Virulence and Drug-Resistance of Staphylococcus aureus Strains Isolated from Venous Ulcers in Polish Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. DNA Isolation
2.3. Polymerase Chain Reaction (PCR) Screening for Resistance Genes
2.4. Susceptibility Testing
2.5. SCCmec Typing
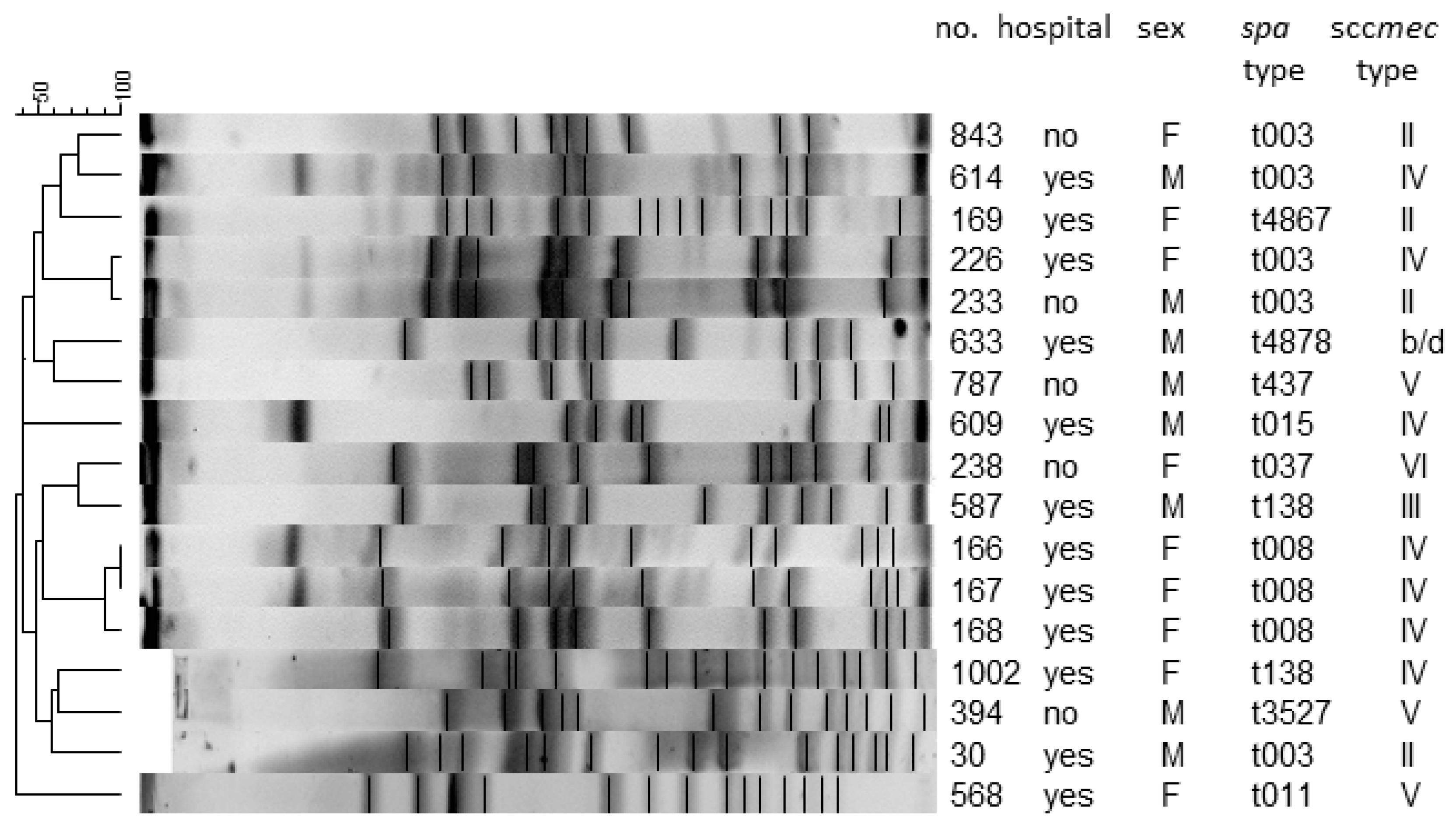
2.6. Pulsed-Field Gel Electrophoresis (PFGE)
2.7. Statistical Methods
2.8. Ethics
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raffetto, J.D. The definition of the venous ulcer. J. Vasc. Surg. 2010, 52, 46S–49S. [Google Scholar] [CrossRef]
- Alavi, A.; Sibbald, R.G.; Phillips, T.J.; Miller, O.F.; Margolis, D.J.; Marston, W.; Woo, K.; Romanelli, M.; Kirsner, R.S. What’ s new: Management of venous leg ulcers Approach to venous leg ulcers. J. Am. Dermatol. 2019, 74, 627–640. [Google Scholar] [CrossRef]
- Nelson, E.A.; Adderley, U. Venous leg ulcers. BMJ Clin. Evid. 2016, 2016, 1902. [Google Scholar]
- Mohd-Zain, Z.; Mohd-Nawi, S.F.A.; Adnan, A.; Kumar, S. Frequency and molecular epidemiology of Panton-Valentine leukocidin gene in Staphylococcus aureus colonising HIV-infected patients. Malays. J. Pathol. 2017, 39, 115–122. [Google Scholar] [PubMed]
- Callam, M.J.; Harper, D.R.; Dale, J.J.; Ruckley, C.V. Chronic ulceration of the leg: Extent of the problem and provision of care. Br. Med. J. 1987, 290, 1855–1856. [Google Scholar] [CrossRef]
- Ruckley, C.V. Socioeconomic impact of chronic venous insufficiency and leg ulcers. Angiology 1997, 48, 67–69. [Google Scholar] [CrossRef]
- Xie, T.; Ye, J.; Rerkasem, K.; Mani, R. The venous ulcer continues to be a clinical challenge: An update. Burn. Trauma 2018, 6, 18. [Google Scholar] [CrossRef]
- Jawień, A.; Szewczyk, M.T.; Kaszuba, A.; Gaciong, Z.A.; Krasiński, Z.; Wroński, J.; Grzela, T.; Koblik, T. Wytyczne Grupy Ekspertów w sprawie gojenia owrzodzeń żylnych goleni. Leczenie Ran 2011, 8, 59–80. [Google Scholar]
- Tomkowski, W.; Kuca, P.; Urbanek, T.; Chmielewski, D.; Krasiński, Z.; Pruszczyk, P.; Windy, J.; Oszkinis, G.; Jawień, A.; Burakowski, J. Venous thromboembolism—recommendations on the prevention, diagnostic approach and management. The 2017 Polish Consensus Statement. Acta Angiol. 2017, 23, 35–71. [Google Scholar] [CrossRef]
- Bryant, R.A. Acute and Chronic Wounds, 2nd ed.; Mosby: St. Louis, MO, USA, 2000. [Google Scholar]
- Agale, S.V. Chronic Leg Ulcers: Epidemiology, Aetiopathogenesis, and Management. Ulcers 2013, 1–9. [Google Scholar] [CrossRef]
- Wertheim, H.F.; Melles, D.C.; Vos, M.C.; van Leeuwen, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Rahim, K.; Saleha, S.; Zhu, X.; Huo, L.; Basit, A.; Franco, O.L. Bacterial Contribution in Chronicity of Wounds. Microb. Ecol. 2016, 73, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Saseedharan, S.; Sahu, M.; Chaddha, R.; Pathrose, E.; Bal, A.; Bhalekar, P.; Sekar, P.; Krishnan, P. Epidemiology of diabetic foot infections in a reference tertiary hospital in India. Braz. J. Microbiol. 2018, 49, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Dunyach-Remy, C.; Essebe, C.N.; Sotto, A.; Lavigne, J.P. Staphylococcus aureus toxins and diabetic foot ulcers: Role in pathogenesis and interest in diagnosis. Toxins 2016, 8, 209. [Google Scholar] [CrossRef]
- Bien, J.; Sokolova, O.; Bozko, P. Characterization of Virulence Factors of Staphylococcus aureus: Novel Function of Known Virulence Factors That are Implicated in Activation of Airway Epithelial Proinflammatory Response. J. Pathog. 2011, 2011, 1–13. [Google Scholar] [CrossRef]
- Wojkowska-Mach, J.; Godman, B.; Glassman, A.; Kurdi, A.; Pilc, A.; Rozanska, A.; Skoczyński, S.; Wałaszek, M.; Bochenek, T. Antibiotic consumption and antimicrobial resistance in Poland; Findings and implications 11 Medical and Health Sciences 1117 Public Health and Health Services. Antimicrob. Resist. Infect. Control 2018, 7. [Google Scholar] [CrossRef]
- ECDC. European Center for Disease Control and Prevention. Annual Surveillance Reports and Protocols-Antimicrobial Consumption. 2015. Available online: https://ecdc.europa.eu/en/antimicrobial-consumption/surveillance-anddisease-%0Adata/report-protocol (accessed on 16 August 2017).
- Pomorska-Wesołowska, M.; Rózańska, A.; Natkaniec, J.; Gryglewska, B.; Szczypta, A.; Dzikowska, M.; Chmielarczyk, A.; Wójkowska-Mach, J. Longevity and gender as the risk factors of methicillin-resistant Staphylococcus aureus infections in southern Poland. BMC Geriatr. 2017, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.M.; Schuenck, R.P.; Malvar, K.L.; Iorio, N.L.P.; Matos, P.D.M.; Olendzki, A.N.; Oelemann, W.M.R.; dos Santos, K.R.N. Staphylococcus aureus, Staphylococcus epidermidis and Staphylococcus haemolyticus: Methicillin-resistant isolates are detected directly in blood cultures by multiplex PCR. Microbiol. Res. 2010, 165, 243–249. [Google Scholar] [CrossRef]
- Sutcliffe, J.; Grebe, T.; Tait-Kamradt, A.; Wondrack, L. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 1996, 40, 2562–2566. [Google Scholar] [CrossRef] [PubMed]
- Anthony, R.M.; Connor, A.M.; Power, E.G.M.; French, G.L. Use of the polymerase chain reaction for rapid detection of high-level mupirocin resistance in staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 1999, 18, 30–34. [Google Scholar] [CrossRef]
- Mcdougal, L.K.; Steward, C.D.; Steward, C.D.; Killgore, G.E.; Chaitram, J.M.; McAllister, S.K.; Tenover, F.C. Pulsed-Field Gel Electrophoresis Typing of Oxacillin-Resistant Staphylococcus aureus Isolates from the United States: Establishing a National Database. J. Clin. Microbiol. 2003, 41, 5113–5120. [Google Scholar] [CrossRef]
- Lipsky, B.A.; Hoey, C. Topical Antimicrobial Therapy for Treating Chronic Wounds. Clin. Infect. Dis. 2009, 49, 1541–1549. [Google Scholar] [CrossRef]
- Pugliese, D.J. Infection in Venous Leg Ulcers: Considerations for Optimal Management in the Elderly. Drugs Aging 2016, 33, 87–96. [Google Scholar] [CrossRef]
- Seth, A.K.; Geringer, M.R.; Hong, S.J.; Leung, K.P.; Mustoe, T.A. Comparative analysis of single-species and polybacterial wound biofilms using a quantitative, in vivo, rabbit ear model. PLoS ONE 2012, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mudge, E.J. Recent accomplishments in wound healing. Int. Wound J. 2015, 12, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Schreml, S.; Szeimies, R.M.; Prantl, L.; Karrer, S.; Landthaler, M.; Babilas, P. Oxygen in acute and chronic wound healing. Br. J. Dermatol. 2010, 163, 257–268. [Google Scholar] [CrossRef]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.D.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Chmielarczyk, A.; Pilarczyk-Zurek, M.; Kamińska, W.; Pobiega, M.; Romaniszyn, D.; Ziółkowski, G.; Wojkowska-Mach, J.; Bulanda, M. Molecular Epidemiology and Drug Resistance of Acinetobacter baumannii Isolated from Hospitals in Southern Poland: ICU as a Risk Factor for XDR Strains. Microb. Drug Resist. 2016, 22, 328–335. [Google Scholar] [CrossRef]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, P.W.; Sax, H.; Wolfensberger, A.; Clack, L.; Kuster, S.P.; Swissnoso. The preventable proportion of healthcare-associated infections 2005–2016: Systematic review and meta-analysis. Infect. Control Hosp. Epidemiol. 2018, 39, 1277–1295. [Google Scholar] [CrossRef] [PubMed]
- Olczak-Pieńkowska, A. Podsumowanie Aktualnych Danych nt. Konsumpcji Antybiotyków w Krajach unii Europejskiej–Dane Europejskiej Sieci Monitorowania Konsumpcji Antybiotyków ESAC-Net (listopad 2017) Zakład Epidemiologii i Mikrobiologii Klinicznej, Narodowy Instytut Leków, Warszawa. Available online: http://antybiotyki.edu.pl/wp-content/uploads/Biuletyn/biuletyn-npoa-2018_1.pdf (accessed on 15 June 2020).
- Mazi, W.; Sangal, V.; Sandstrom, G.; Saeed, A.; Yu, J. Evaluation of spa-typing of methicillin-resistant Staphylococcus aureus using high-resolution melting analysis. Int. J. Infect. Dis. 2015, 38, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Sabat, A.J.; Budimir, A.; Nashev, D.; Sá-Leão, R.; van Dijl, J.M.; Laurent, F.; Grundmann, H.; Friedrich, A.W.; ESCMID Study Group of Epidemiological Mark-ers (ESGEM). Overview of molecular typing methods for outbreak detection and epi-demiological surveillance. Euro. Surveill. 2013, 18, 20380. [Google Scholar] [CrossRef] [PubMed]
- Go-pal Rao, G.; Batura, R.; Nicholl, R.; Coogan, F.; Patel, B.; Bassett, P.; Kearns, A.M. Outbreak re-port of investigation and control of an outbreak of Panton-Valentine Leukocidin-positive methicillin-sensitive Staphylococcus aureus (PVL-MSSA) infection in neo-nates and mothers. BMC Infect. Dis. 2019, 19, 178. [Google Scholar] [CrossRef]
- Chmielarczyk, A.; Pomorska-Wesołowska, M.; Szczypta, A.; Romaniszyn, D.; Pobiega, M.; Wójkowska-Mach, J. Molecular analysis of meticillin-resistant Staphylococcus aureus strains isolated from different types of infections from patients hospitalized in 12 regional, non-teaching hospitals in southern Poland. J. Hosp. Infect. 2017, 95, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Chantelau, E.; Tanudjaja, T.; Altenhöfer, F.; Ersanli, Z.; Lacigova, S.; Metzger, C. Antibiotic treatment for uncomplicated neuropathic forefoot ulcers in diabetes: A controlled trial. Diabet. Med. 1996, 13, 156–159. [Google Scholar] [CrossRef]
- Öien, R.F.; Forssell, H.W. Ulcer healing time and antibiotic treatment before and after the introduction of the Registry of Ulcer Treatment: An improvement project in a national quality registry in Sweden. BMJ Open 2013, 3, 1–6. [Google Scholar] [CrossRef]
- Schultz, G.; Bjarnsholt, T.; James, G.A.; Leaper, D.J.; McBain, A.J.; Malone, M.; Stoodley, P.; Swanson, T.; Tachi, M.; Wolcott, R.D.; et al. Consensus guidelines for the identification and treatment of biofilms in chronic nonhealing wounds. Wound Repair Regen. 2017, 25, 744–757. [Google Scholar] [CrossRef]
- Gardner, S.E.; Frantz, R.A.; Saltzman, C.L.; Hillis, S.L.; Park, H.; Scherubel, M. Diagnostic validity of three swab techniques for identifying chronic wound infection. Wound Repair Regen. 2006, 14, 548–557. [Google Scholar] [CrossRef]
- Pellizzer, G.; Strazzabosco, M.; Presi, S.; Furlan, F.; Lora, L.; Benedetti, P.; Bonato, M.; Erle, G.; De Lalla, F. Deep tissue biopsy vs. superficial swab culture monitoring in the microbiological assessment of limb-threatening diabetic foot infection. Diabet. Med. 2001, 18, 822–827. [Google Scholar] [CrossRef]
- Miller, C.N.; Carville, K.; Newall, N.; Kapp, S.; Lewin, G.; Karimi, L.; Santamaria, N. Assessing bacterial burden in wounds: Comparing clinical observation and wound swabs. Int. Wound J. 2011, 8, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Kelly, F. Reliability in Wound Swabbing. Br. J. Nurs. 2003, 12, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Malone, M.; Mayer, D.; Salisbury, A.-M.; Schultz, G. Role of anaerobes in polymicrobial communities and biofilms complicating diabetic foot ulcers. Int. Wound J. 2018, 15, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Zenilman, J.M.; Melendez, J.H.; Shirtliff, M.E.; Agostinho, A.; James, G.; Stewart, P.S.; Mongodin, E.F.; Rao, D.; Rickard, A.H.; et al. The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen. 2011, 19, 532–541. [Google Scholar] [CrossRef] [PubMed]
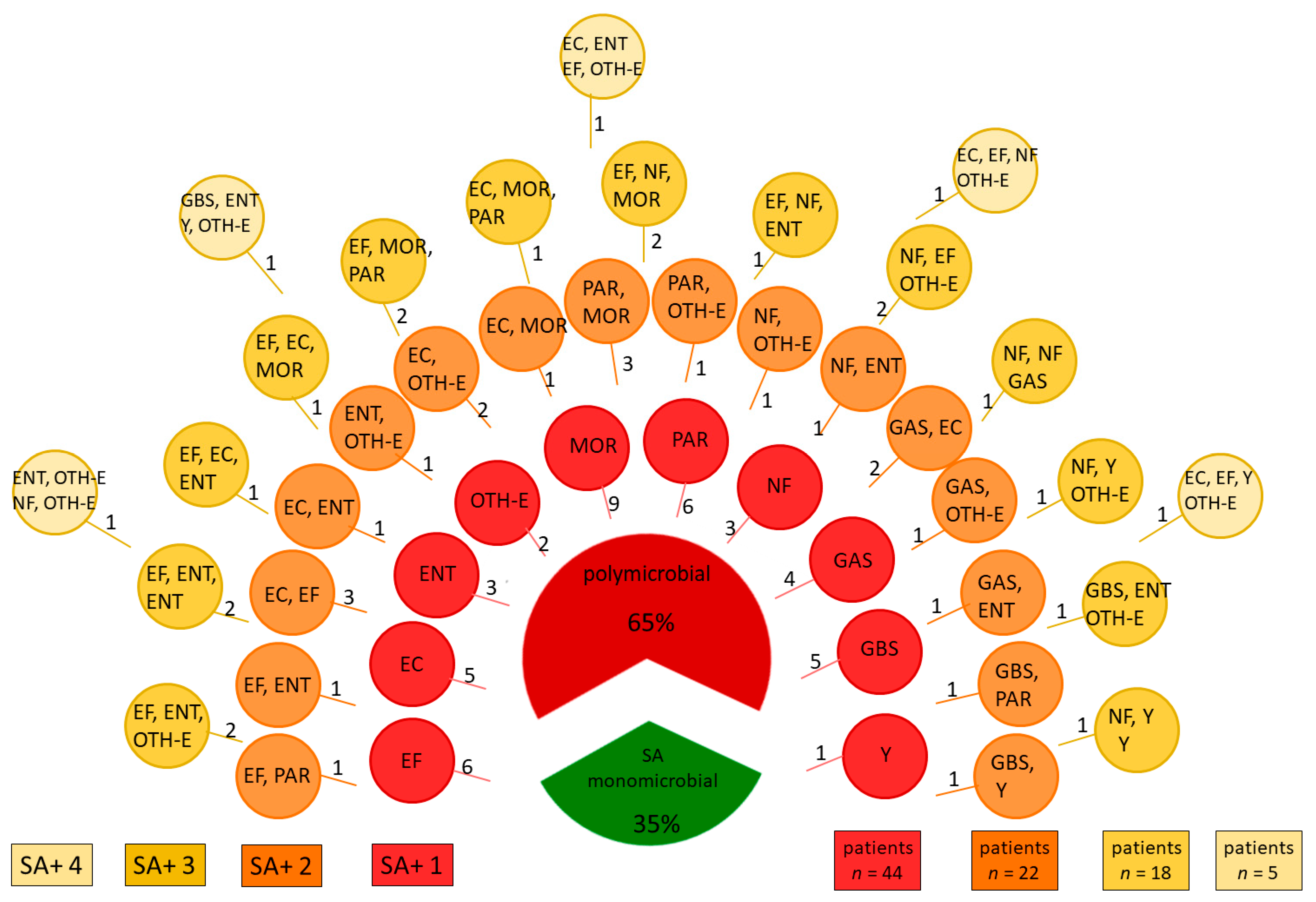
| Identified Pathogen | n |
|---|---|
| Enterococcus faecalis (EF) | 30 |
| Streptococcus agalactiae (GBS) | 10 |
| Streptococcus pyogenes (GAS) | 9 |
| Escherichia coli (EC) | 19 |
| Enterobacter spp (ENT) | 19 |
| OTH-E: Other Bacteria from Enterobacteriaceae Family | |
| Klebsiella spp. | 12 |
| Serratia spp. | 3 |
| Citrobacter spp. | 6 |
| Morganellaceae Family (MOR) | |
| Proteus spp. | 17 |
| Morganella morgani | 3 |
| Providentia regretti | 1 |
| Pseudomonas aeruginosa (PAR) | 15 |
| Other Non-Fermenting Bacteria (NF) | |
| Acinetobacter spp. | 13 |
| Stenotrophomonas maltophilia | 4 |
| Candida spp. (Y) | 7 |
| Characteristics of the Study Group | Total n = 143 | Sensitivity to Antibiotics | OR (95% CI) | |
|---|---|---|---|---|
| Yes n = 70 | No n = 73 | |||
| Age (Years) by Categories n [%] | ||||
| ≤59 years | 45 (31.5) | 23 (33.3) | 22 (30.1) | 0.8 (0.36–1.73) |
| 60–75 years | 58 (40.6) | 33 (47.8) | 25 (34.2) | ref. |
| 76–80 years | 16 (11.2) | 6 (8.7) | 10 (13.7) | 0.5 (0.15–1.41) |
| ≥81 years | 24 (16.8) | 7 (10.1) | 16 (21.9) | 0.3 (0.12–0.93) |
| Gender n [%] | ||||
| Female | 76 (53.2) | 34 (48.6) | 42 (57.5) | 0.7 (0.36–1.35) |
| Male (ref.) | 67 (46.9) | 36 (51.4) | 31 (42.5) | |
| Ambulatory Care n [%] | ||||
| yes | 48 (32.2) | 29 (41.4) | 19 (26.0) | 2.0 (0.99–4.08) |
| no | 95 (66.4) | 41 (58.6) | 54 (74.0) | |
| Hospital Stay n [%] | ||||
| surgical wards | 61 (42.7) | 26 (37.1) | 35 (47.8) | 0.9 (0.40–2.19) |
| non-surgical wards or LTCF | 34 (23.8) | 15 (21.4) | 19 (13.3) | |
| The Presence of Resistance Genes n [;%], YES | ||||
| mecA | 17 (11.9) | n/a | 17 (11.9) | n/a |
| ermA | 8 (5.6) | n/a | 8 (5.6) | n/a |
| ermB | 2 (1.4) | n/a | 2 (1.4) | n/a |
| msr | 1 (0.7) | n/a | 1 (0.7) | n/a |
| The Presence of Various Virulence Factors Genes n [;%], YES | ||||
| lukE | 98 (68.5) | 45 (64.3) | 53 (72.6) | 0.7 (0.33–1.38) |
| tsst-1 | 17 (11.9) | 9 (12.9) | 8 (11.0) | 1.2 (0.43–3.30) |
| pvl | 2 (1.4) | 1 (1.4) | 1 (1.4) | 1.0 (0.06–17.01) |
| etA/B | 23 (16.1) | 10 (14.3) | 13 (17.8) | 0.8 (0.31–1.89) |
| Antimicrobial Category | Antimicrobial Agent | Total n = 143 | Hospital Stay | OR (95%CI) | |
|---|---|---|---|---|---|
| Yes, n = 95 | No, n = 48 | ||||
| Aminoglycosides | Gentamicin | 24 (16.8) | 16 (16.8) | 8 (16.7) | 0.9 (0.38–2.45) |
| Amikacin | 29 (20.2) | 19 (20.0) | 10 (20.8) | 2.1 (0.71–5.89) | |
| Tobramycin | 39 (27.3) | 27 (28.4) | 12 (25.0) | 1.1 (0.51–2.49) | |
| Anti-MRSA cephalosporins | Ceftaroline | 0 (0) | 0 (0.0) | 0 (0.0) | n/a |
| Fluoroquinolones | Ciprofloxacin | 25 (17.5) | 20 (21.1) | 5 (10.4) | 2.2 (0.76–6.26) |
| Moxifloxacin | 18 (12.6) | 14 (14.7) | 4 (8.3) | 0.5 (0.23–1.33) | |
| Folate pathway inhibitors | Trimethoprim/Sulfamethoxazole | 8 (5.6) | 6 (6/3) | 2 (4.2) | 1.5 (0.29–7.65) |
| Lincosamides | Clindamycin | 29 (20.2) | 18 (18.9) | 11 (22.9) | 0.7 (0.32–1.74) |
| Macrolides | Erythromycin | 31 (21.6) | 17 (17.9) | 14 (29.2) | 0.6 (0.27–1.43) |
| Glycopeptides | Vancomycin | 0 (0) | 0 (0.0) | 0 (0.0) | n/a |
| Glycylcyclines | Tigecycline | 0 (0) | 0 (0.0) | 0 (0.0) | n/a |
| Oxazolidinones | Linezolid | 0 (0) | 0 (0.0) | 0 (0.0) | n/a |
| Phenicols | Chloramphenicol | 16 (11.2) | 13 (13.7) | 3 (6.2) | 0.3 (0.13–0.69) |
| Streptogramins | Quinupristin-dalfopristin | 0 (0) | 0 (0.0) | 0 (0.0) | n/a |
| Tetracyclines | Tetracycline | 35 (24.5) | 27 (28.4) | 8 (16.7) | 1.9 (0.78–4.56) |
| Doxycycline | 19 (13.3) | 13 (13.7) | 6 (12.5) | 1.1 (0.37–2.99) | |
| Others (O) | Mupirocin | 8 (5.6) | 5 (5.3) | 3 (6.2) | 0.8 (0.18–3.49) |
| Multidrug Resistance n (;%) | |||||
| MRSA, yes | 17 (11.9) | 12 (12.6) | 5 (10.4) | 1.2 (0.39–3.59) | |
| MLSB, yes | 28 (19.6) | 16 (16.8) | 12 (25.0) | 9.1 (1.17–71.02) | |
| Non-Susceptible to Antimicrobial Categories n (;%) | |||||
| 0 categories (fully susceptible) | 70 (49) | 41 (43.2) | 29 (60.4) | 0.4 (0.22–0.92) | |
| 1 category | 26 (18.2) | 23 (24.2) | 3 (6.2) | 4.8 (1.38–17.06) | |
| Aminoglycosides | 9 (6.3) | 7 (7.4) | 2 (4.2) | ||
| Tetracyclines | 9 (6.3) | 9 (9.5) | 0 (0.0) | ||
| Phenicols | 4 (2.8) | 4 (4.2) | 0 (0.0) | ||
| Fluoroquinolones | 2 (1.4) | 2 (2.1) | 0 (0.0) | ||
| Lincosamides | 1 (0.7) | 1 (1.1) | 0 (0.0) | ||
| Macrolides | 1 (0.7) | 0 (0.0) | 1 (2.1) | ||
| 2 categories | 19 (13.3) | 11 (11.6) | 8 (16.7) | 0.6 (0.23–1.67) | |
| Macrolides + Lincosamides | 5 (3.5) | 2 (2.1) | 3 (6.3) | ||
| Tetracyclines + Phenicols | 3 (2.1) | 2 (2.1) | 1 (2.1) | ||
| Aminoglycosides + Fluoroquinolones | 3 (2.1) | 3 (3.2) | 0 (0.0) | ||
| Tetracyclines + Fluoroquinolones | 2 (1.4) | 2 (2.1) | 0 (0.0) | ||
| Aminoglycosides + Tetracycline | 2 (1.4) | 1 (1.1) | 1 (2.1) | ||
| Aminoglycosides A+ Phenicols | 2 (1.4) | 1 (1.1) | 1 (2.1) | ||
| Tetracyclines + Macrolides | 1 (0.7) | 0 (0.0) | 1 (2.1) | ||
| Aminoglycosides + Macrolides | 1 (0.7) | 0 (0.0) | 1 (2.1) | ||
| 3 categories or more | 28 (19.6) | 19 (20.0) | 8 (16.7) | 1.2 (0.48–2.96) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gajda, M.; Załugowicz, E.; Pomorska-Wesołowska, M.; Bochenek, T.; Gryglewska, B.; Romaniszyn, D.; Chmielarczyk, A.; Wójkowska-Mach, J. Virulence and Drug-Resistance of Staphylococcus aureus Strains Isolated from Venous Ulcers in Polish Patients. Int. J. Environ. Res. Public Health 2021, 18, 4662. https://doi.org/10.3390/ijerph18094662
Gajda M, Załugowicz E, Pomorska-Wesołowska M, Bochenek T, Gryglewska B, Romaniszyn D, Chmielarczyk A, Wójkowska-Mach J. Virulence and Drug-Resistance of Staphylococcus aureus Strains Isolated from Venous Ulcers in Polish Patients. International Journal of Environmental Research and Public Health. 2021; 18(9):4662. https://doi.org/10.3390/ijerph18094662
Chicago/Turabian StyleGajda, Mateusz, Emilia Załugowicz, Monika Pomorska-Wesołowska, Tomasz Bochenek, Barbara Gryglewska, Dorota Romaniszyn, Agnieszka Chmielarczyk, and Jadwiga Wójkowska-Mach. 2021. "Virulence and Drug-Resistance of Staphylococcus aureus Strains Isolated from Venous Ulcers in Polish Patients" International Journal of Environmental Research and Public Health 18, no. 9: 4662. https://doi.org/10.3390/ijerph18094662
APA StyleGajda, M., Załugowicz, E., Pomorska-Wesołowska, M., Bochenek, T., Gryglewska, B., Romaniszyn, D., Chmielarczyk, A., & Wójkowska-Mach, J. (2021). Virulence and Drug-Resistance of Staphylococcus aureus Strains Isolated from Venous Ulcers in Polish Patients. International Journal of Environmental Research and Public Health, 18(9), 4662. https://doi.org/10.3390/ijerph18094662







