Computer-Aided Diagnosis of Coal Workers’ Pneumoconiosis in Chest X-ray Radiographs Using Machine Learning: A Systematic Literature Review
Abstract
1. Introduction
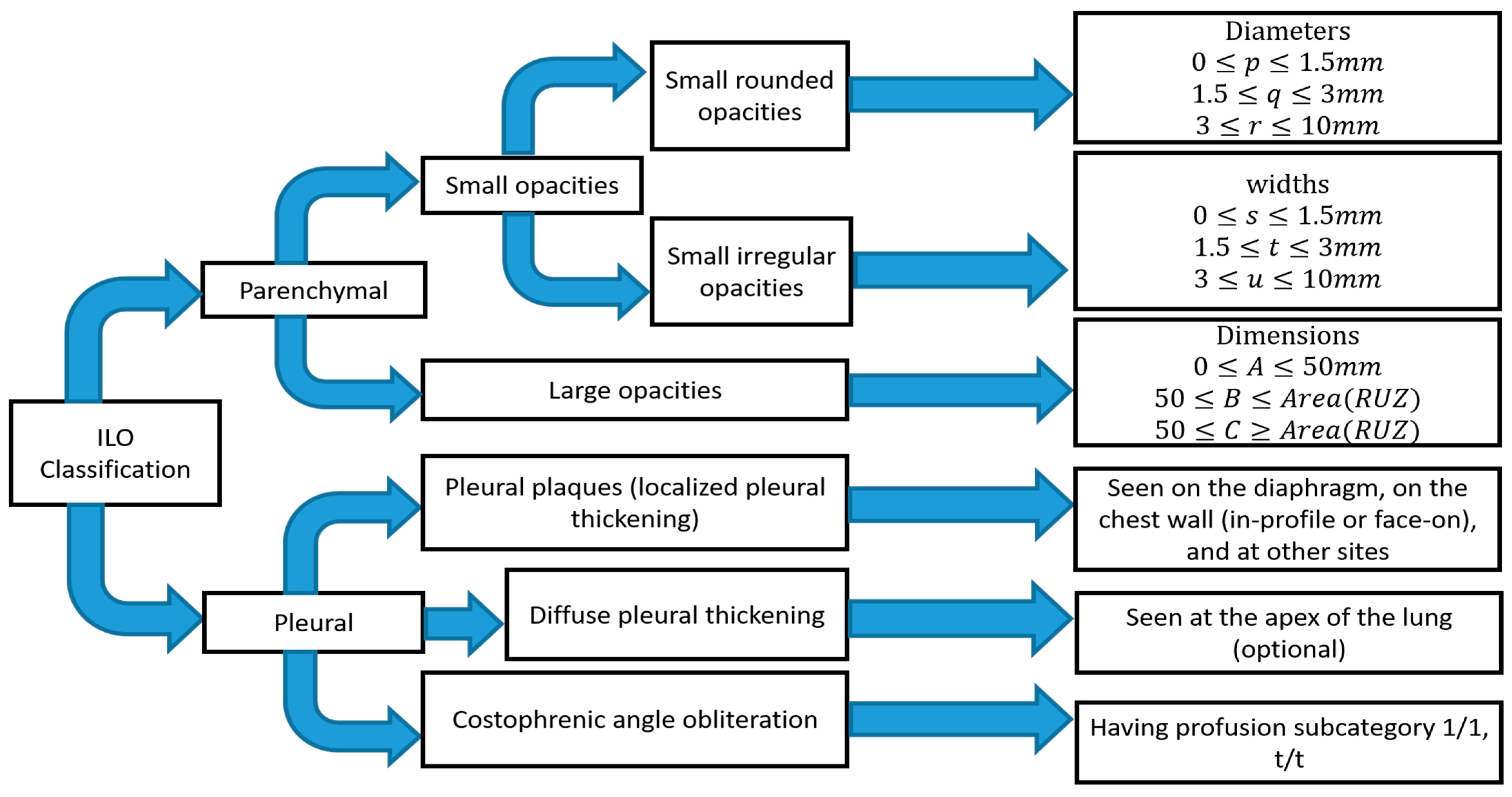
1.1. Standard Classification of Pneumoconiosis
2. Method
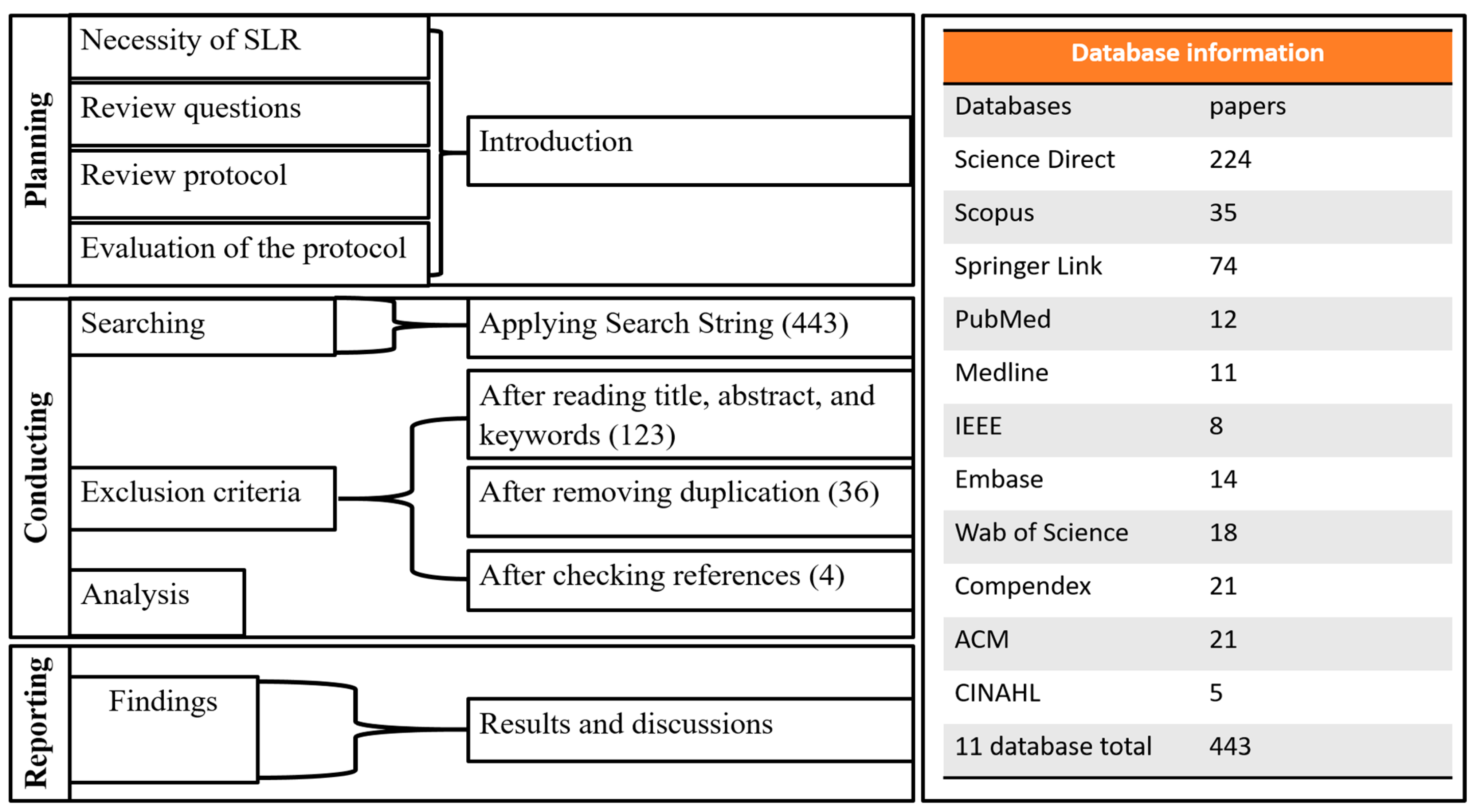
2.1. Search Strategy and Database Selection
2.2. Study Selection Criteria
3. Study Results
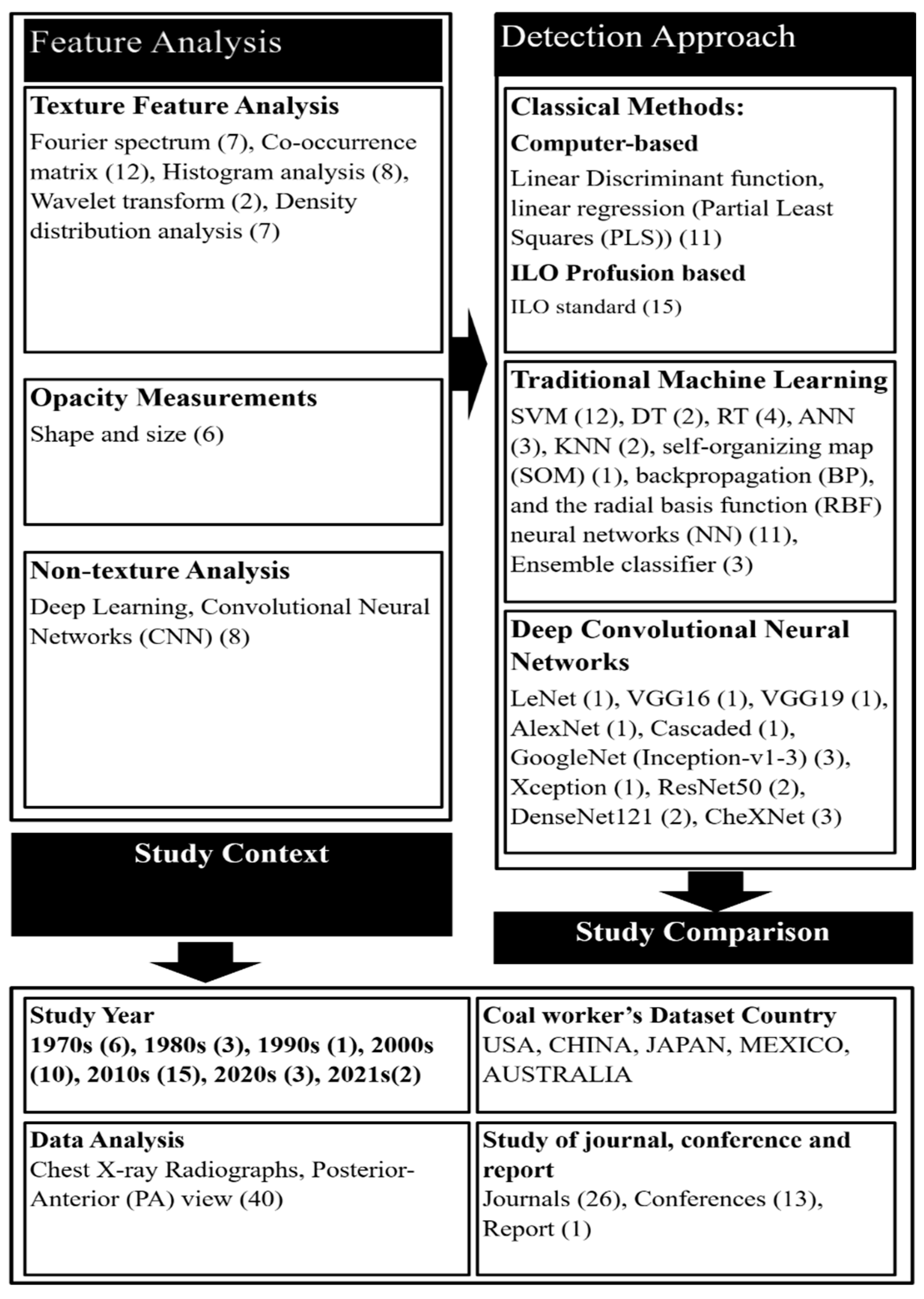
4. Analysis of Returned Articles
4.1. Datasets
4.1.1. Fourier Spectrum Analysis
4.1.2. Co-Occurrence Matrix Analysis
4.1.3. Histogram Analysis
4.1.4. Wavelet Analysis
4.1.5. Density Distribution Analysis
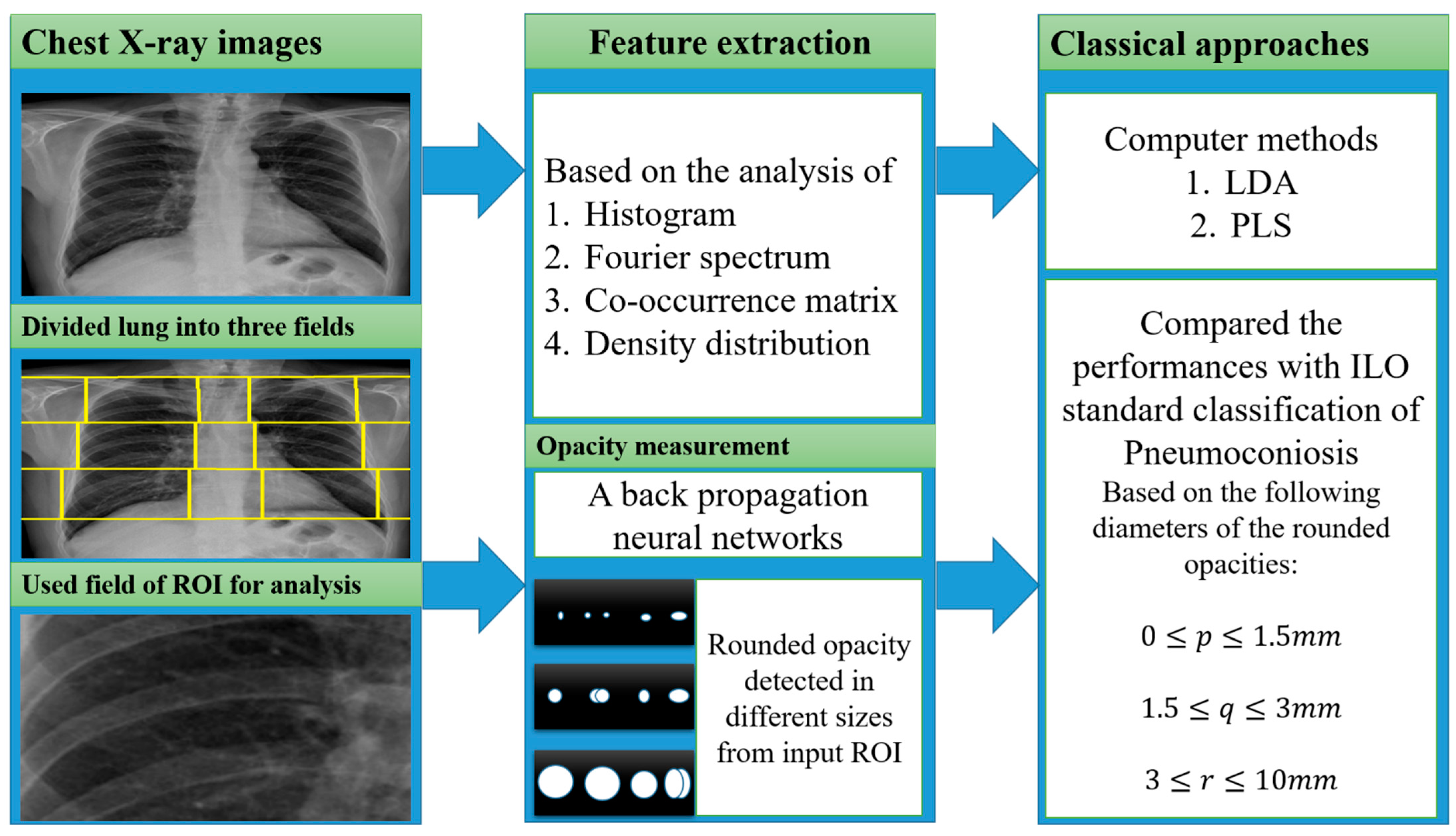
4.2. Opacity Measurement
4.3. Non-Texture Analysis
4.4. Detection Approach of CWP
4.4.1. Classical Methods
4.4.2. Traditional Machine Learning
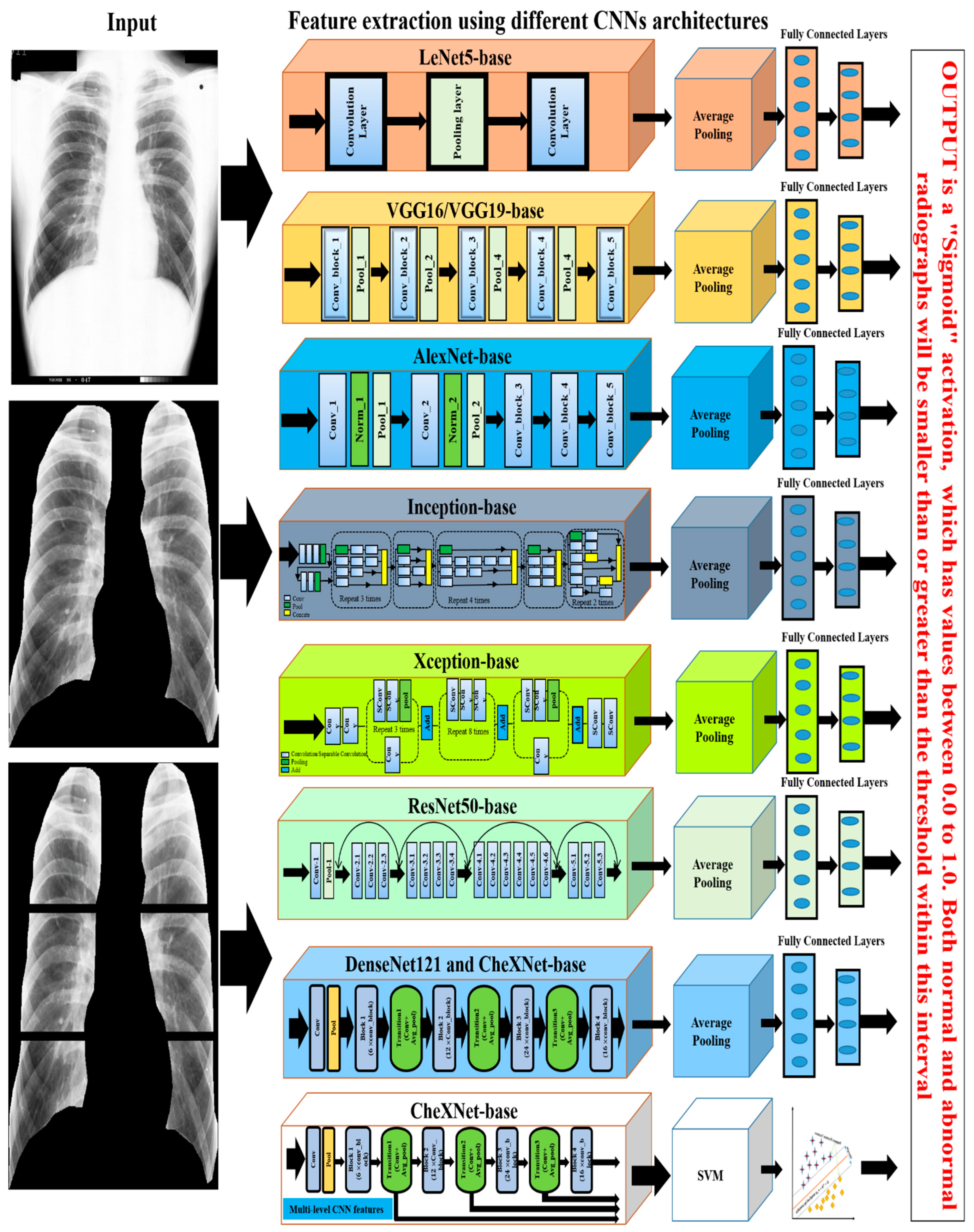
4.4.3. CNN-Based
5. Study Limitations and Future Directions for Research
5.1. Direction 1: Combination of All Private Datasets
5.2. Direction 2: Apply Deep Transfer Learning
5.3. Direction 3: Apply SVM on the Deep CNN Feature
5.4. Direction 4: Apply Ensemble Learning
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cullinan, P.; Reid, P. Pneumoconiosis. Prim. Care Respir. J. 2013, 22, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Fishwick, D.; Barber, C. Pneumoconiosis. Medicine 2012, 40, 310–313. [Google Scholar] [CrossRef]
- Schenker, M.B.; Pinkerton, K.E.; Mitchell, D.; Vallyathan, V.; Elvine-Kreis, B.; Green, F.H. Pneumoconiosis from Agricultural Dust Exposure among Young California Farmworkers. Environ. Health Perspect. 2009, 117, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Leggat, P.A. 24 Years of Pneumoconiosis Mortality Surveillance in Australia. J. Occup. Health 2006, 48, 309–313. [Google Scholar] [CrossRef]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Zosky, G.; Hoy, R.F.; Silverstone, E.J.; Brims, F.J.; Miles, S.; Johnson, A.R.; Gibson, P.; Yates, D.H. Coal workers’ pneumoconiosis: An Australian perspective. Med. J. Aust. 2016, 204, 414–418. [Google Scholar] [CrossRef]
- Hall, N.B.; Blackley, D.J.; Halldin, C.N.; Laney, A.S. Current Review of Pneumoconiosis Among US Coal Miners. Curr. Environ. Health Rep. 2019, 2019, 1–11. [Google Scholar] [CrossRef]
- Castranova, V.; Vallyathan, V. Silicosis and coal workers’ pneumoconiosis. Environ. Health Perspect. 2000, 108, 675–684. [Google Scholar] [CrossRef]
- Joy, G.J.; Colinet, J.F. Coal Mine Respirable Dust Control View project Dust control with canopy air curtain View project Mınıng engıneerıng Coal workers’ pneumoconiosis prevalence disparity between Australia and the United States. Min. Eng. Mag. 2012, 64, 71. [Google Scholar]
- Doi, K. Computer-Aided Diagnosis in Medical Imaging: Historical Review, Current Status and Future Potential. Comput. Med Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef]
- Li, Q.; Nishikawa, R.M. (Eds.) Computer-Aided Detection and Diagnosis in Medical Imaging; Taylor & Francis: Oxford, UK, 2015. [Google Scholar]
- Chen, C.-M.; Chou, Y.-H.; Tagawa, N.; Do, Y. Computer-Aided Detection and Diagnosis in Medical Imaging. Comput. Math. Methods Med. 2013, 2013, 1–2. [Google Scholar] [CrossRef]
- Halalli, B.; Makandar, A. Computer Aided Diagnosis—Medical Image Analysis Techniques. In Breast Imaging; InTech: London, UK, 2018. [Google Scholar]
- Han, L.; Gao, Q.; Yang, J.; Wu, Q.; Zhu, B.; Zhang, H.; Ding, B.; Ni, C. Survival Analysis of Coal Workers’ Pneumoconiosis (CWP) Patients in a State-Owned Mine in the East of China from 1963 to 2014. Int. J. Environ. Res. Public Health 2017, 14, 489. [Google Scholar] [CrossRef] [PubMed]
- Beer, C.; Kolstad, H.A.; Søndergaard, K.; Bendstrup, E.; Heederik, D.; Olsen, K.E.; Omland, Ø.; Petsonk, E.; Sigsgaard, T.; Sherson, D.L.; et al. A systematic review of occupational exposure to coal dust and the risk of interstitial lung diseases. Eur. Clin. Respir. J. 2017, 4, 1264711. [Google Scholar] [CrossRef]
- Yang, H.-Y. Prediction of pneumoconiosis by serum and urinary biomarkers in workers exposed to asbestos-contaminated minerals. PLoS ONE 2019, 14, e0214808. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.; Wang, L.; Au, W.; Su, M. Prevalence of coal workers’ pneumoconiosis in China: A systematic analysis of 2001–2011 studies. Int. J. Hyg. Environ. Health 2013, 217, 46–51. [Google Scholar] [CrossRef]
- Coggon, D.; Taylor, A.N. Coal mining and chronic obstructive pulmonary disease: A review of the evidence. Thorax 1998, 53, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Laney, A.S.; Petsonk, E.L.; Attfield, M.D. Pneumoconiosis among underground bituminous coal miners in the United States: Is silicosis becoming more frequent? Occup. Environ. Med. 2010, 67, 652–656. [Google Scholar] [CrossRef]
- Gulumian, M.; Borm, P.J.A.; Vallyathan, V.; Castranova, V.; Donaldson, K.; Nelson, G.; Murray, J. Mechanistically Identified Suitable Biomarkers of Exposure, Effect, and Susceptibility for Silicosis and Coal-Worker’S Pneumoconiosis: A Comprehensive Review. J. Toxicol. Environ. Health Part B 2006, 9, 357–395. [Google Scholar] [CrossRef]
- Hall, N.B.; Halldin, C.N.; Blackley, D.J.; Laney, A.S. Assessment of pneumoconiosis in surface coal miners after implementation of a national radiographic surveillance program, United States, 2014–2019. Am. J. Ind. Med. 2020, 63, 1104–1108. [Google Scholar] [CrossRef]
- Baur, X.; Sanyal, S.; Abraham, J.L. Mixed-dust pneumoconiosis: Review of diagnostic and classification problems with presentation of a work-related case. Sci. Total Environ. 2018, 652, 413–421. [Google Scholar] [CrossRef]
- Ayaaba, E.; Liu, Y.; Li, Y.; Han, L.; Yedu, Q.; Chunhui, N. Measures to control the prevalence of pneumoconiosis in coal mining: A review of the literature. Int. J. Transl. Med. Res. Public Health 2017, 1, 4–13. [Google Scholar] [CrossRef]
- Luo, Y.; Qi, X.; Pang, J.; Wang, J.; Wang, C. Omics Approaches for Exploring Pneumoconiosis: A Review. Biomed. Environ. Sci. 2021, 34, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Perret, J.L.; Plush, B.; Lachapelle, P.; Hinks, T.S.; Walter, C.; Clarke, P.; Irving, L.; Brady, P.; Dharmage, S.; Stewart, A. Coal mine dust lung disease in the modern era. Respirology 2017, 22, 662–670. [Google Scholar] [CrossRef]
- Han, L.; Han, R.; Ji, X.; Wang, T.; Yang, J.; Yuan, J.; Wu, Q.; Zhu, B.; Zhang, H.; Ding, B.; et al. Prevalence Characteristics of Coal Workers’ Pneumoconiosis (CWP) in a State-Owned Mine in Eastern China. Int. J. Environ. Res. Public Health 2015, 12, 7856–7867. [Google Scholar] [CrossRef]
- Jones, J.; Hancox, J. Reticular Interstitial Pattern, Radiology Reference Article, Radiopaedia.org. Available online: https://radiopaedia.org/articles/reticular-interstitial-pattern (accessed on 7 August 2020).
- Oikonomou, A.; Prassopoulos, P. Mimics in chest disease: Interstitial opacities. Insights into Imaging 2012, 4, 9–27. [Google Scholar] [CrossRef] [PubMed]
- Nickson, C. Pulmonary Opacities on Chest X-ray, LITFL-CCC Differential Diagnosis. 2019. Available online: https://litfl.com/pulmonary-opacities-on-chest-x-ray/ (accessed on 6 August 2020).
- Occupational Safety and Health Series No. 22 (Rev. 2011), “Guidelines for the use of the ILO International Classification of Radiographs of Pneumoconioses, Revised Edition 2011”. 2011. Available online: http://www.ilo.org/global/topics/safety-and-health-at-work/resources-library/publications/WCMS_168260/lang--en/index.htm (accessed on 7 August 2020).
- Chong, S.; Lee, K.S.; Chung, M.J.; Han, J.; Kwon, O.J.; Kim, T.S. Pneumoconiosis: Comparison of Imaging and Pathologic Findings. RadioGraphics 2006, 26, 59–77. [Google Scholar] [CrossRef]
- Sun, J.; Weng, D.; Jin, C.; Yan, B.; Xu, G.; Jin, B.; Xia, S.; Chen, J. The Value of High Resolution Computed Tomography in the Diagnostics of Small Opacities and Complications of Silicosis in Mine Machinery Manufacturing Workers, Compared to Radiography. J. Occup. Health 2008, 50, 400–405. [Google Scholar] [CrossRef]
- Ngatu, N.R.; Suzuki, S.; Kusaka, Y.; Shida, H.; Akira, M.; Suganuma, N. Effect of a Two-hour Training on Physicians’ Skill in Interpreting Pneumoconiotic Chest Radiographs. J. Occup. Health 2010, 52, 294–301. [Google Scholar] [CrossRef]
- Murray, V.; Pattichis, M.S.; Davis, H.; Barriga, E.S.; Soliz, P. Multiscale AM-FM analysis of pneumoconiosis X-ray images. In Proceedings of the International Conference on Image Processing (ICIP), Cairo, Egypt, 7–10 November 2009; pp. 4201–4204. [Google Scholar] [CrossRef]
- Van Ginneken, B.B.; Katsuragawa, S.; Romeny, B.B.T.H.; Doi, K.; Viergever, M.M. Automatic detection of abnormalities in chest radiographs using local texture analysis. IEEE Trans. Med Imaging 2002, 21, 139–149. [Google Scholar] [CrossRef]
- Kondo, H.; Kouda, T. Detection of pneumoconiosis rounded opacities using neural network. In Proceedings of the Annual Conference of the North American Fuzzy Information Processing Society—NAFIPS, Vancouver, BC, Canada, 25–28 July 2001; Volume 3, pp. 1581–1585. [Google Scholar] [CrossRef]
- Kondo, H.; Kouda, T. Computer-aided diagnosis for pneumoconiosis using neural network. Int. J. Biomed. Soft Comput. Hum. Sci. Off. J. Biomed. Fuzzy Syst. Assoc. 2001, 7, 13–18. [Google Scholar]
- Kondo, H.; Zhang, L.; Koda, T. Computer Aided Diagnosis for Pneumoconiosis Radiograps Using Neural Network. Int. Arch. Photogramm. Remote Sens. 2000, 33, 453–458. [Google Scholar]
- Ledley, R.S.; Huang, H.; Rotolo, L.S. A texture analysis method in classification of coal workers’ pneumoconiosis. Comput. Biol. Med. 1975, 5, 53–67. [Google Scholar] [CrossRef]
- Katsuragawa, S.; Doi, K.; MacMahon, H.; Nakamori, N.; Sasaki, Y.; Fennessy, J.J. Quantitative computer-aided analysis of lung texture in chest radiographs. RadioGraphics 1990, 10, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hasegawa, J.-I.; Toriwaki, J.-I. Quantitative diagnosis of pneumoconiosis based on recognition of small rounded opacities in chest X-ray images. In Proceedings of the International Conference on Pattern Recognition, Rome, Italy, 14 May–17 November 1988; pp. 462–464. [Google Scholar] [CrossRef]
- Kobatake, H.; Oh’Ishi, K.; Miyamichi, J. Automatic diagnosis of pneumoconiosis by texture analysis of chest X-ray images. In Proceedings of the IEEE International Conference on Acoustics, Speech, and Signal Processing; Institute of Electrical and Electronics Engineers, Dalas, TX, USA, 6–9 April 1987; Volume 12, pp. 610–613. [Google Scholar] [CrossRef]
- Savol, A.M.; Li, C.C.; Hoy, R.J. Computer-aided recognition of small rounded pneumoconiosis opacities in chest X-rays. IEEE Trans. Pattern Anal. Mach. Intell. 1980, PAMI-2, 479–482. [Google Scholar] [CrossRef]
- Turner, A.F.; Kruger, R.P.; Thompson, W.B. Automated computer screening of chest radiographs for pneumoconiosis. Investig. Radiol. 1976, 11, 258–266. [Google Scholar] [CrossRef]
- Jagoe, J.R.; Paton, K.A. Measurement of Pneumoconiosis by Computer. IEEE Trans. Comput. 1976, C-25, 95–97. [Google Scholar] [CrossRef]
- Jagoe, J.R.; Paton, K.A. Reading chest radiographs for pneumoconiosis by computer. Occup. Environ. Med. 1975, 32, 267–272. [Google Scholar] [CrossRef]
- Hall, E.L.; Crawford, W.O.; Roberts, F.E. Computer Classification of Pneumoconiosis from Radiographs of Coal Workers. IEEE Trans. Biomed. Eng. 1975, BME-22, 518–527. [Google Scholar] [CrossRef]
- Kruger, R.P.; Thompson, W.B.; Turner, A.F. Computer Diagnosis of Pneumoconiosis. IEEE Trans. Syst. Man Cybern. 1974, 4, 40–49. [Google Scholar] [CrossRef]
- Arzhaeva, Y.; Wang, D.; Devnath, L.; Amirgholipour, S.K.; McBean, R.; Hillhouse, J.; Luo, S.; Meredith, D.; Newbigin, K. Development of Automated Diagnostic Tools for Pneumoconiosis Detection from Chest X-ray Radiographs; The Final Report Prepared for Coal Services Health and Safety Trust; Coal Services Health and Safety Trust: Sydney, Australia, 2019. [Google Scholar]
- Okumura, E.; Kawashita, I.; Ishida, T. Computerized Classification of Pneumoconiosis on Digital Chest Radiography Artificial Neural Network with Three Stages. J. Digit. Imaging 2017, 30, 413–426. [Google Scholar] [CrossRef]
- Abe, K.; Minami, M.; Miyazaki, R.; Tian, H. Application of a Computer-aid Diagnosis of Pneumoconiosis for CR X-ray Images. J. Biomed. Eng. Med Imaging 2014, 1, 606. [Google Scholar] [CrossRef]
- Zhu, B.; Luo, W.; Li, B.; Chen, B.; Yang, Q.; Xu, Y.; Wu, X.; Chen, H.; Zhang, K. The development and evaluation of a computerized diagnosis scheme for pneumoconiosis on digital chest radiographs. Biomed. Eng. Online 2014, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Okumura, E.; Kawashita, I.; Ishida, T. Development of CAD based on ANN analysis of power spectra for pneumoconiosis in chest radiographs: Effect of three new enhancement methods. Radiol. Phys. Technol. 2014, 7, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Abe, K. Computer-Aided Diagnosis of Pneumoconiosis X-ray Images Scanned with a Common CCD Scanner. Autom. Control Intell. Syst. 2013, 1, 24. [Google Scholar] [CrossRef][Green Version]
- Cai, C.X.; Zhu, B.Y.; Chen, H. Computer-aided diagnosis for pneumoconiosis based on texture analysis on digital chest radiographs. Appl. Mech. Mater. 2013, 241, 244–247. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, H.; Chen, B.; Xu, Y.; Zhang, K. Support Vector Machine Model for Diagnosing Pneumoconiosis Based on Wavelet Texture Features of Digital Chest Radiographs. J. Digit. Imaging 2013, 27, 90–97. [Google Scholar] [CrossRef]
- Masumoto, Y.; Kawashita, I.; Okura, Y.; Nakajima, M.; Okumura, E.; Ishida, T. Computerized Classification of Pneumoconiosis Radiographs Based on Grey Level Co-occurrence Matrices. Jpn. J. Radiol. Technol. 2011, 67, 336–345. [Google Scholar] [CrossRef]
- Okumura, E.; Kawashita, I.; Ishida, T. Computerized Analysis of Pneumoconiosis in Digital Chest Radiography: Effect of Artificial Neural Network Trained with Power Spectra. J. Digit. Imaging 2010, 24, 1126–1132. [Google Scholar] [CrossRef]
- Yu, P.; Xu, H.; Zhu, Y.; Yang, C.; Sun, X.; Zhao, J. An Automatic Computer-Aided Detection Scheme for Pneumoconiosis on Digital Chest Radiographs. J. Digit. Imaging 2010, 24, 382–393. [Google Scholar] [CrossRef]
- Nakamura, M.; Abe, K.; Minami, M. Extraction of Features for Diagnosing Pneumoconiosis from Chest Radiographs Obtained with a CCD Scanner. J. Digit. Inf. Manag. 2010, 8, 147–152. [Google Scholar]
- Sundararajan, R.; Xu, H.; Annangi, P.; Tao, X.; Sun, X.; Mao, L. A multiresolution support vector machine based algorithm for pneumoconiosis detection from chest radiographs. In Proceedings of the 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Rotterdam, The Netherlands, 14–17 April 2010; pp. 1317–1320. [Google Scholar]
- Nakamura, M.; Abe, K.; Minami, M. Quantitative evaluation of pneumoconiosis in chest radiographs obtained with a CCD scanner. In Proceedings of the 2nd International Conference on the Applications of Digital Information and Web Technologies, ICADIWT 2009, London, UK, 4–6 August 2009; pp. 646–651. [Google Scholar] [CrossRef]
- Yu, P.; Zhao, J.; Xu, H.; Yang, C.; Sun, X.; Chen, S.; Mao, L. Computer Aided Detection for Pneumoconiosis Based on Histogram Analysis. In Proceedings of the 2009 1st International Conference on Information Science and Engineering, ICISE 2009, Nanjing, China, 26–28 December 2009; pp. 3625–3628. [Google Scholar] [CrossRef]
- Yu, P.; Zhao, J.; Xu, H.; Sun, X.; Mao, L. Computer Aided Detection for Pneumoconiosis Based on Co-Occurrence Matrices Analysis. In Proceedings of the 2009 2nd International Conference on Biomedical Engineering and Informatics, Tianjin, China, 17–19 October 2009; pp. 1–4. [Google Scholar] [CrossRef]
- Pattichis, M.; Christodoulou, C.; James, D.; Ketai, L.; Soliz, P. A screening system for the assessment of opacity profusion in chest radiographs of miners with pneumoconiosis. In Proceedings of the IEEE Southwest Symposium on Image Analysis and Interpretation, Sante Fe, NM, USA, 7–9 April 2002; pp. 130–133. [Google Scholar] [CrossRef]
- Soliz, P.; Pattichis, M.; Ramachandran, J.; James, D.S. Computer-assisted diagnosis of chest radiographs for pneumoconioses. In Proceedings of the Medical Imaging 2001: Image Processing, San Diago, CA, USA, 17–22 February 2001; Volume 4322, pp. 667–675. [Google Scholar] [CrossRef]
- Zhang, L.; Rong, R.; Li, Q.; Yang, D.M.; Yao, B.; Luo, D.; Zhang, X.; Zhu, X.; Luo, J.; Liu, Y.; et al. A deep learning-based model for screening and staging pneumoconiosis. Sci. Rep. 2021, 11, 2201. [Google Scholar] [CrossRef] [PubMed]
- Devnath, L.; Luo, S.; Summons, P.; Wang, D. Automated detection of pneumoconiosis with multilevel deep features learned from chest X-Ray radiographs. Comput. Biol. Med. 2020, 129, 104125. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Arzhaeva, Y.; Devnath, L.; Qiao, M.; Amirgholipour, S.; Liao, Q.; McBean, R.; Hillhouse, J.; Luo, S.; Meredith, D.; et al. Automated Pneumoconiosis Detection on Chest X-Rays Using Cascaded Learning with Real and Synthetic Radiographs. In Proceedings of the 2020 Digital Image Computing: Techniques and Applications (DICTA), Melbourne, Australia, 29 November–1 December 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Wang, X.; Yu, J.; Zhu, Q.; Li, S.; Zhao, Z.; Yang, B.; Pu, J. Potential of deep learning in assessing pneumoconiosis depicted on digital chest radiography. Occup. Environ. Med. 2020, 77, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Devnath, L.; Luo, S.; Summons, P.; Wang, D. Performance Comparison of Deep Learning Models for Black Lung Detection on Chest X-ray Radiographs. In Proceedings of the ACM International Conference Proceeding Series; Association for Computing Machinery: New York, NY, USA, 2020; pp. 152–154. [Google Scholar] [CrossRef]
- Devnath, L.; Luo, S.; Summons, P.; Wang, D. An accurate black lung detection using transfer learning based on deep neural networks. In Proceedings of the International Conference Image and Vision Computing New Zealand, Dunedin, New Zealand, 2–4 December 2019. [Google Scholar] [CrossRef]
- Zheng, R.; Deng, K.; Jin, H.; Liu, H.; Zhang, L. An Improved CNN-Based Pneumoconiosis Diagnosis Method on X-ray Chest Film. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), LNCS; Springer: New York, NY, USA, 2019; Volume 11956 LNCS, pp. 647–658. [Google Scholar]
- Wirth, M.A. Texture Analysis; University of Guelph: Guelph, ON, Canada, 2004. [Google Scholar]
- Armi, L.; Fekri-Ershad, S. Texture image analysis and texture classification methods—A review. Int. Online J. Image Process. Pattern Recognit. 2019, 2, 1–29. [Google Scholar]
- Haralick, R.M. Statistical image texture analysis. In Handbook of Pattern Recognition and Image Processing; Academic Press: Cambridge, MA, USA, 1986; pp. 247–279. [Google Scholar]
- Tomita, F.; Tsuji, S. Statistical Texture Analysis. In Computer Analysis of Visual Textures; Springer: New York, NY, USA, 1990; pp. 13–36. [Google Scholar]
- Srinivasan, G.N.; Shobha, G. Statistical Texture Analysis. Int. J. Comput. Inf. Eng. 2008, 2, 4268–4273. [Google Scholar]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- He, D.-C.; Wang, L. Texture features based on texture spectrum. Pattern Recognit. 1991, 24, 391–399. [Google Scholar] [CrossRef]
- Hu, X.; Ensor, A. Fourier Spectrum Image Texture Analysis. In Proceedings of the International Conference Image and Vision Computing New Zealand, Auckland, New Zealand, 19–21 November 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man. Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef]
- Monash Centre for Occupational and Environmental Health. Review of Respiratory Component of the Coal Mine Workers’ Health Scheme for the Queensland Department of Natural Resources and Mines Final Report; Monash University: Monash, Australia, 2016. [Google Scholar]
- Black Lung Case Confirmed in NSW—Australian Mining. Available online: https://www.australianmining.com.au/news/black-lung-case-confirmed-in-nsw/ (accessed on 2 July 2020).
- Vallyathan, V.; Landsittel, D.P.; Petsonk, E.L.; Kahn, J.; Parker, J.E.; Osiowy, K.T.; Green, F.H. The Influence of Dust Standards on the Prevalence and Severity of Coal Worker’s Pneumoconiosis at Autopsy in the United States of America. Arch. Pathol. Lab. Med. 2011, 135, 1550–1556. [Google Scholar] [CrossRef]
- Materka, A.; Strzelecki, M. Texture Analysis Methods—A Review; COST B11 Report; Technical University of Lodz, Institute of Electronics: Lodz, Poland, 1998. [Google Scholar]
- Agwu, K.K.; Ohagwu, C.C. Histogram-Based Texture Characterization and Classification of Brain Tissues in Non-Contrast CT Images of Stroke Patients. In Pattern Recognition—Analysis and Applications; InTech: London, UK, 2016. [Google Scholar]
- Sattar, F.; Floreby, L.; Salomonsson, G.; Lovstrom, B. Image enhancement based on a nonlinear multiscale method. IEEE Trans. Image Process. 1997, 6, 888–895. [Google Scholar] [CrossRef]
- Toet, A. Multiscale color image enhancement. Pattern Recognit. Lett. 1992, 13, 167–174. [Google Scholar] [CrossRef]
- Scheunders, P.; Livens, S.; Van de Wouwer, G.; Vautrot, P.; Van Dyck, D. Wavelet-based Texture Analysis. Int. J. Comput. Sci. Inf. Manag. 1997, 1, 22–34. [Google Scholar]
- Arivazhagan, S.; Ganesan, L. Texture classification using wavelet transform. Pattern Recognit. Lett. 2003, 24, 1513–1521. [Google Scholar] [CrossRef]
- Livens, S. Wavelets for texture analysis, an overview. In Proceedings of the Sixth International Conference on Image Processing and Its Applications, Dublin, Ireland, 14–17 July 1997; pp. 581–585. [Google Scholar] [CrossRef]
- Chang, T.; Kuo, C.-C. Texture analysis and classification with tree-structured wavelet transform. IEEE Trans. Image Process. 1993, 2, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Kouda, T.; Kondo, H. Automatic Detection of Interstitial Lung Disease using Neural Network. Int. J. Fuzzy Log. Intell. Syst. 2002, 2, 15–19. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Goodfellow, L.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Nielsen, M. Neural Networks and Deep Learning; Determination Press: San Francisco, CA, USA, 2015; Volume 2018. [Google Scholar]
- Zhang, B.; Qi, S.; Monkam, P.; Li, C.; Yang, F.; Yao, Y.-D.; Qian, W. Ensemble Learners of Multiple Deep CNNs for Pulmonary Nodules Classification Using CT Images. IEEE Access 2019, 7, 110358–110371. [Google Scholar] [CrossRef]
- Shih, F.Y. Image Processing and Pattern Recognition: Fundamentals and Techniques; John Wiley and Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the Advances in Neural Information Processing Systems 25 (NIPS 2012), Stateline, NV, USA, 3–8 December 2012; pp. 1097–1105. [Google Scholar]
- He, K. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Chollet, F. Building Powerful Image Classification Models Using Very Little Data, Keras Blog. 2016. Available online: http://deeplearning.lipingyang.org/wp-content/uploads/2016/12/Building-powerful-image-classification-models-using-very-little-data.pdf (accessed on 24 April 2020).
- Zhu, J.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A Survey on Deep Transfer Learning. In Proceedings of the 27th International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October 2018; pp. 270–279. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Torrey, L.; Shavlik, J. Transfer Learning: Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques; IGI Global: Hershey, PA, USA, 2010. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition, Published as a Conference Paper at ICLR. 2015. Available online: https://arxiv.org/abs/1409.1556 (accessed on 26 April 2020).
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Huang, G.; Liu, Z.; Van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
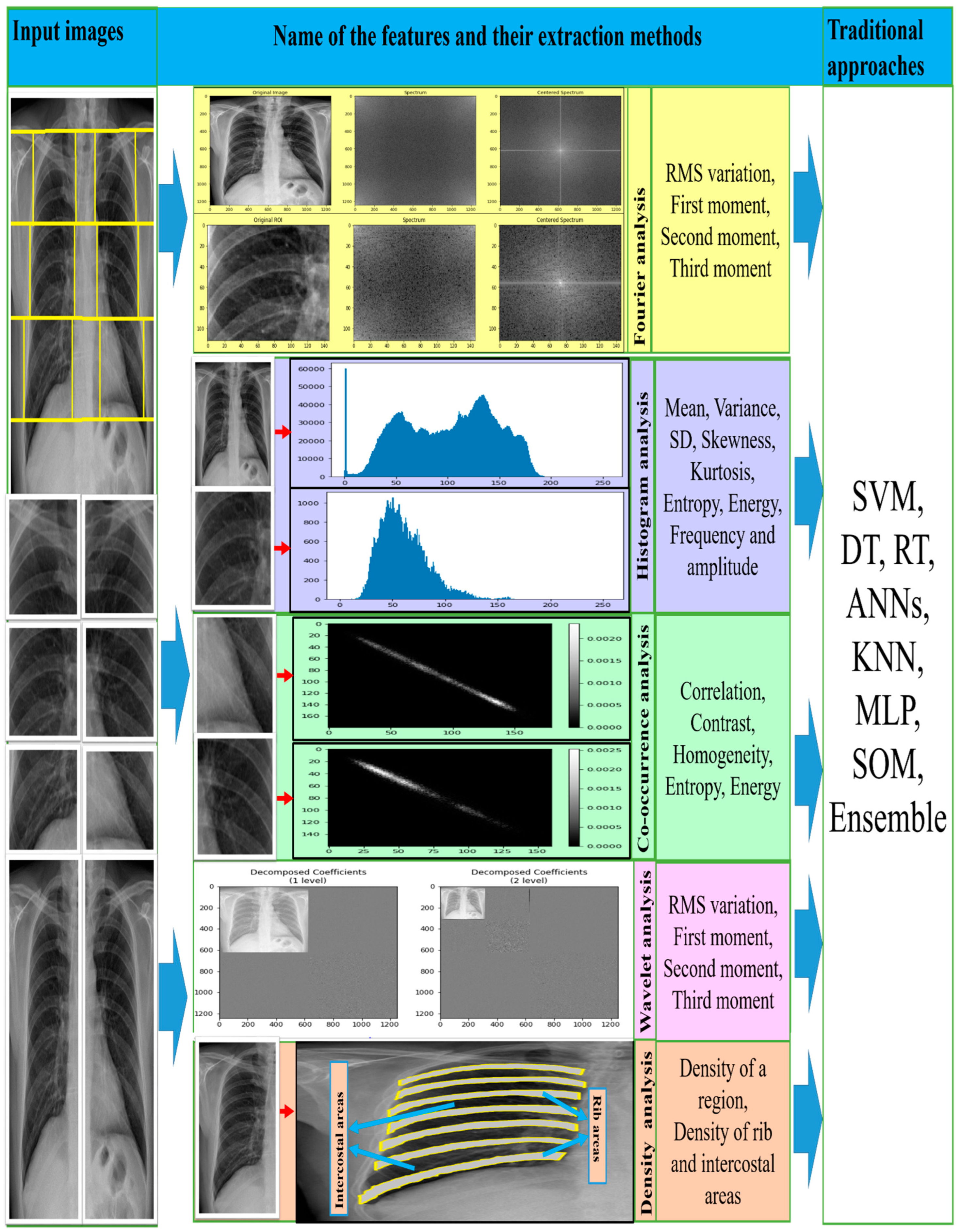
| Feature Types | Features Name | Descriptions |
|---|---|---|
| Fourier spectrum-based | RMS variation | A measurement of the magnitude of lung texture |
| First moment | Central tendency of lung texture | |
| Second moment | A measure of dispersion from the overall central tendency | |
| Third moment | A measure of the nature (coarse or fine) of the lung texture | |
| Co-occurrence matrix-based | Correlation | Measurement of the relationship from different angles or directions between each pair of pixels on the image. Most of them used directions such as 0°, 45°, 90° and 135° |
| Contrast or inertia | Contrast measurements of pixel intensity (greyscale tone or colour tone) using a pixel and its neighbor across the whole image | |
| Homogeneity | Measures the proximity of the pairs of pixels across the diagonal of the co-occurrence matrix. It should be elevated if the greyscale levels of all diagonal entries is similar | |
| Entropy | Measures spatial disturbances in pixel intensity relations which could be responsible for the image abnormality | |
| Energy | Shows the uniformity of the intensity relationships of the pixels by measuring the number of repeated pairs. The higher value of energy means the bigger homogeneity presents in the texture | |
| Histogram-based | Mean | A measure of the colour intensity of each pixel on which the image brightness depends |
| Variance | A measure of the breadth of the histogram indicates the deviation of the grey levels from the mean value | |
| SD | A scalar value computed from the image array that shows the lower or higher contrast of the colour intensities | |
| Skewness | The positive and negative asymmetry represents the degree of distortion of the histogram in relation to the mean intensity distribution, giving an idea about the image of a surface | |
| kurtosis | It is a measure of the degree of sharpness of the histogram relative to the mean intensity distribution | |
| Entropy | Entropy measures the random nature of the distribution of coefficient values on intensity distributions. It provides high readings with an image of more intensity levels | |
| Energy | The energy characteristic measures the uniform distribution of the intensity levels. It provides high readings with an image of fewer intensity levels | |
| Wavelet transform-based | Energy | A wavelet coefficient is calculated from the distribution of grey level intensity in the sub-band images on a successive scale. The different energy levels of the sub-bands provide the differences in texture patterns |
| Density distribution-based | Density of a region | Measures how many pixels are contained in a particular region. The rapidly changing density of a region indicates the profusion of opacities |
| Density of rib areas | Measures the mean of the pixel densities obtained from all the rib areas. The higher contrast occurs when the opacities appear around the edges of the ribs. | |
| Density of intercostal areas | Measures the average pixel densities for all intercostal areas. A higher contrast occurs when the opacities appear around the edges between the intercostal and rib areas |
| Year and Country of Data | Ref No. | Feature Analysis Method | Classical Approaches | Number of CWP CXR | Evaluation Performance |
|---|---|---|---|---|---|
| Accuracy | |||||
| 2009 (M) | [34] | Histogram analysis | Computer and ILO standard | 11 | AUC > 80.00% |
| 2002 | [35] | Opacity measurement | NN and ILO standard-based | 1 | - |
| 2001 | [36] | Opacity measurement | NN and ILO standard-based | 1 | |
| 2001 | [37] | Opacity measurement | NN and ILO standard-based | 1 | - |
| 2000 | [38] | Opacity measurement | NN and ILO standard-based | 1 | - |
| 1997 (U) | [39] | Fourier spectrum | Computer and ILO standard-based | 68 | - |
| 1990 (J) | [40] | Fourier spectrum | Computer and ILO standard-based | - | |
| 1988 (J) | [41] | Opacity measurement | Computer and ILO standard-based | 9 | 81.0% |
| 1987 (J) | [42] | Co-occurrence matrix, density distribution | Computer and ILO standard-based | 11 | 81.8% |
| 1980 (U) | [43] | Opacity measurement | Computer and ILO standard-based | 3 | 67% |
| 1976 (U) | [44] | Fourier spectrum | Computer and ILO standard-based | 141 | 82.9% |
| 1976 (U) | [45] | Density Distribution | Computer and ILO standard-based | 36 | 80.5% |
| 1975 (U) | [46] | Density Distribution | Computer and ILO standard-based | 36 | 80.5% |
| 1975 (U) | [47] | Histogram analysis | Computer and ILO standard-based | 38 | 84.0% |
| 1974 | [48] | Fourier spectrum, co-occurrence matrix | Computer and ILO standard-based | 141 | 88.0% |
| Year and Country of D | Ref No. | Feature Analysis Method | Traditional Machine Learning Approaches | Number of CWP CXR | Evaluation Performance | |||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Specificity | Recall | AUC | |||||
| 2019 (A) | [49] | Histogram analysis | SVM, MLP, NN | 71 | SVM = 73.17% | 92.31% | 73.30% | |
| MLP = 71.11% | 72.00% | 70.00% | ||||||
| NN = 83.00% | 85.00% | 82.00% | ||||||
| 2017 (J) | [50] | Fourier spectrum, co-occurrence matrix, histogram analysis | ANN | 46 | - | Category 1 = 38.2% | - | Category 1 = 73.0% |
| Category 2 = 52.5% | Category 2 = 79.0% | |||||||
| Category 3 = 60.1% | Category 3 = 85.0% | |||||||
| 2014 (J) | [51] | Density distribution | SVM, RT, NN | 15 right-lung | - | - | RT = 93.2% | - |
| NN = 93.2% | ||||||||
| SVM = 93.2% | ||||||||
| 2014 (C) | [52] | Wavelet analysis | SVM and ensemble | 40 | 90.5% | 93.3% | 84.9% | 96.1% |
| 2014 (J) | [53] | Fourier spectrum, co-occurrence matrix | ANN | 15 | - | - | - | 93.0% |
| 2013 (J) | [54] | Density Distribution | SVM, RT, NN | 12 right-lung | - | - | RT = 91.67% | |
| NN = 91.67% | ||||||||
| SVM = 100.0% | ||||||||
| 2013 (C) | [55] | Co-occurrence matrix, histogram analysis | ANN | 40 | 79.3% | 70.6% | 91.7% | 85.8% |
| 2013 (C) | [56] | Wavelet analysis | SVM and DT | 40 | SVM = 87.2% | SVM = 90.6% | SVM = 80.0% | SVM = 94.0% |
| DT = 83.2% | DT = 89.4% | DT = 70.0% | DT = 86.0% | |||||
| 2011 (J) | [57] | Co-occurrence matrix | SVM | 68 | 69.7% | - | - | - |
| 2011 (J) | [58] | Fourier spectrum, co-occurrence matrix | ANN | 12 | - | - | - | 97.2% |
| 2011 (C) | [59] | Co-occurrence matrix, histogram analysis | SVM and ensemble | 250 | 88.9% | 87.7% | 92.0% | 97.8% |
| 2010 (J) | [60] | Density distribution | SVM, RT, NN | 6 right-lung | - | - | - | - |
| 2010 (C) | [61] | Co-occurrence matrix, histogram analysis | SVM and Classifiers ensemble | 259 | 92.83% | 90.25% | 96.65% | - |
| 2009 (J) | [62] | Density distribution | SVM, RT, NN | 6 right-lung | - | - | - | - |
| 2009 (C) | [63] | Histogram analysis | SVM | 196 | 94.1% | 94.6% | 93.6% | |
| 2009 (C) | [64] | Co-occurrence matrix | SVM | 59 | 95.15% | 94.2% | 95.6% | |
| 2002 (M) | [65] | Co-occurrence and spatial dependence matrix analysis | SOM, NN, KNN | 74 | SOM = 71.0% | - | - | |
| NN = 75.0% | ||||||||
| KNN = 72.0% | ||||||||
| 2001 (C) | [66] | Co-occurrence matrix | NN | 212 | 86.8% | - | - | - |
| Year and Country of Data | Ref No. | Feature Analysis Method | Deep Learning Approaches | Number of CWP CXR | Evaluation Performance | |||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Specificity | Recall | AUC | |||||
| 2021(C) | [67] | Non-texture CNN | ResNet | 512 | 92.70% | - | - | - |
| 2021(A) | [68] | Non-texture CNN | CheXNet | 71 | 92.68% | 83.33% | 100% | 97.05% |
| 2020(A) | [69] | Non-texture CNN | Cascaded Learning, CheXNet | 71 | Cascaded = 90.24% | 88.46% | 93.33% | - |
| CheXNet = 78.05% | 80.77% | 73.33% | ||||||
| 2020 (C) | [70] | Non-texture CNN | InceptionV3 | 923 | - | 93.30% | 62.30% | 87.80% |
| 2020 (A) | [71] | Non-texture CNN | VGG16, VGG19, ResNet, InceptionV3, Xception, DenseNet, CheXNet | 71 | VGG16 = 82.93% | 80.00% | 84.62% | - |
| VGG19 = 80.49% | 80.00% | 80.77% | ||||||
| ResNet = 85.37% | 80.00% | 88.46% | ||||||
| InceptionV3 = 87.80% | 86.67% | 88.46% | ||||||
| Xception = 85.37% | 93.33% | 80.77% | ||||||
| DenseNet = 82.93% | 80.00% | 84.62% | ||||||
| CheXNet = 85.37% | 93.33% | 80.77% | ||||||
| 2019 (A) | [49] | Non-texture CNN | 15 layers CNN | 71 | 90.24% | 89.29% | 90.74% | - |
| 2019 (A) | [72] | Non-texture CNN | DenseNet, CheXNet | 71 | CheXNet = 85.37% | 80.00% | 88.46% | - |
| DenseNet = 80.49% | 73.33% | 84.62% | ||||||
| 2019 (C) | [73] | Non-texture CNN | LeNet, AleXNet, InceptionV1, InceptionV2, GoogleNetCF | 1600 | GoogleNetCF = 93.88% | - | - | - |
| InceptionV1 = 91.60% | ||||||||
| InceptionV2 = 90.70% | ||||||||
| AleXNet = 87.90% | ||||||||
| LeNet = 71.6% | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Devnath, L.; Summons, P.; Luo, S.; Wang, D.; Shaukat, K.; Hameed, I.A.; Aljuaid, H. Computer-Aided Diagnosis of Coal Workers’ Pneumoconiosis in Chest X-ray Radiographs Using Machine Learning: A Systematic Literature Review. Int. J. Environ. Res. Public Health 2022, 19, 6439. https://doi.org/10.3390/ijerph19116439
Devnath L, Summons P, Luo S, Wang D, Shaukat K, Hameed IA, Aljuaid H. Computer-Aided Diagnosis of Coal Workers’ Pneumoconiosis in Chest X-ray Radiographs Using Machine Learning: A Systematic Literature Review. International Journal of Environmental Research and Public Health. 2022; 19(11):6439. https://doi.org/10.3390/ijerph19116439
Chicago/Turabian StyleDevnath, Liton, Peter Summons, Suhuai Luo, Dadong Wang, Kamran Shaukat, Ibrahim A. Hameed, and Hanan Aljuaid. 2022. "Computer-Aided Diagnosis of Coal Workers’ Pneumoconiosis in Chest X-ray Radiographs Using Machine Learning: A Systematic Literature Review" International Journal of Environmental Research and Public Health 19, no. 11: 6439. https://doi.org/10.3390/ijerph19116439
APA StyleDevnath, L., Summons, P., Luo, S., Wang, D., Shaukat, K., Hameed, I. A., & Aljuaid, H. (2022). Computer-Aided Diagnosis of Coal Workers’ Pneumoconiosis in Chest X-ray Radiographs Using Machine Learning: A Systematic Literature Review. International Journal of Environmental Research and Public Health, 19(11), 6439. https://doi.org/10.3390/ijerph19116439










