Occurrence of Antibiotic-Resistant Staphylococcus spp. in Orange Orchards in Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Orange Orchards and Recruitment
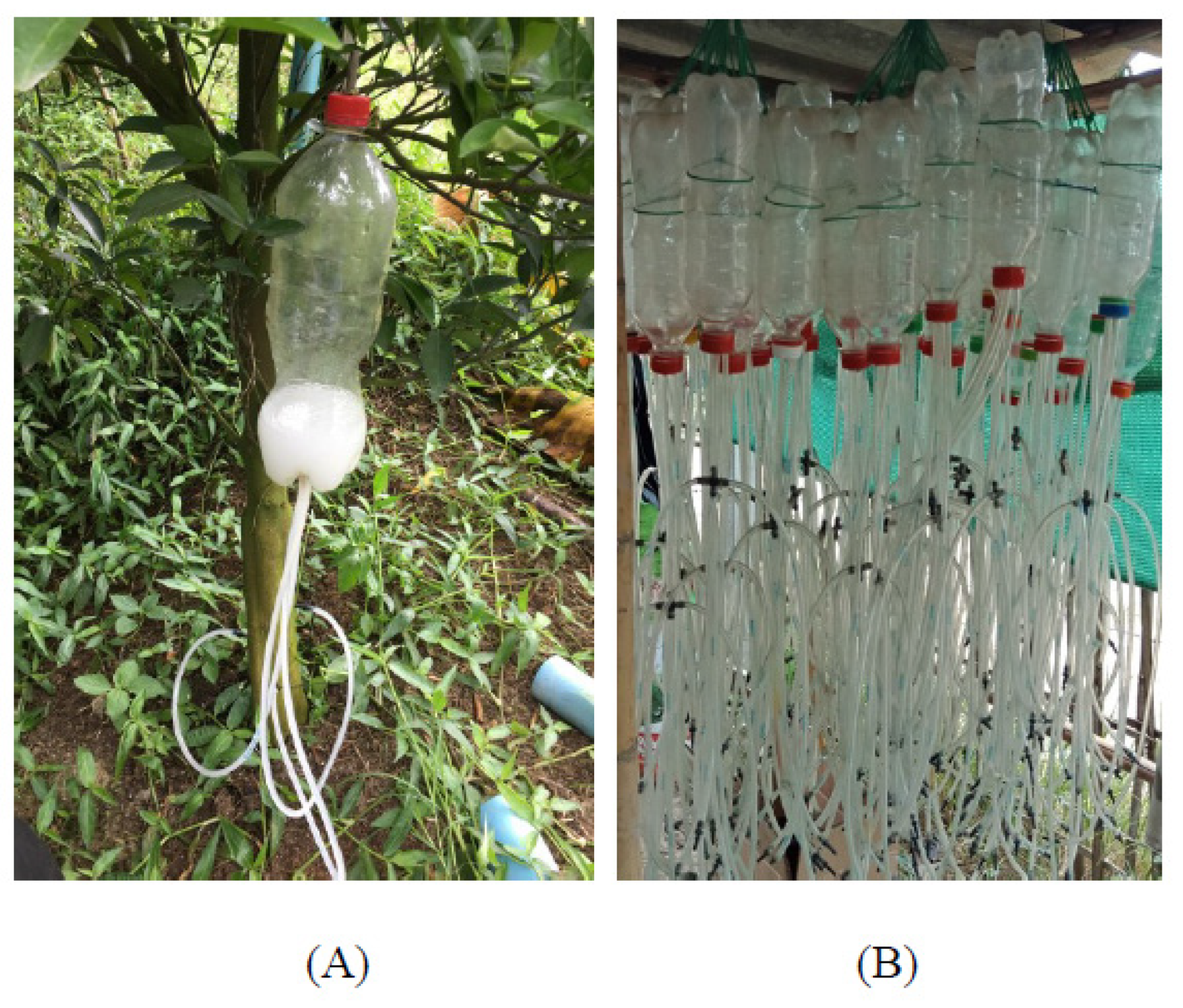
2.2. Environmental Sample Collection
2.3. Worker Samples Collection
2.4. Bacterial Isolates
2.5. Identification of Isolates
2.6. Antimicrobial Susceptibility Testing
2.7. Data Analysis
3. Results
3.1. Demographics
3.2. Identification of Staphylococcus Species Isolated
3.3. Antimicrobial Susceptibility
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Fact Sheet: Antibiotic Resistance. 2018. Available online: http://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 4 November 2021).
- Interagency Coordination Group on Antimicrobial Resistance. No Time to Wait: Securing the Future from Drug-Resistant Infections. Report to the Secretary-General of the United Nations. 2019. Available online: https://www.who.int/publications/i/item/no-time-to-wait-securing-the-future-from-drug-resistant-infections (accessed on 4 November 2021).
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States 2019. 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 4 November 2021).
- Aslam, M. Antimicrobial resistance and genetic profiling of Escherichia coli from a commercial beef packing plant. J. Food Prot. 2006, 69, 1508–1513. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.B.O.; D’Souza, D.H.; Jaykus, L.; Ferket, P.R.; Sheldon, B.W. Genotypes, serotypes, and antibiotic resistance profiles of Salmonella isolated from commercial North Carolina turkey farms. J. Food Prot. 2007, 70, 1328–1333. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Diarra, M.S.; Rempel, H. Antimicrobial resistance genes in Escherichia coli isolates recovered from a commercial beef processing plant. J. Food Prot. 2009, 72, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Edrington, T.S.; Long, M.; Ross, T.T.; Thomas, J.D.; Callaway, T.R.; Anderson, R.C.; Craddock, F.; Salisbury, M.W.; Nisbet, D.J. Prevalence and antimicrobial resistance profiles of Escherichia coli O157: H7 and Salmonella isolated from feedlot lambs. J. Food Prot. 2009, 72, 1713–1717. [Google Scholar] [CrossRef] [PubMed]
- Jouini, A.; Slama, K.B.; Sáenz, Y.; Klibi, N.; Costa, D.; Vinué, L.; Zarazaga, M.; Boudabous, A.; Torres, C. Detection of multiple-antimicrobial resistance and characterization of the implicated genes in Escherichia coli isolates from foods of animal origin in Tunis. J. Food Prot. 2009, 72, 1082–1088. [Google Scholar] [CrossRef]
- Taylor, P.; Reeder, R. Antibiotic use on crops in low and middle-income countries based on recommendations made by agricultural advisors. CABI Agric. Biosci. 2020, 1, 1–14. [Google Scholar] [CrossRef]
- US Environmental Protection Agency. Proposed Registration Decision for the New Use of the Active Ingredient Streptomycin Sulfate on Citrus Crop Group 10–10. 2018. Available online: https://www.regulations.gov/document?D=EPA-HQ-OPP-2016-0067-0023 (accessed on 4 November 2021).
- Vidaver, A.K. Uses of antimicrobials in plant agriculture. Clin. Infect. Dis. 2002, 34, S107–S110. [Google Scholar] [CrossRef]
- Qiao, M.; Ying, G.G.; Singerd, A.C.; Zhu, Y.G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef]
- McKenna, M. Antibiotics set to flood Florida’s troubled orange orchards. Nature 2019, 567, 302–303. [Google Scholar] [CrossRef]
- Schwarz, R.E.; van Vuuren, S.P. Decreases in fruit greening of sweet orange by trunk injections with tetracyclines. Plant Dis. Rep. 1970, 55, 747–750. [Google Scholar]
- Zhang, M.; Powell, C.A.; Guo, Y.; Doud, M.S.; Duan, Y.A. Graft-based chemotherapy method for screening effective molecules and rescuing Huanglongbing-affected citrus plants. Phytopathology 2012, 102, 567–574. [Google Scholar] [CrossRef][Green Version]
- Zhang, M.; Guo, Y.; Powell, C.A.; Doud, M.S.; Yang, C.; Duan, Y. Effective antibiotics against ‘Candidatus Liberibacter asiaticus’ in HLB-affected citrus plants identified via the graft-based evaluation. PLoS ONE 2014, 9, e111032. [Google Scholar] [CrossRef] [PubMed]
- Public Health Ministry of Thailand. Landscape of Antimicrobial Resistance. 2015. Available online: https://www.fda.moph.go.th/sites/drug/SitePages/AMR.aspx (accessed on 4 November 2021).
- Chanvatik, S.; Donnua, S.; Lekagul, A.; Kaewkhankhaeng, W.; Vongmongkol, V.; Athipunyakom, P.; Khamlar, S.; Prommintara, M.; Tangcharoensathien, V. Antibiotic use in mandarin production (Citrus reticulata Blanco) in major mandarin producing areas in Thailand: A survey assessment. PLoS ONE 2019, 14, e0225172. [Google Scholar] [CrossRef] [PubMed]
- Sommanustweechai, A.; Chanvatik, S.; Sermsinsiri, V.; Sivilaikul, S.; Patcharanarumol, W.; Yeunga, S.; Tangcharoensathienb, V. Antibiotic distribution channels in Thailand: Results of key-informant interviews, reviews of drug regulations and database searches. Bull. World Health Organ. 2018, 96, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Dandachi, I.; Sokhn, E.S.; Dahdouh, E.A.; Azar, E.; El-Bazzal, B.; Rolain, J.M.; Daoud, Z. Prevalence and Characterization of Multi-Drug-Resistant Gram-Negative Bacilli Isolated From Lebanese Poultry: A Nationwide Study. Front. Microbiol. 2018, 9, 550. [Google Scholar] [CrossRef]
- Beresin, G.A.; Wright, J.M.; Rice, G.E.; Jagai, J.S. Swine exposure and methicillin-resistant Staphylococcus aureus infection among hospitalized patients with skin and soft tissue infections in Illinois: A ZIP code-level analysis. Environ. Res. 2017, 159, 46–60. [Google Scholar] [CrossRef]
- Kittl, S.; Brodard, I.; Heim, D.; Andina, P.; Fister, P.; Overesch, G. Methicillin-resistant Staphylococcus aureus strains in Swiss pigs and their relation to isolates from farmers and veterinarians. Appl. Environ. Microbiol. 2020, 86, e01865-19. [Google Scholar] [CrossRef]
- Foster, T. Staphylococcus. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. Available online: https://www.ncbi.nlm.nih.gov/books/NBK8448/ (accessed on 20 December 2021).
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef]
- Heilmann, C.; Ziebuhr, W.; Becker, K. Are coagulase-negative staphylococci virulent? Clin. Microbiol. Infect. 2019, 25, 1071–1080. [Google Scholar] [CrossRef]
- Guo, Y.; Song, G.; Sun, M.; Wang, J.; Wang, Y. Prevalence and Therapies of antibiotic-resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2020, 10, 107. [Google Scholar] [CrossRef]
- Ming-Xiang, Z.; Rong-Rong, Z.; Wen-Jun, W.; Ning-Jie, Z.; Wen-En, L.; Fu-Ping, H.; Xue-Gong, F. Antimicrobial resistance and molecular epidemiological characteristics of clinical isolates of Staphylococcus aureus in Changsha area. Chin. Med. J. 2012, 125, 2289–2294. [Google Scholar]
- Charu, A.; Jyoti, K.C.; Shreya, N. Prevalence of Methicillin-Resistant Staphylococcus aureus in Shrines. Int. J. Microbiol. 2020, 2020, 7981648. [Google Scholar]
- Lim, C.; Takahashi, E.; Hongsuwan, M.; Wuthiekanun, V.; Thamlikitkul, V.; Hinjoy, S.; Day, N.P.J.; Peacock, S.J.; Limmathurotsakul, D. Epidemiology and burden of multidrug resistant bacterial infection in a developing country. Elife 2016, 5, e18082. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, W.; Korzeniewska, E.; Harnisz, M.; Hubeny, J.; Buta, M.; Rolbiecki, D. The prevalence of drug-resistant and virulent Staphylococcus spp. in a municipal wastewater treatment plant and their spread in the environment. Environ. Int. 2020, 143, 105914. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Diversity and genetic lineages of environmental staphylococci: A surface water overview. FEMS Microbiol. Ecol. 2020, 96, fiaa191. [Google Scholar] [CrossRef]
- Sharafati Chaleshtori, R.; Sharafati Chaleshtori, F.; Karimi, A. Antibiotic resistance pattern of Staphylococcus strains isolated from orange and apple juices in Shahre-kord, Iran. Pak. J. Med. Sci. 2010, 26, 615–618. [Google Scholar]
- Alipour, M.; Hajiesmaili, R.; Talebjannat, M.; Yahyapour, Y. Identification and antimicrobial resistance of Enterococcus spp. isolated from the river and coastal waters in northern Iran. Sci. World J. 2014, 2014, 287458. [Google Scholar] [CrossRef] [PubMed]
- Ismaïl, R.; Aviat, F.; Valérie, M.; Bayon, I.L.; Gay-Perret, P.; Kutnik, M.; Fédérighi, M. Methods for Recovering Microorganisms from Solid Surfaces Used in the Food Industry: A Review of the Literature. Int. J. Environ. Res. Public Health 2013, 10, 6169–6183. [Google Scholar] [CrossRef]
- Kaur, K.; Kahlon, R.S. Prevalence of Antimicrobial Resistance in Staphylococcus aureus Isolated from Ready to Eat Foods, Hand Swabs and Utensil Swabs of Street Vendors Selling Food on Wheels. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2424–2431. [Google Scholar] [CrossRef]
- Miller, J.M.; Holmes, H.T. Specimen Collection, Transport, and Storage. In Manual of Clinical Microbiology, 7th ed.; Murray, P.R., Baron, E.J., Eds.; American Society for Microbiology: Washington, DC, USA, 1999; pp. 33–104. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing: CLSI Supplement M100, 29th ed.; Clinical and Laboratory Standard Institute: Malvern, PA, USA, 2018; pp. 54–62. [Google Scholar]
- Buchanan, R.E.; Gibbons, N.E. Bergey’s Manual of Determinative Bacteriology, 8th ed.; The Williams and Wilkins Co.: Baltimore, MD, USA, 1974; pp. 483–489. [Google Scholar]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Verweij, P.E.; Kema, G.H.; Zwaan, B.; Melchers, W.J. Triazole fungicides and the selection of resistance to medical triazoles in the opportunistic mould Aspergillus fumigatus. Pest Manag. Sci. 2013, 69, 165–170. [Google Scholar] [CrossRef]
- Xu, Z.; Shah, H.N.; Misra, R.; Chen, J.; Zhang, W.; Liu, Y.; Cutler, R.R.; Mkrtchyan, H.V. The prevalence, antibiotic resistance and mecA characterization of coagulase negative staphylococci recovered from non-healthcare settings in London, UK. Antimicrob. Resist. Infect. Control. 2018, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lu, Q.; Cheng, Y.; Wen, G.; Luo, Q.; Shao, H.; Zhang, T. High concentration of coagulase-negative staphylococci carriage among bioaerosols of henhouses in Central China. BMC Microbiol. 2020, 20, 21. [Google Scholar] [CrossRef] [PubMed]
- Nasaj, M.; Saeidi, Z.; Tahmasebi, H.; Dehbashi, S.; Reza, A.M. Prevalence and distribution of resistance and enterotoxins/enterotoxin-like genes in different clinical isolates of coagulase-negative Staphylococcus. Eur. J. Med. Res. 2020, 25, 48. [Google Scholar] [CrossRef] [PubMed]
- Teeraputon, S.; Santanirand, P.; Wongchai, T.; Songjang, W.; Lapsomthob, N.; Jaikrasun, D.; Toonkaew, S.; Tophon, P. Prevalence of methicillin resistance and macrolide–lincosamide–streptogramin B resistance in Staphylococcus haemolyticus among clinical strains at a tertiary-care hospital in Thailand. New Microbes New Infect. 2017, 19, 28–33. [Google Scholar] [CrossRef]

| Staphylococcus Species | Orange Orchard | Other Fruit Orchard | Total | |||
|---|---|---|---|---|---|---|
| Env. | Worker | Env. | Worker | Env. | Worker | |
| Staphylococcus epidermidis | - | 3 h,h,n | - | 1 n | - | 4 |
| Staphylococcus arlettae | 2 w,w | 1 n | - | - | 2 | 1 |
| Staphylococcus haemolyticus | 1 os | 1 n | - | - | 1 | 1 |
| Staphylococcus saprophyticus | 1 s | - | - | - | 1 | - |
| Susceptibility * | |||
|---|---|---|---|
| Staphylococcal Isolate | Ampicillin | Erythromycin | Tetracycline |
| Staphylococcus epidermidis 01 008h1g2 | R | S | S |
| Staphylococcus epidermidis 008h1g3 | R | S | S |
| Staphylococcus epidermidis 009n1g1 | S | S | S |
| Staphylococcus epidermidis 016h1g2 | S | S | S |
| Staphylococcus arlettae 001wg6 | R | R | S |
| Staphylococcus arlettae 003wg1 | R | R | S |
| Staphylococcus arlettae 006n2g1 | S | R | S |
| Staphylococcus haemolyticus 001n2b1 | S | S | S |
| Staphylococcus haemolyticus 002worg2 | S | R | S |
| Staphylococcus saprophyticus 004soig1 | S | R | S |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rattanapunya, S.; Deethae, A.; Woskie, S.; Kongthip, P.; Matthews, K.R. Occurrence of Antibiotic-Resistant Staphylococcus spp. in Orange Orchards in Thailand. Int. J. Environ. Res. Public Health 2022, 19, 246. https://doi.org/10.3390/ijerph19010246
Rattanapunya S, Deethae A, Woskie S, Kongthip P, Matthews KR. Occurrence of Antibiotic-Resistant Staphylococcus spp. in Orange Orchards in Thailand. International Journal of Environmental Research and Public Health. 2022; 19(1):246. https://doi.org/10.3390/ijerph19010246
Chicago/Turabian StyleRattanapunya, Siwalee, Aomhatai Deethae, Susan Woskie, Pornpimol Kongthip, and Karl R. Matthews. 2022. "Occurrence of Antibiotic-Resistant Staphylococcus spp. in Orange Orchards in Thailand" International Journal of Environmental Research and Public Health 19, no. 1: 246. https://doi.org/10.3390/ijerph19010246
APA StyleRattanapunya, S., Deethae, A., Woskie, S., Kongthip, P., & Matthews, K. R. (2022). Occurrence of Antibiotic-Resistant Staphylococcus spp. in Orange Orchards in Thailand. International Journal of Environmental Research and Public Health, 19(1), 246. https://doi.org/10.3390/ijerph19010246









