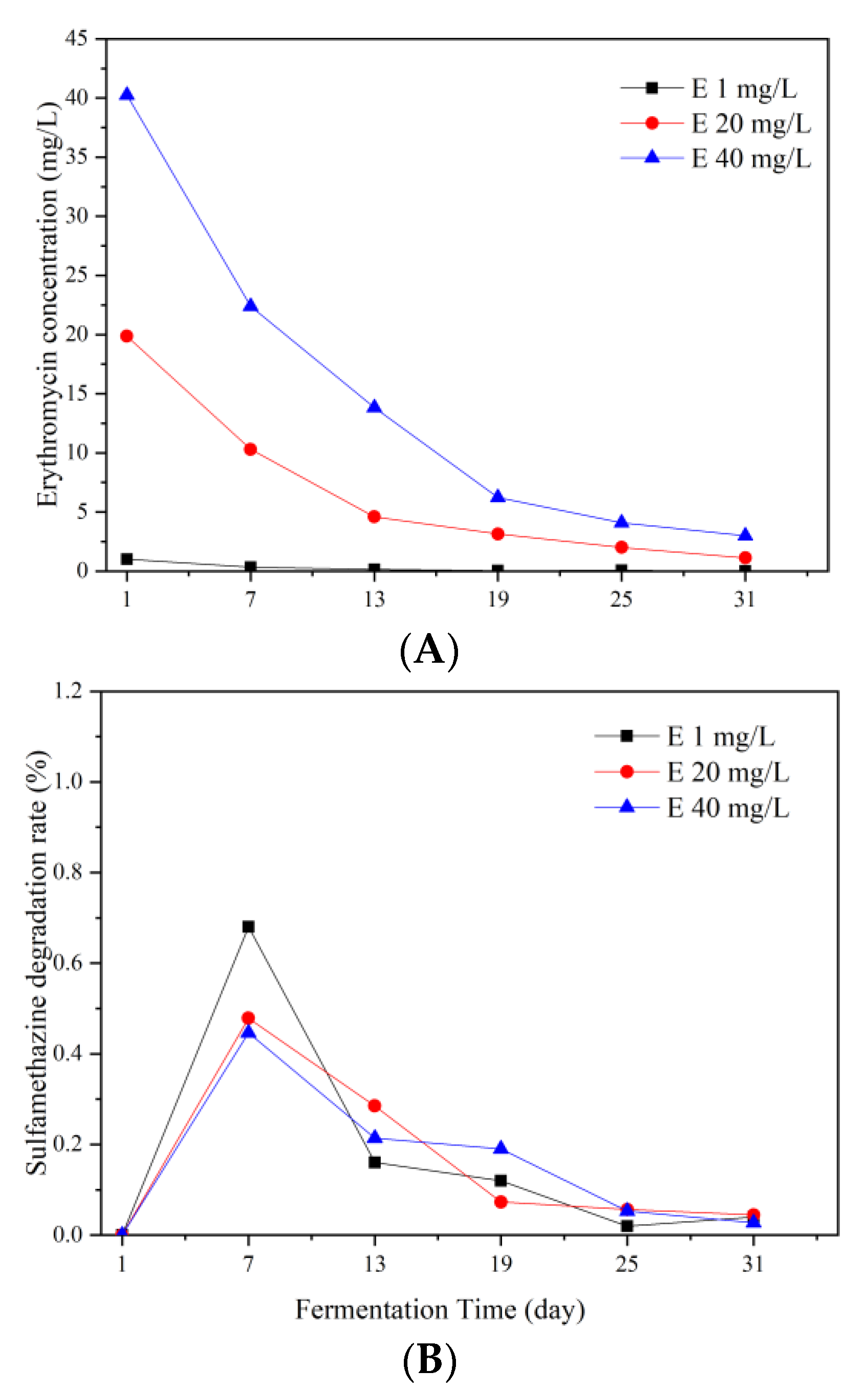
3.1. Erythromycin Removal
3.2. Impacts of Erythromycin on Biogas Properties
3.2.1. Biogas Yields
3.2.2. CH4 Yields
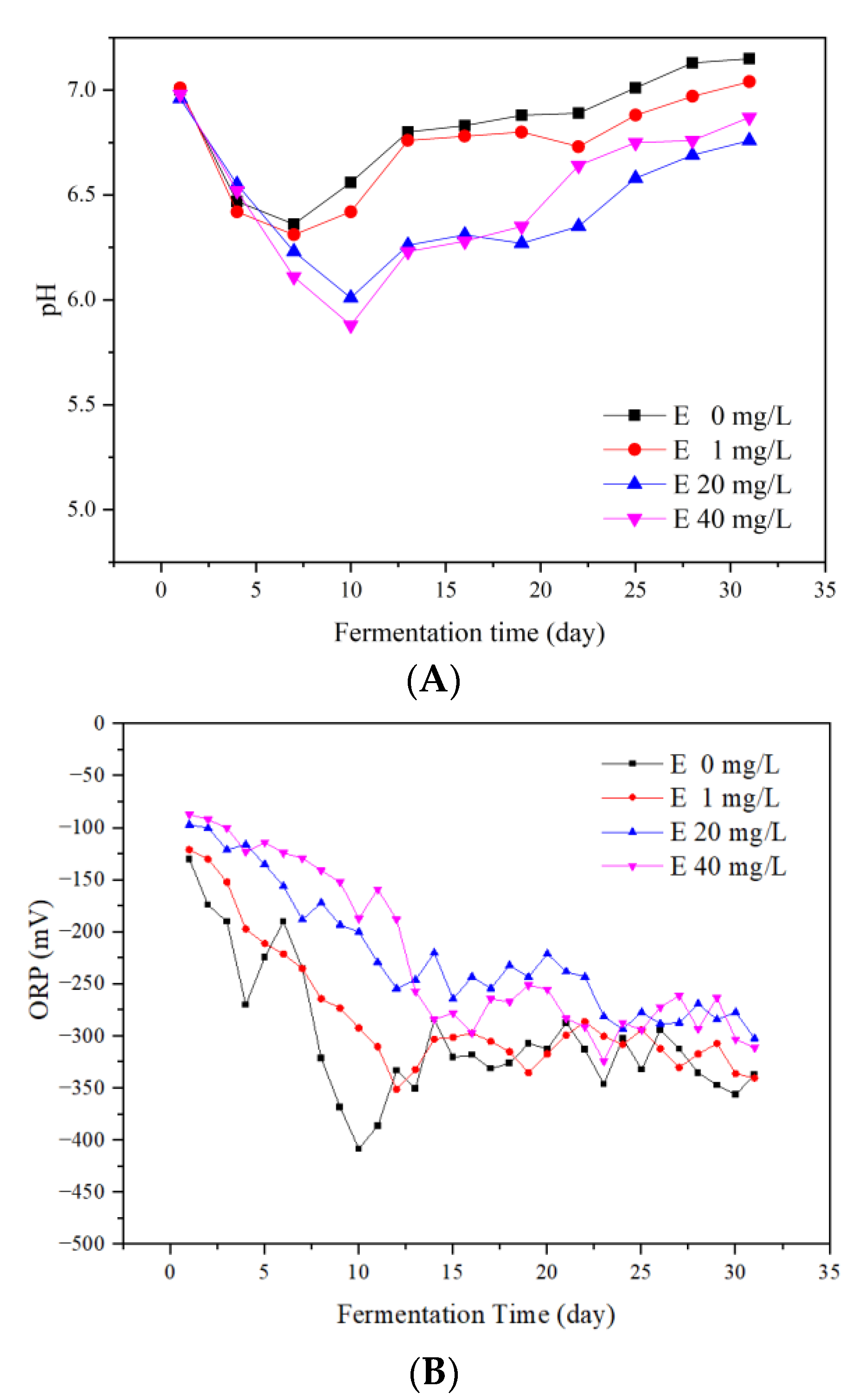
3.3. Impact of Erythromycin on Process Stability
3.3.1. The pH Values
3.3.2. ORP
3.4. Substrate Biodegradation
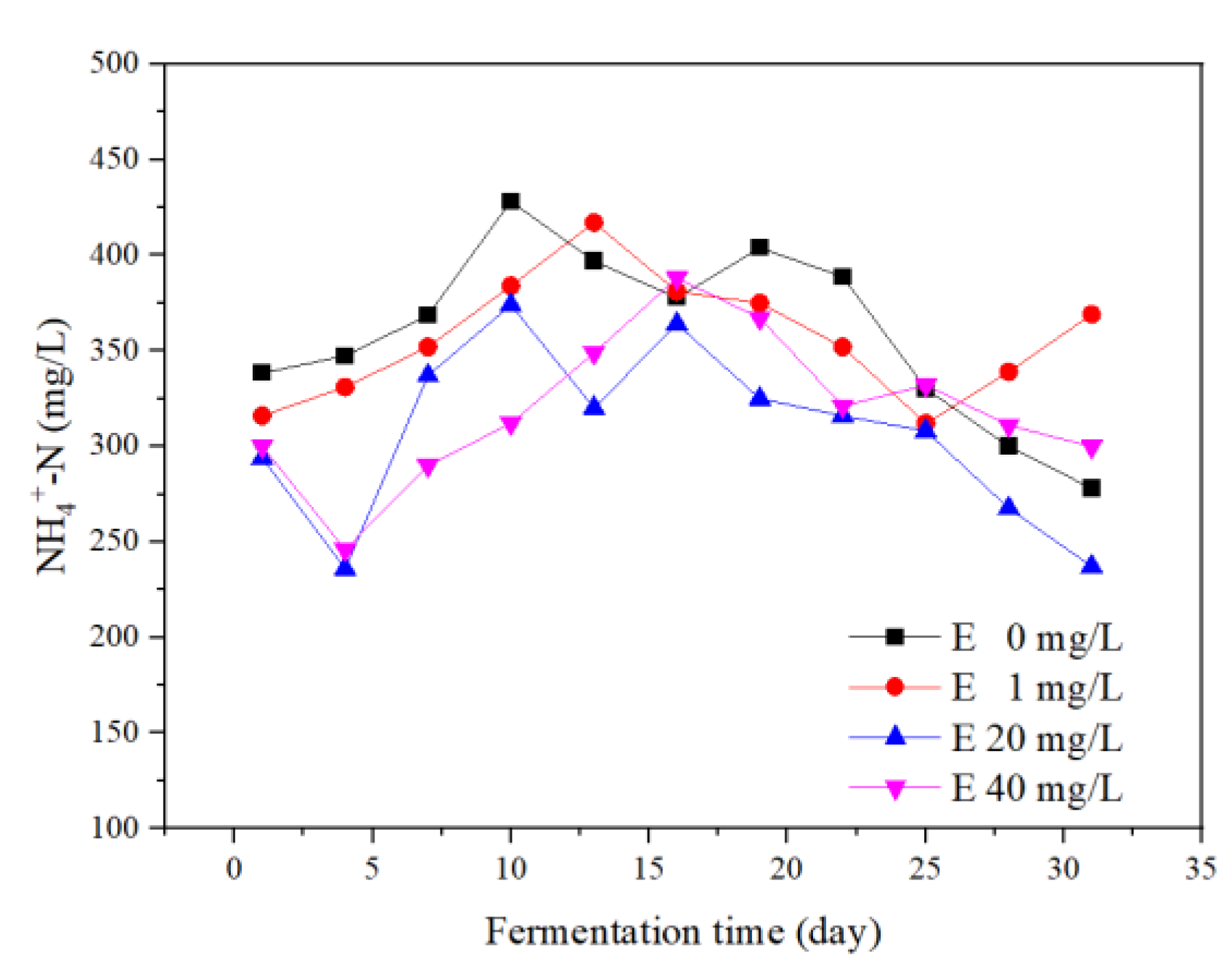
3.4.1. Variations of NH4+–N Concentrations
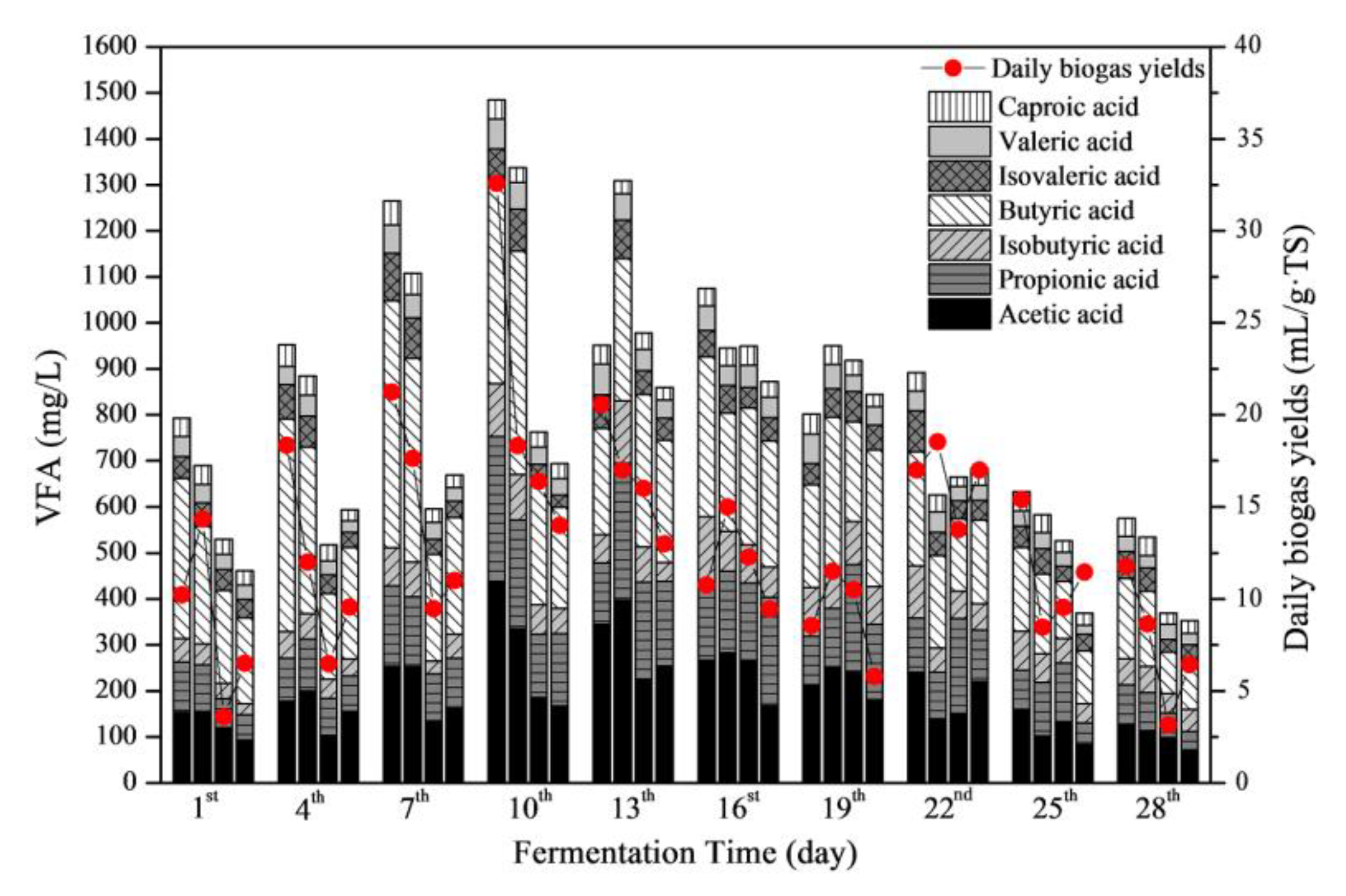
3.4.2. Responses of VFAs
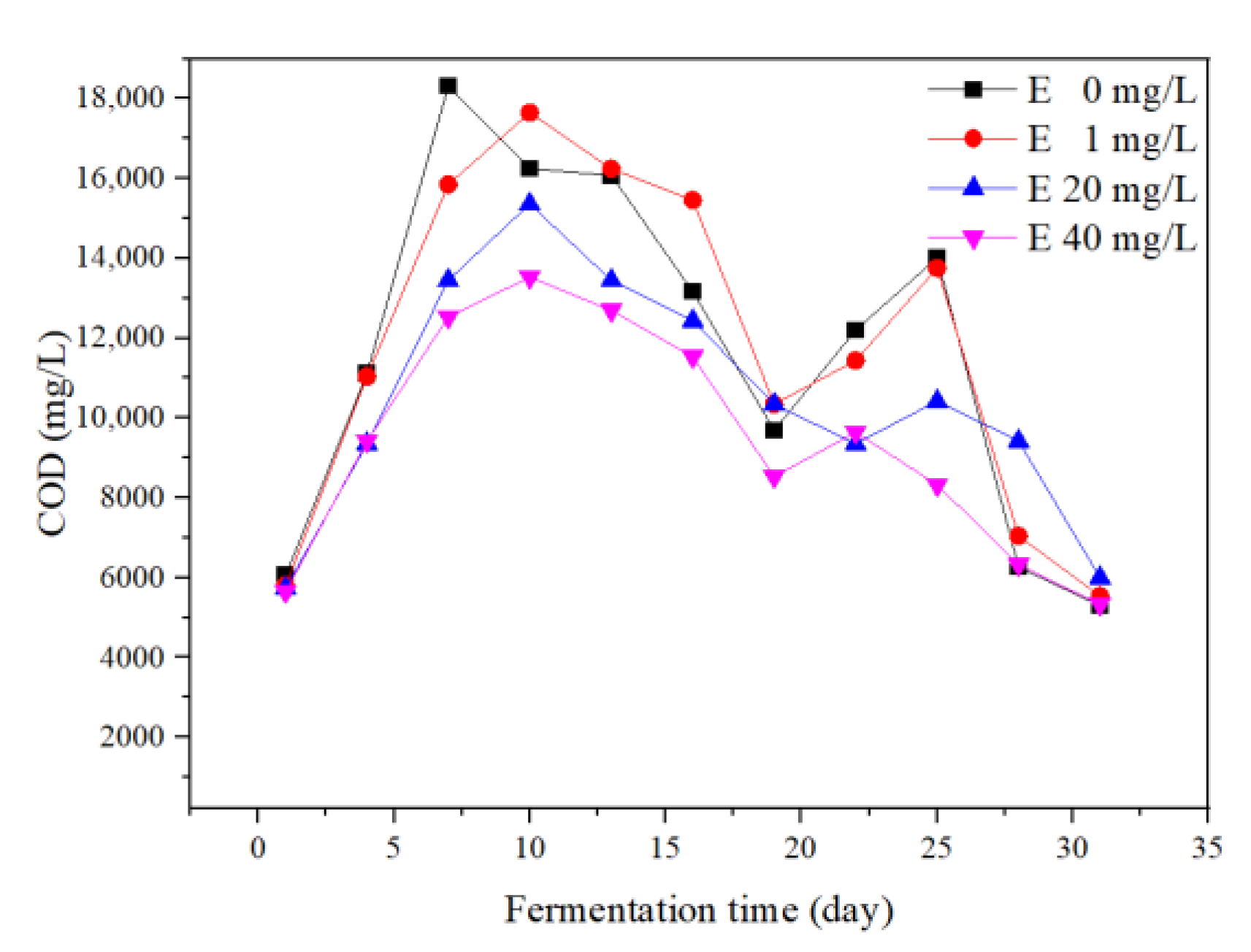
3.4.3. COD
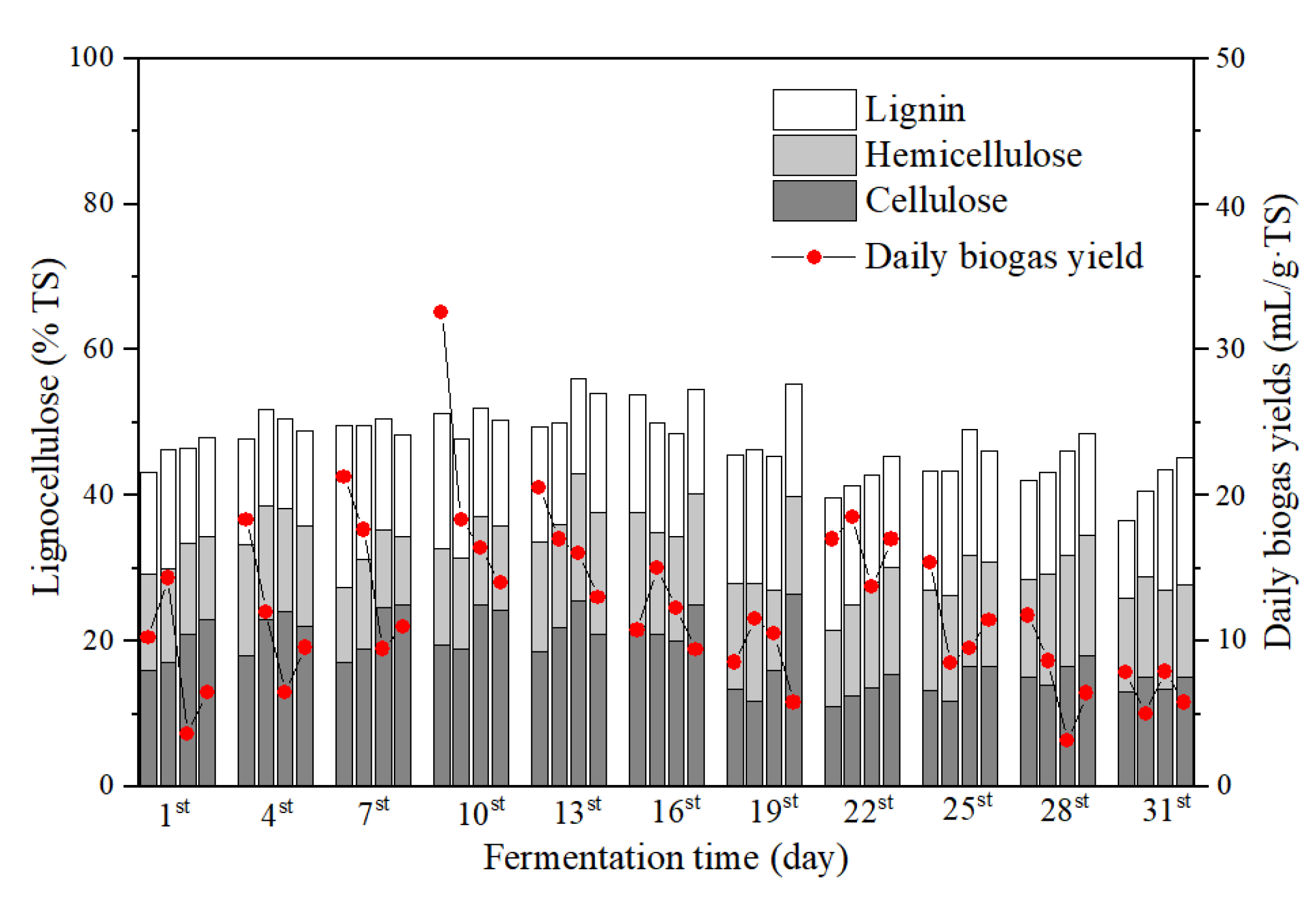
3.4.4. Degradation of Lignocelluloses
3.5. Responses of Enzyme Activity
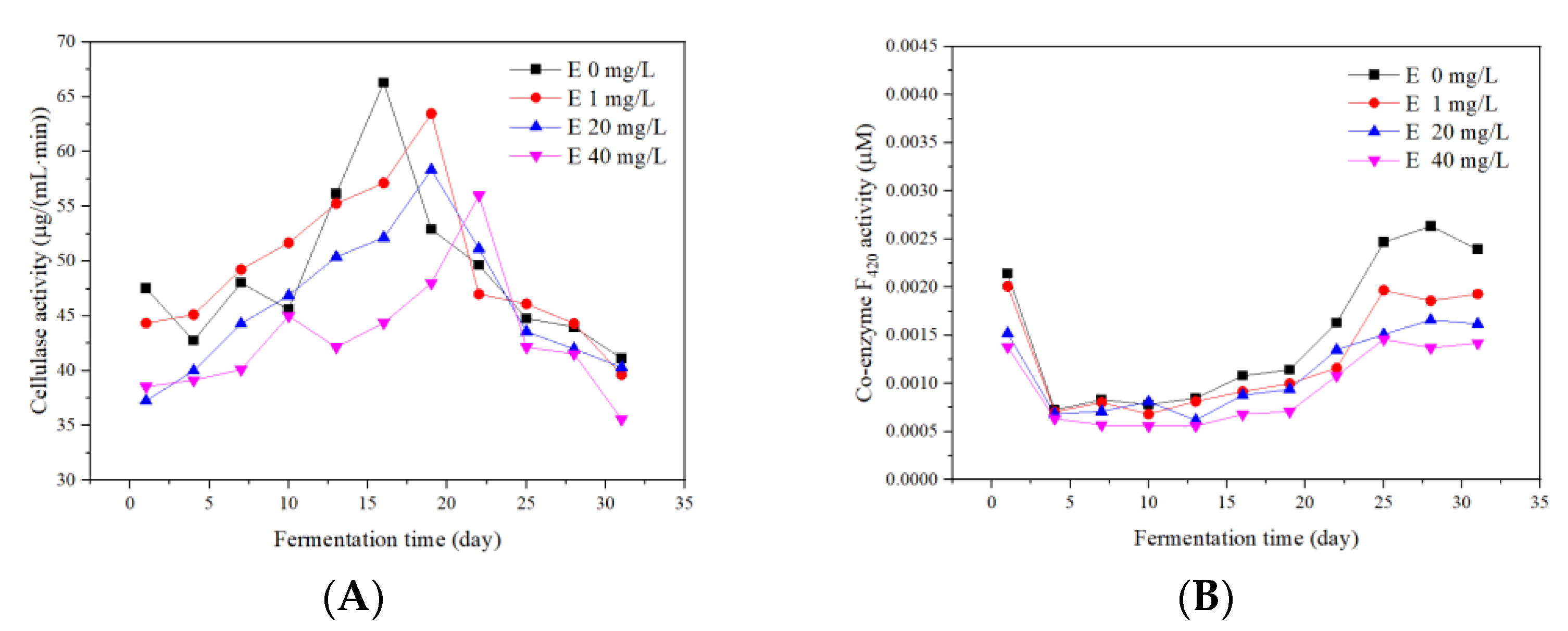
3.5.1. Cellulase
3.5.2. Coenzyme F420
3.6. Impacts of Erythromycin on the Microbial Communities
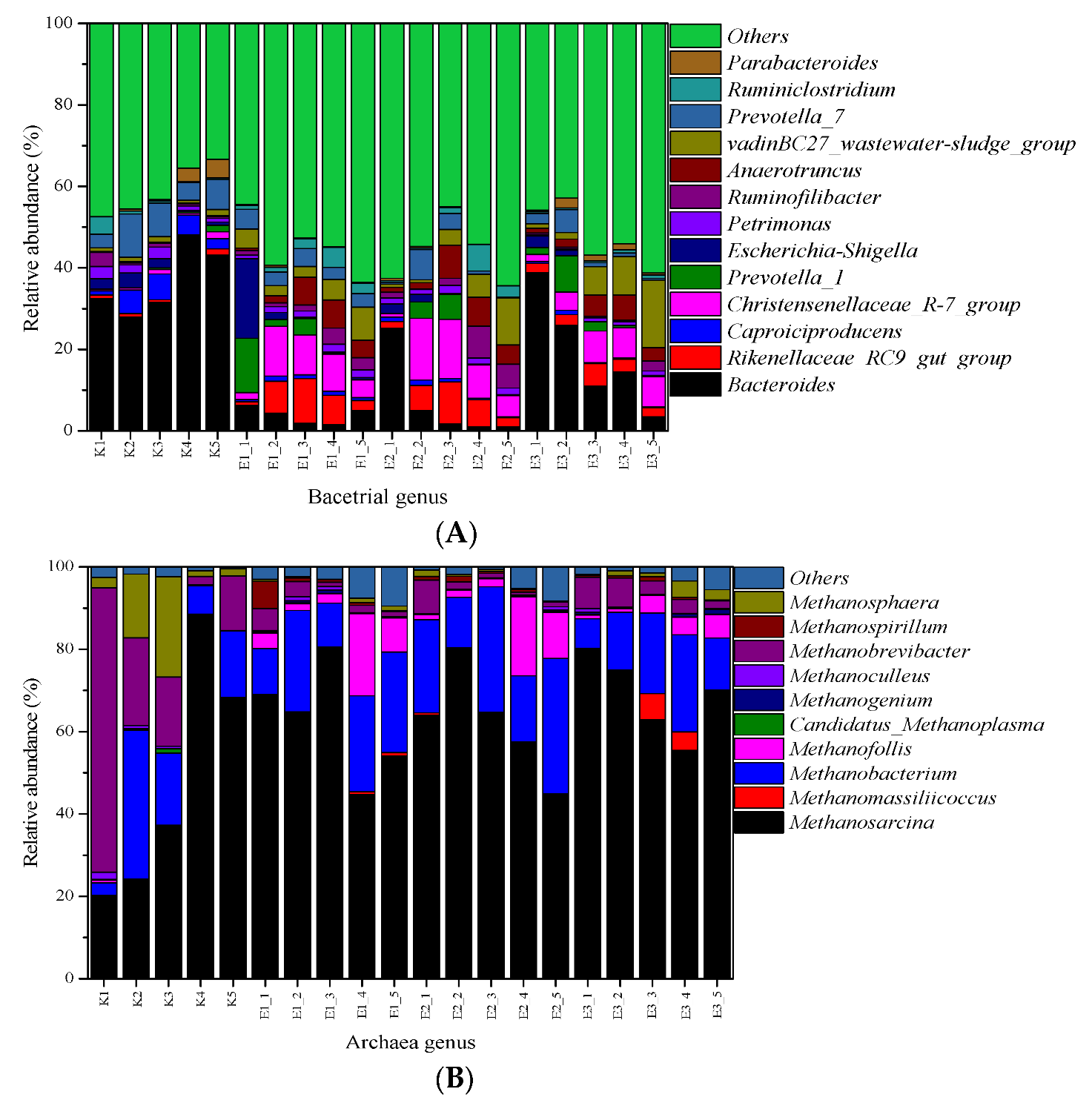
3.6.1. Bacterial Communities
3.6.2. Archaeal Communities
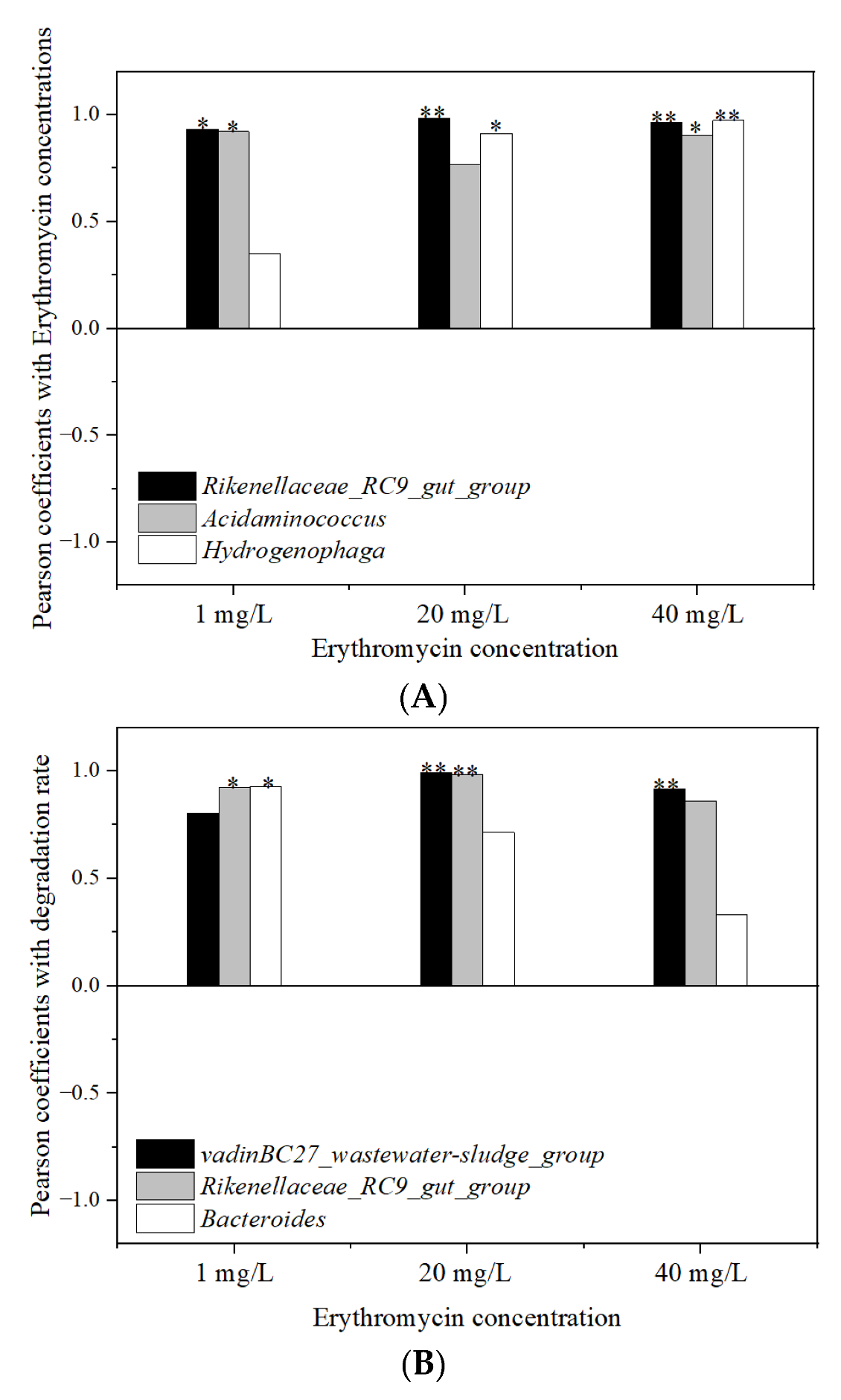
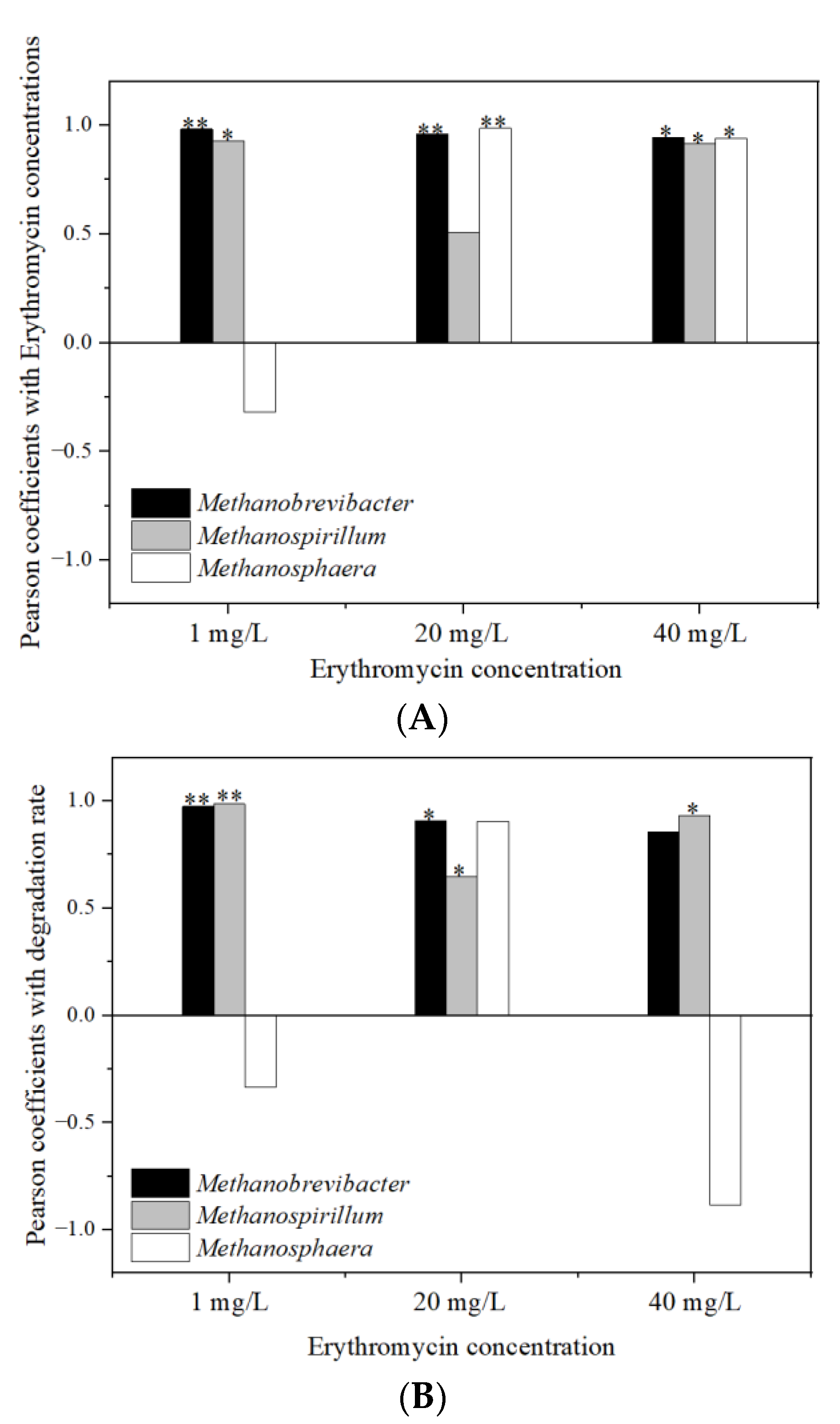
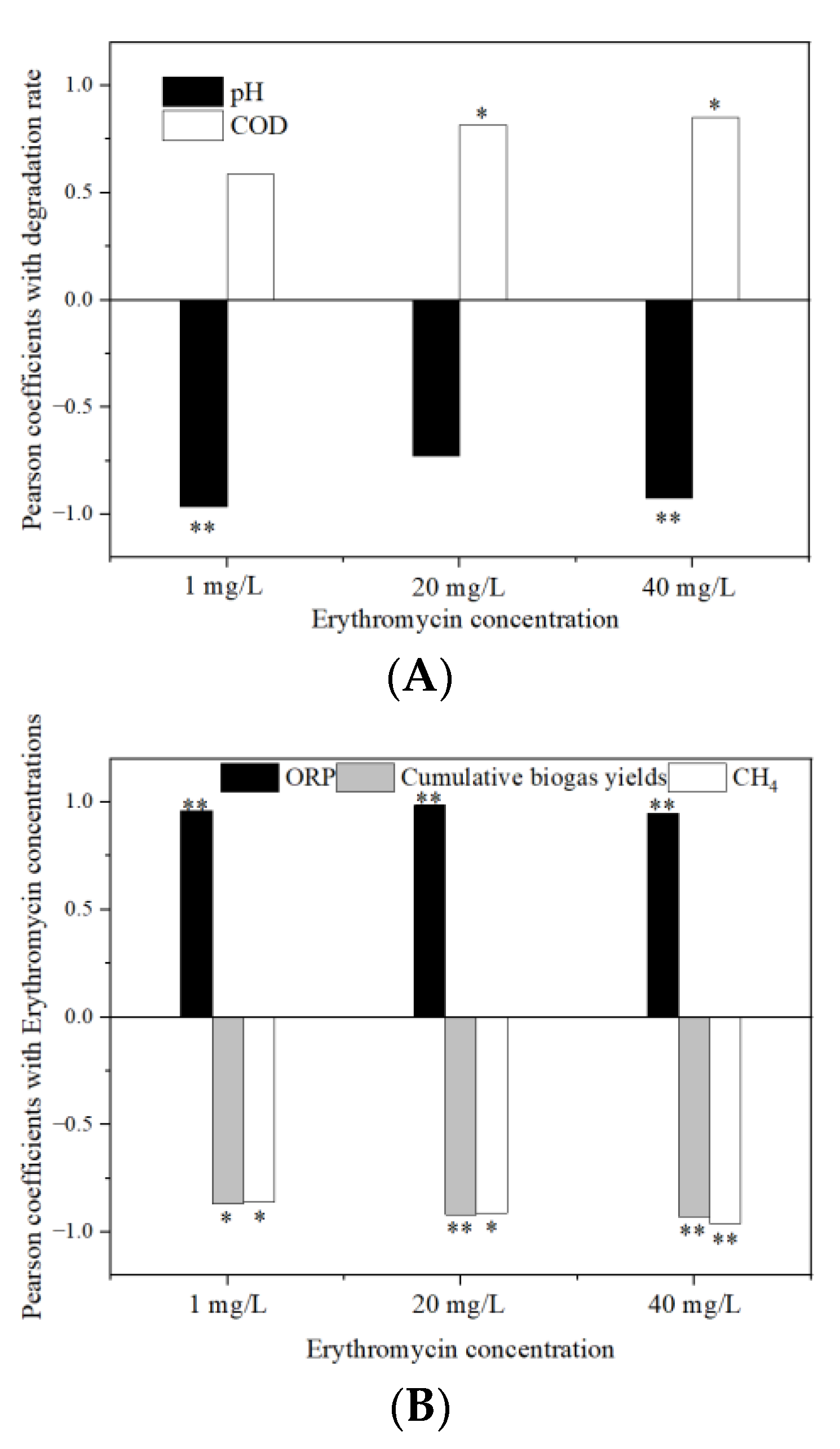
3.7. Relationship Analysis between the Erythromycin Removal and Fermentation Parameters
3.7.1. Relationship with Microbial Community
3.7.2. Relationship with Other Fermentation Parameters
3.8. Implications of This Study
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, P.; Ding, Y.; Li, H.; Xagoraraki, I. Occurrence of pharmaceuticals in a municipal wastewater treatment plant: Mass balance and removal processes. Chemosphere 2012, 88, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Campagnolo, E.R.; Johnson, K.R.; Karpati, A.; Rubin, C.S.; Kolpin, D.W.; Meyer, M.T.; Esteban, J.E.; Currier, R.W.; Smith, K.; Thu, K.M.; et al. Antimicrobial residues in animal waste and water resources proximal to large-scale swine and poultry feeding operations. Sci. Total Environ. 2002, 299, 89–95. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, X.; Zhang, Y.; Gao, H. Potential toxicity of sulfanilamide antibiotic: Binding of sulfamethazine to human serum albumin. Sci. Total Environ. 2012, 432, 269–274. [Google Scholar] [CrossRef]
- Ghosh, S.; Ramsden, S.J.; LaPara, T.M. The role of anaerobic digestion in controlling the release of tetracycline resistance genes and class 1 integrons from municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 2009, 84, 791–796. [Google Scholar] [CrossRef]
- Diehl, D.L.; LaPara, T.M. Effect of temperature on the fate of genes encoding tetracycline resistance and the integrase of class 1 integrons within anaerobic and aerobic digesters treating municipal wastewater solids. Environ. Sci. Technol. 2010, 44, 9128–9133. [Google Scholar] [CrossRef]
- Poels, J.; Van Assche, P.; Verstraete, W. Effects of disinfectants and antibiotics on the anaerobic digestion of piggery waste. Agric. Wastes 1984, 9, 239–247. [Google Scholar] [CrossRef]
- Loftin, K.A.; Henny, C.; Adams, C.D.; Surampali, R.; Mormile, M.R. Inhibition of microbial metabolism in anaerobic lagoons by selected sulfonamides, tetracyclines, lincomycin, and tylosin tartrate. Environ. Toxicol. Chem. 2005, 24, 782–788. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, J.; Liu, J.; Yu, D.; Zhong, H.; Wang, Y.; Chen, M.; Tong, J.; Wei, Y. Effects of chlortetracycline, Cu and their combination on the performance and microbial community dynamics in swine manure anaerobic digestion. J. Environ. Sci. 2018, 67, 206–215. [Google Scholar] [CrossRef]
- Feng, L.; Casas, M.E.; Ottosen, L.D.M.; Møller, H.B.; Bester, K. Removal of antibiotics during the anaerobic digestion of pig manure. Sci. Total Environ. 2017, 603–604, 219–225. [Google Scholar] [CrossRef]
- Aydin, S.; Ince, B.; Cetecioglu, Z.; Arikan, O.; Ozbayram, E.G.; Shahi, A.; Ince, O. Combined effect of erythromycin, tetracycline and sulfamethoxazole on performance of anaerobic sequencing batch reactors. Bioresour. Technol. 2015, 186, 207–214. [Google Scholar] [CrossRef]
- Mironov, V.A.; Sergienko, O.V.; Nastasyak, I.N.; Danilenko, V.N. Biogenesis and Regulation of Biosynthesis of Erythromycins in Saccharopolyspora erythraea. Appl. Biochem. Microbiol. 2004, 40, 531–541. [Google Scholar] [CrossRef]
- Deng, W.; Zhang, A.; Chen, S.; He, X.; Jin, L.; Yu, X.; Yang, S.; Li, B.; Fan, L.; Ji, L.; et al. Heavy metals, antibiotics and nutrients affect the bacterial community and resistance genes in chicken manure composting and fertilized soil. J. Environ. Manag. 2020, 257, 109980. [Google Scholar] [CrossRef] [PubMed]
- Alenzi, A.; Hunter, C.; Spencer, J.; Roberts, J.; Craft, J.; Pahl, O.; Escudero, A. Pharmaceuticals effect and removal, at environmentally relevant concentrations, from sewage sludge during anaerobic digestion. Bioresour. Technol. 2021, 319, 124102. [Google Scholar] [CrossRef]
- Wang, M.; Ren, P.; Wang, Y.; Cai, C.; Liu, H.; Dai, X. Erythromycin stimulates rather than inhibits methane production in anaerobic digestion of antibiotic fermentation dregs. Sci. Total Environ. 2022, 807, 151007. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, P.; Gajdács, M. Writing a strong scientific paper in medicine and the biomedical sciences: A checklist and recommendations for early career researchers. Biol. Futur. 2021, 72, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Van Houten, R.T.; Borger, A.R.; Eikelboom, D.H.; Fan, Y. Minimization of excess sludge production for biological wastewater treatment. Water Res. 2003, 37, 4453–4467. [Google Scholar] [CrossRef]
- Zhang, H.; Han, X.; Tian, Y.; Li, Y.; Yang, K.; Hao, H.; Chai, Y.; Xu, X. Process analysis of anaerobic fermentation of Phragmites australis straw and cow dung exposing to elevated chromium (VI) concentrations. J. Environ. Manag. 2018, 224, 414–424. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, H.; Zheng, L.; Li, S.; Hao, H.; Huang, H. Effect of Zn addition on the Cd-containing anaerobic fermentation process: Biodegradation and microbial communities. Int. J. Environ. Res. Public Health 2019, 16, 2998. [Google Scholar] [CrossRef] [Green Version]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef]
- Yang, Z.; Xu, R.; Zheng, Y.; Chen, T.; Zhao, L.; Li, M. Characterization of extracellular polymeric substances and microbial diversity in anaerobic co-digestion reactor treated sewage sludge with fat, oil, grease. Bioresour. Technol. 2016, 212, 164–173. [Google Scholar] [CrossRef]
- Jiang, M.; Song, S.; Fu, H.; Cai, C.; Liu, H. Research of feasibility for anaerobic digestion of erythromycin mycelial dreg and degradation rules of erythromycin. Environ. Prot. Sci. 2017, 43, 62–67. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, L.; Li, Y.; Qiu, C. Inhibition of methanogenium by erythromycin and its domestation. Environ. Sci. 2013, 34, 1540–1544. [Google Scholar] [CrossRef]
- Liu, A.; Xu, S.; Lu, C.; Peng, P.; Zhang, Y.; Feng, D.; Liu, Y. Anaerobic fermentation by aquatic product wastes and other auxiliary materials. Clean Technol. Environ. Policy 2014, 16, 415–421. [Google Scholar] [CrossRef]
- Bitton, G. Bulking and Foaming in Activated Sludge Plants. In Wastewater Microbiology; Wiley: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Lee, S. Relationship between Oxidation Reduction Potential (ORP) and Volatile Fatty Acid (VFA) Production in the Acid-Phase Anaerobic Digestion Process. Master’s Thesis, University of Canterbury, Christchurch, New Zealand, 2008. [Google Scholar] [CrossRef]
- Tian, Y.; Yang, K.; Li, S.; Zhang, H.; Han, X.; Hao, H.; Chai, Y.; Xu, X. How does copper stress influence the relationships among the anaerobic co-fermentation parameters? J. Renew. Sustain. Energy 2018, 10, 053107. [Google Scholar] [CrossRef]
- Sawayama, S.; Tada, C.; Tsukahara, K.; Yagishita, T. Effect of ammonium addition on methanogenic community in a fluidized bed anaerobic digestion. J. Biosci. Bioeng. 2004, 97, 65–70. [Google Scholar] [CrossRef]
- Wang, C.; Yang, C.; Chen, Z. Rumen microbial volatile fatty acids in relation to oxidation reduction potential and electricity generation from straw in microbial fuel cells. Biomass Bioenergy 2012, 37, 318–329. [Google Scholar] [CrossRef]
- Beevi, B.S.; Jose, P.P.; Madhu, G. Effect of total solid concentration on anaerobic digestion of the organic fraction of municipal solid waste. Int. J. Sci. Res. Publ. 2013, 3, 1–5. [Google Scholar]
- Triolo, J.M.; Sommer, S.G.; Møller, H.B.; Weisbjerg, M.R.; Jiang, X.Y. A new algorithm to characterize biodegradability of biomass during anaerobic digestion: Influence of lignin concentration on methane production potential. Bioresour. Technol. 2011, 102, 9395–9402. [Google Scholar] [CrossRef]
- Mudhoo, A.; Kumar, S. Effects of heavy metals as stress factors on anaerobic digestion processes and biogas production from biomass. Int. J. Environ. Sci. Technol. 2013, 10, 1383–1398. [Google Scholar] [CrossRef] [Green Version]
- Bayer, E.A.; Lamed, R.; Himmel, M.E. The potential of cellulases and cellulosomes for cellulosic waste management. Curr. Opin. Biotechnol. 2007, 18, 237–245. [Google Scholar] [CrossRef]
- Merlin Christy, P.; Gopinath, L.R.; Divya, D. A review on anaerobic decomposition and enhancement of biogas production through enzymes and microorganisms. Renew. Sustain. Energy Rev. 2014, 34, 167–173. [Google Scholar] [CrossRef]
- Cheng, Y.-H.; Sang, S.-X.; Huang, H.-Z.; Liu, X.-J.; Ouyang, J.-B. Variation of coenzyme F420 activity and methane yield in landfill simulation of organic waste. J. China Univ. Min. Technol. 2007, 17, 403–408. [Google Scholar] [CrossRef]
- Kretsinger, H.R.; Uversky, N.V.; Permyakov, A.E. Encyclopedia of Metalloproteins; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Harper, S.R.; Pohland, F.G. Recent developments in hydrogen management during anaerobic biological wastewater treatment. Biotechnol. Bioeng. 1986, 28, 585–602. [Google Scholar] [CrossRef]
- Van Haandel, A.C. Influence of the digested COD concentration on the alkalinity requirement in anaerobic digesters. Water Sci. Technol. 1994, 30, 23–34. [Google Scholar] [CrossRef]
- Martens, E.C.; Chiang, H.C.; Gordon, J.I. Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host Microbe 2008, 4, 447–457. [Google Scholar] [CrossRef] [Green Version]
- Theuerl, S.; Klang, J.; Heiermann, M.; De Vrieze, J. Marker microbiome clusters are determined by operational parameters and specific key taxa combinations in anaerobic digestion. Bioresour. Technol. 2018, 263, 128–135. [Google Scholar] [CrossRef]
- Ruan, Y.; Shen, L.; Zou, Y.; Qi, Z.; Yin, J.; Jiang, J.; Guo, L.; He, L.; Chen, Z.; Tang, Z.; et al. Comparative genome analysis of Prevotella intermedia strain isolated from infected root canal reveals features related to pathogenicity and adaptation. BMC Genom. 2015, 16, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Petersen, L.M.; Bautista, E.J.; Nguyen, H.; Hanson, B.M.; Chen, L.; Lek, S.H.; Sodergren, E.; Weinstock, G.M. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome 2017, 5, 1–13. [Google Scholar] [CrossRef]
- Kim, B.-C.; Jeon, B.; Kim, S.-I.; Kim, H.; Um, Y.; Sang, B.-I. Caproiciproducens galactitolivorans gen. Nov., sp. nov., a bacterium capable of producing caproic acid from galactitol, isolated from a wastewater treatment plant. Int. J. Syst. Evol. Microbiol. 2015, 65, 4902–4908. [Google Scholar] [CrossRef]
- Fosses, A.; Maté, M.; Franche, N.; Liu, N.; Denis, Y.; Borne, R.; De Philip, P.; Fierobe, H.P.; Perret, S. A seven-gene cluster in Ruminiclostridium cellulolyticum is essential for signalization, uptake and catabolism of the degradation products of cellulose hydrolysis. Biotechnol. Biofuels 2017, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Morotomi, M.; Nagai, F.; Watanabe, Y. Description of Christensenella minuta gen. nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2011, 62, 144–149. [Google Scholar] [CrossRef]
- Su, X.; Tian, Q.; Zhang, J.; Yuan, X.; Shi, X.; Guo, R.; Qiu, Y. Acetobacteroides hydrogenigenes gen. nov., Sp. nov., An anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int. J. Syst. Evol. Microbiol. 2014, 64, 2986–2991. [Google Scholar] [CrossRef]
- Xu, M.; Chen, X.; Qiu, M.; Zeng, X.; Xu, J.; Deng, D.; Sun, G.; Li, X.; Guo, J. Bar-coded pyrosequencing reveals the responses of PBDE-degrading microbial communities to electron donor amendments. PLoS ONE 2012, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lawson, P.; Song, Y.; Liu, C.; Molitoris, D.; Vaisanen, M.-L.; Collins, M.; Finegold, S. Anaerotruncus colihominis gen. nov., sp. nov., from human faeces. Int. J. Syst. Evol. Microbiol. 2004, 54, 413–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galagan, J.E.; Nusbaum, C.; Roy, A.; Endrizzi, M.G.; Macdonald, P.; Fitzhugh, W.; Calvo, S.; Engels, R.; Smirnov, S.; Atnoor, D.; et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002, 12, 532–542. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; He, Q.; Ma, Y.; Wang, X.; Peng, X. Dynamics of microbial community in a mesophilic anaerobic digester treating food waste: Relationship between community structure and process stability. Bioresour. Technol. 2015, 189, 113–120. [Google Scholar] [CrossRef]
- Balch, W.E.; Fox, G.E.; Magrum, L.J.; Woese, C.R.; Wolfe, R.S. Methanogens: Reevaluation of a unique biological group. Microbiol. Rev. 1979, 43, 260–296. [Google Scholar] [CrossRef] [PubMed]
- Yudina, T.G.; Bryukhanov, A.L.; Netrusov, A.I. Susceptibility of archaea to the antibiotic effect of the parasporal inclusion proteins from different bacillus thuringiensis subspecies. Microbiology 2004, 73, 19–23. [Google Scholar] [CrossRef]
- Flint, H.; Edward, B.; Rincon, M.; Lamed, R.; White, B. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008, 6, 121–131. [Google Scholar] [CrossRef]


| Characteristics | Cow Dung | Corn Stover |
|---|---|---|
| TS (% dry weight) | 15.15 ± 0.41 | 91.24 ± 0.12 |
| VS (% TS) | 78.66 ± 0.06 | 86.34 ± 0.02 |
| TN (% TS) | 0.36 ± 0.01 | 0.34 ± 0.01 |
| TOC (% TS) | 25.42 ± 0.00 | 34.68 ± 0.00 |
| Ratio of C/N | 70.22 | 102.30 |
| Cellulose (% TS) | 26.92 ± 0.00 | 28.88 ± 0.00 |
| Hemicellulose (% TS) | 22.34 ± 0.00 | 24.92 ± 0.00 |
| Lignin (% TS) | 19.98 ± 0.00 | 21.30 ± 0.00 |
| Mg (mg/kg) | 8900.00 | 1400.00 |
| Ca (mg/kg) | 8000.00 | 1000.00 |
| K (mg/kg) | 1400.00 | 4200.00 |
| Zn (mg/kg) | <5.00 | 7.10 |
| Fe (mg/kg) | <5.00 | 170.00 |
| Ni (mg/kg) | 13.00 | 0.14 |
| Co (mg/kg) | <0.03 | - |
| Cu (mg/kg) | <5.00 | 9.00 |
| Mn (mg/kg) | <5.00 | 17.00 |
| S (% TS) | 0.22 | 0.10 |
| Coliform bacteria (MPN/g) | 4.6 × 103 | - |
| Mortality rate of ascaris eggs (%) | 100 | - |
| Erythromycin Concentrations (mg/L) | Lignin (% TS) | Hemicellulose (% TS) | Cellulose (% TS) | Total Lignocellulose (% TS) |
|---|---|---|---|---|
| 0 | 16.07 ± 1.00 | 13.45 ± 0.53 | 16.14 ± 0.93 | 45.66 ± 1.58 |
| 1 | 16.88 ± 1.25 | 13.99 ± 0.42 | 15.51 ± 0.64 | 46.38 ± 1.16 |
| 20 | 19.66 ± 1.41 * | 13.71 ± 0.62 | 14.91 ± 0.57 | 48.30 ± 1.18 |
| 40 | 21.07 ± 1.25 * | 13.63 ± 0.68 | 14.86 ± 0.38 | 49.56 ± 1.10 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Yin, M.; Li, S.; Zhang, S.; Han, G. The Removal of Erythromycin and Its Effects on Anaerobic Fermentation. Int. J. Environ. Res. Public Health 2022, 19, 7256. https://doi.org/10.3390/ijerph19127256
Zhang H, Yin M, Li S, Zhang S, Han G. The Removal of Erythromycin and Its Effects on Anaerobic Fermentation. International Journal of Environmental Research and Public Health. 2022; 19(12):7256. https://doi.org/10.3390/ijerph19127256
Chicago/Turabian StyleZhang, Huayong, Meixiao Yin, Shusen Li, Shijia Zhang, and Guixuan Han. 2022. "The Removal of Erythromycin and Its Effects on Anaerobic Fermentation" International Journal of Environmental Research and Public Health 19, no. 12: 7256. https://doi.org/10.3390/ijerph19127256
APA StyleZhang, H., Yin, M., Li, S., Zhang, S., & Han, G. (2022). The Removal of Erythromycin and Its Effects on Anaerobic Fermentation. International Journal of Environmental Research and Public Health, 19(12), 7256. https://doi.org/10.3390/ijerph19127256







