Metabolic Alterations in Myotonic Dystrophy Type 1 and Their Correlation with Lipin
Abstract
1. Introduction
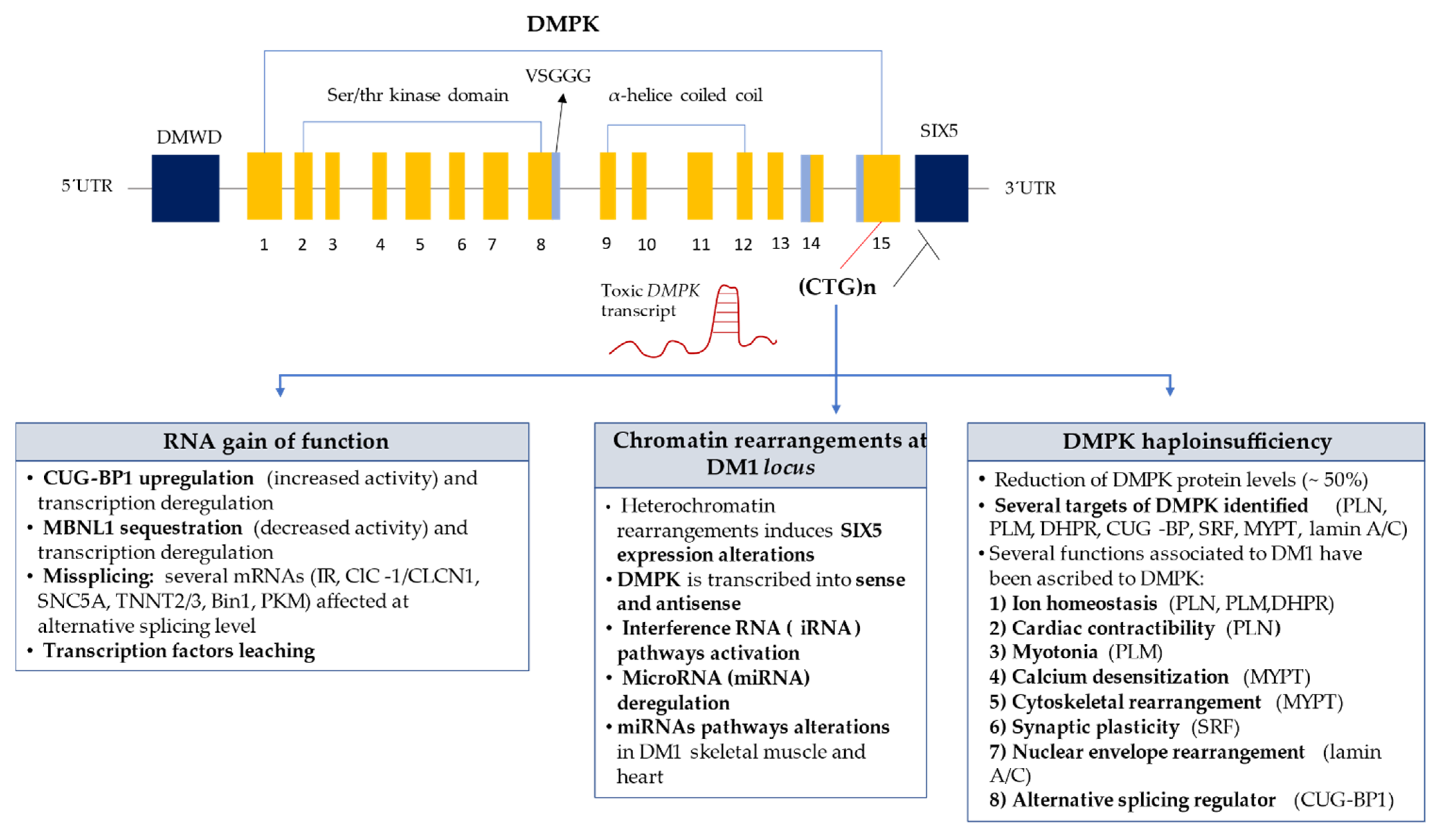
2. Epidemiology and Molecular Characteristics of DM1
3. Physiological and Systemic Features of DM1
4. DM1 Underlying Molecular Mechanisms
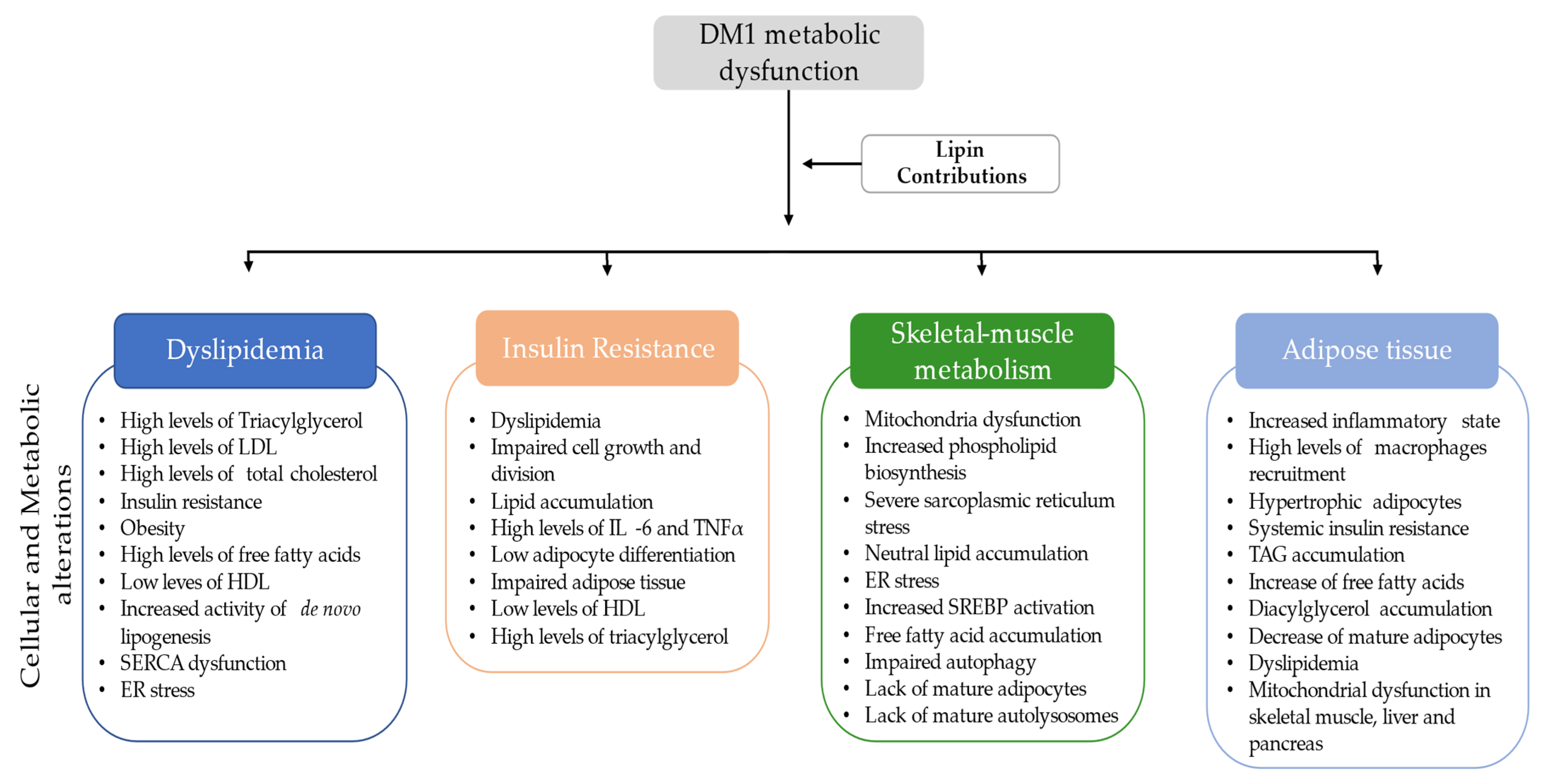
5. Metabolic Dysfunction in DM1
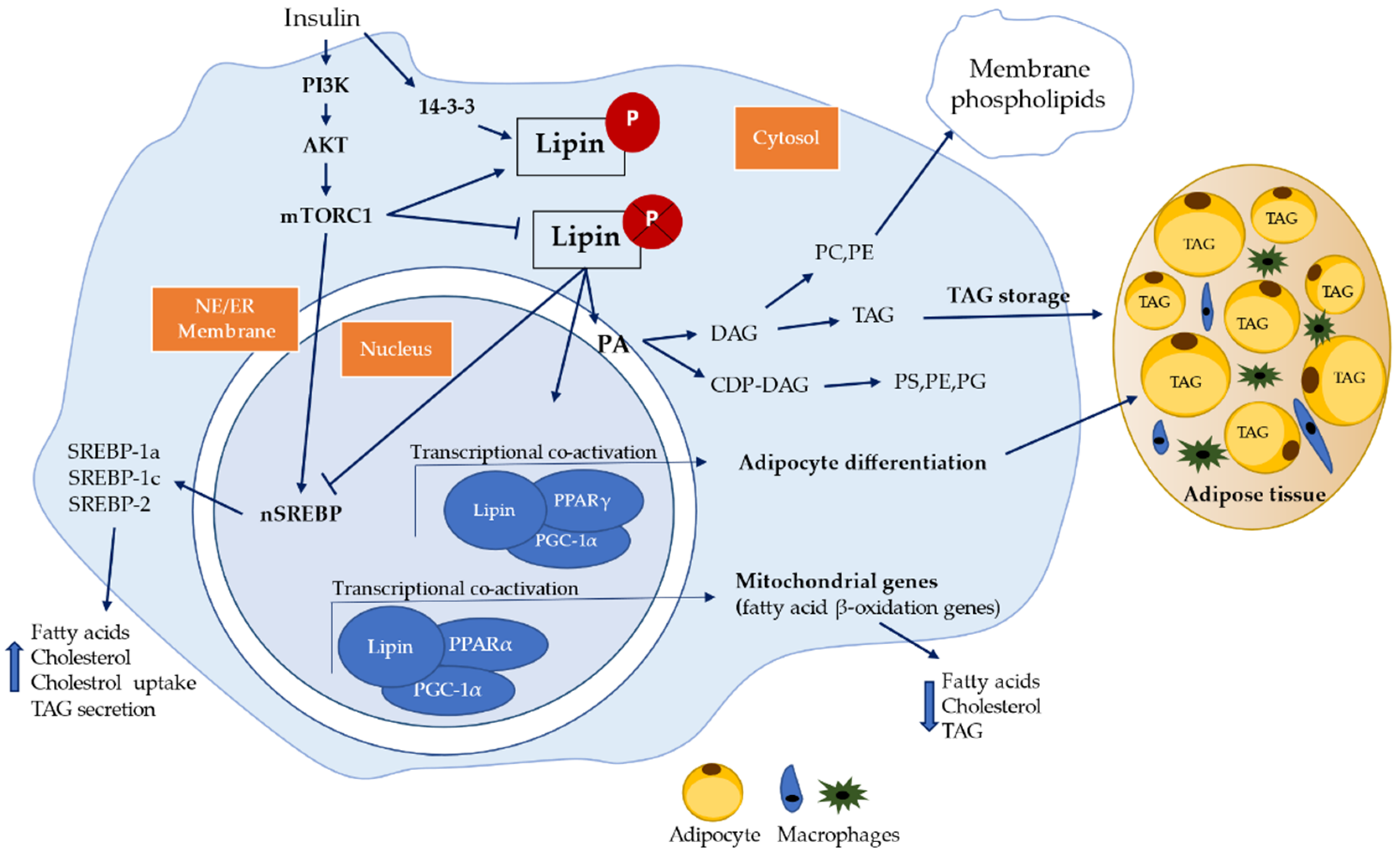
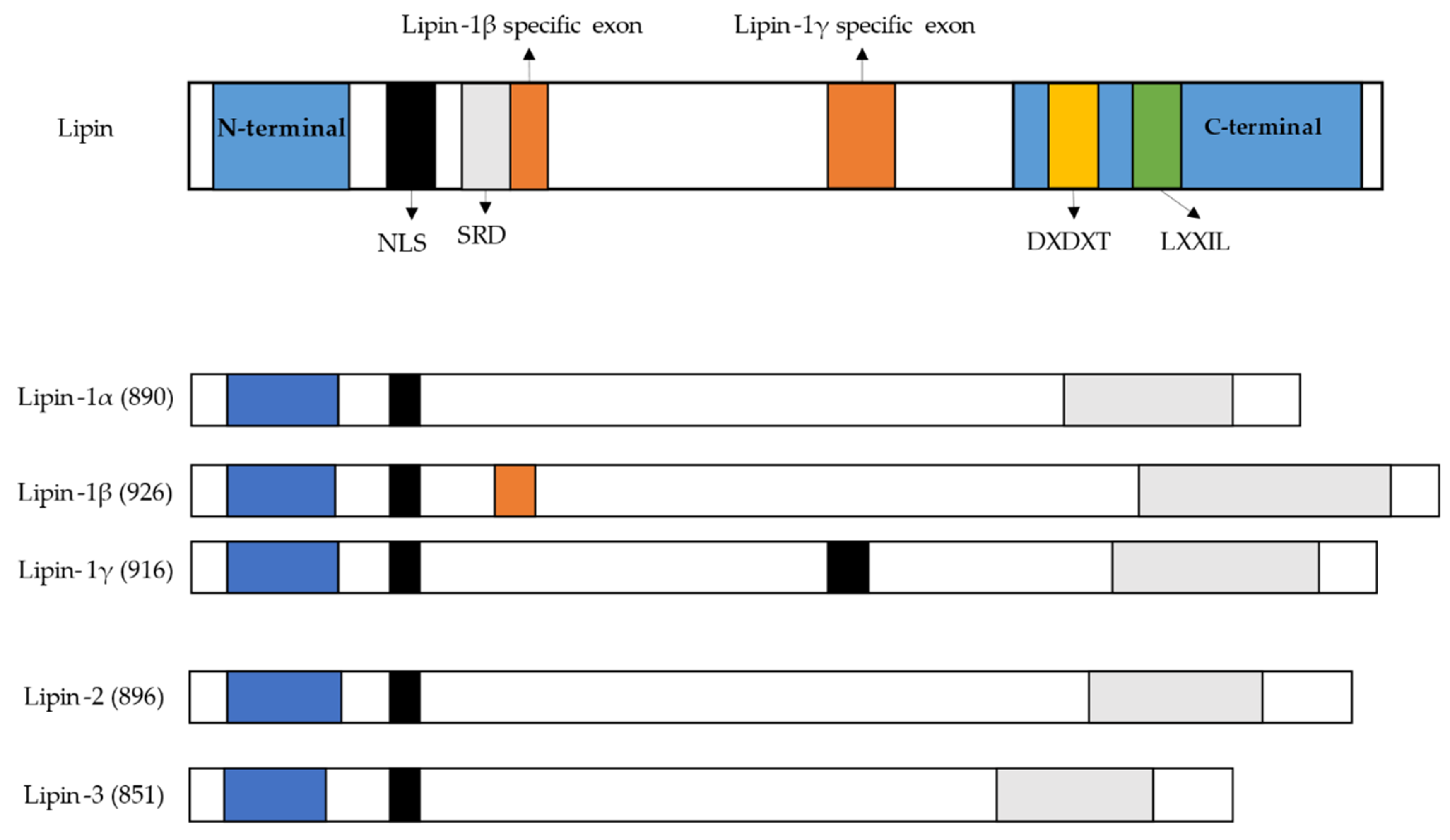
6. Lipin Protein Family
6.1. Dyslipidemia and Lipin
6.2. Insulin Resistance Association with Lipin
6.3. Skeletal Muscle Metabolism and the Lipin Role
6.4. Lipin Association with Adipose Tissue
7. Future Perspectives
8. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, N.E. Myotonic Muscular Dystrophies. Contin. Lifelong Learn. Neurol. 2019, 25, 1682–1695. [Google Scholar] [CrossRef]
- Bozovic, I.; Peric, S.; Pesovic, J.; Bjelica, B.; Brkusanin, M.; Basta, I.; Bozic, M.; Sencanic, I.; Marjanovic, A.; Brankovic, M.; et al. Myotonic Dystrophy Type 2–Data from the Serbian Registry. J. Neuromuscul. Dis. 2018, 5, 461–469. [Google Scholar] [CrossRef]
- Vanacore, N.; Rastelli, E.; Antonini, G.; Bianchi, M.L.E.; Botta, A.; Bucci, E.; Casali, C.; Costanzi-Porrini, S.; Giacanelli, M.; Gibellini, M.; et al. An Age-Standardized Prevalence Estimate and a Sex and Age Distribution of Myotonic Dystrophy Types 1 and 2 in the Rome Province, Italy. Neuroepidemiology 2016, 46, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, R.; Hernández-Hernández, O.; Magaña, J.J.; González-Ramírez, R.; García-López, E.S.; Cisneros, B. Altered nuclear structure in myotonic dystrophy type 1-derived fibroblasts. Mol. Biol. Rep. 2015, 42, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Thornton, C.A. Myotonic Dystrophy. Neurol. Clin. 2014, 32, 705–719. [Google Scholar] [CrossRef]
- Magaña, J.J.; Surez-Snchez, R.; Leyva-Garca, N.; Cisneros, B.; Hernndez-Hernndez, O. Myotonic Dystrophy Protein Kinase: Structure, Function and Its Possible Role in the Pathogenesis of Myotonic Dystrophy Type 1. In Advances in Protein Kinases; InTech: London, UK, 2012. [Google Scholar]
- Cho, D.H.; Tapscott, S.J. Myotonic dystrophy: Emerging mechanisms for DM1 and DM2. Biochim. Biophys. Acta Mol. Basis Dis. 2007, 1772, 195–204. [Google Scholar] [CrossRef]
- Nieuwenhuis, S.; Okkersen, K.; Widomska, J.; Blom, P.; ’t Hoen, P.A.C.; van Engelen, B.; Glennon, J.C. Insulin Signaling as a Key Moderator in Myotonic Dystrophy Type 1. Front. Neurol. 2019, 10. [Google Scholar] [CrossRef]
- Takada, H. Lipid Metabolism in Myotonic Dystrophy. In Myotonic Dystrophy; Springer: Singapore, 2018; pp. 161–170. [Google Scholar]
- Okuno, H.; Okuzono, H.; Hayase, A.; Kumagai, F.; Tanii, S.; Hino, N.; Okada, Y.; Tachibana, K.; Doi, T.; Ishimoto, K. Lipin-1 is a novel substrate of protein phosphatase PGAM5. Biochem. Biophys. Res. Commun. 2019, 509, 886–891. [Google Scholar] [CrossRef]
- Csaki, L.S.; Dwyer, J.R.; Li, X.; Nguyen, M.H.K.; Dewald, J.; Brindley, D.N.; Lusis, A.J.; Yoshinaga, Y.; de Jong, P.; Fong, L.; et al. Lipin-1 and lipin-3 together determine adiposity in vivo. Mol. Metab. 2014, 3, 145–154. [Google Scholar] [CrossRef]
- Chen, Y.; Rui, B.-B.; Tang, L.-Y.; Hu, C.-M. Lipin Family Proteins—Key Regulators in Lipid Metabolism. Ann. Nutr. Metab. 2015, 66, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Péterfy, M.; Phan, J.; Reue, K. Alternatively Spliced Lipin Isoforms Exhibit Distinct Expression Pattern, Subcellular Localization, and Role in Adipogenesis. J. Biol. Chem. 2005, 280, 32883–32889. [Google Scholar] [CrossRef] [PubMed]
- Finck, B.N.; Gropler, M.C.; Chen, Z.; Leone, T.C.; Croce, M.A.; Harris, T.E.; Lawrence, J.C.; Kelly, D.P. Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metab. 2006, 4, 199–210. [Google Scholar] [CrossRef]
- Bhatt, J.M. The Epidemiology of Neuromuscular Diseases. Neurol. Clin. 2016, 34, 999–1021. [Google Scholar] [CrossRef] [PubMed]
- Theadom, A.; Rodrigues, M.; Roxburgh, R.; Balalla, S.; Higgins, C.; Bhattacharjee, R.; Jones, K.; Krishnamurthi, R.; Feigin, V. Prevalence of Muscular Dystrophies: A Systematic Literature Review. Neuroepidemiology 2014, 43, 259–268. [Google Scholar] [CrossRef]
- LaPelusa, A.; Kentris, M. Muscular Dystrophy. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Esposito, F.; Cè, E.; Rampichini, S.; Monti, E.; Limonta, E.; Fossati, B.; Meola, G. Electromechanical delays during a fatiguing exercise and recovery in patients with myotonic dystrophy type 1. Eur. J. Appl. Physiol. 2017, 117, 551–566. [Google Scholar] [CrossRef]
- Ashizawa, T.; Gagnon, C.; Groh, W.J.; Gutmann, L.; Johnson, N.E.; Meola, G.; Moxley, R.; Pandya, S.; Rogers, M.T.; Simpson, E.; et al. Consensus-based care recommendations for adults with myotonic dystrophy type 1. Neurol. Clin. Pract. 2018, 8, 507–520. [Google Scholar] [CrossRef]
- Wansink, D.G.; van Herpen, R.E.M.A.; Coerwinkel-Driessen, M.M.; Groenen, P.J.T.A.; Hemmings, B.A.; Wieringa, B. Alternative Splicing Controls Myotonic Dystrophy Protein Kinase Structure, Enzymatic Activity, and Subcellular Localization. Mol. Cell. Biol. 2003, 23, 5489–5501. [Google Scholar] [CrossRef]
- Lee, J.E.; Cooper, T.A. Pathogenic mechanisms of myotonic dystrophy. Biochem. Soc. Trans. 2009, 37, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Bush, E.W.; Helmke, S.M.; Birnbaum, R.A.; Perryman, M.B. Myotonic Dystrophy Protein Kinase Domains Mediate Localization, Oligomerization, Novel Catalytic Activity, and Autoinhibition †. Biochemistry 2000, 39, 8480–8490. [Google Scholar] [CrossRef] [PubMed]
- De Antonio, M.; Dogan, C.; Hamroun, D.; Mati, M.; Zerrouki, S.; Eymard, B.; Katsahian, S.; Bassez, G. Unravelling the myotonic dystrophy type 1 clinical spectrum: A systematic registry-based study with implications for disease classification. Rev. Neurol. 2016, 172, 572–580. [Google Scholar] [CrossRef]
- Sansone, V.A. The Dystrophic and Nondystrophic Myotonias. Contin. Lifelong Learn. Neurol. 2016, 22, 1889–1915. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.; Hilton-Jones, D. Myotonic dystrophy. Curr. Opin. Neurol. 2014, 27, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Yum, K.; Wang, E.T.; Kalsotra, A. Myotonic dystrophy: Disease repeat range, penetrance, age of onset, and relationship between repeat size and phenotypes. Curr. Opin. Genet. Dev. 2017, 44, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Passeri, E.; Bugiardini, E.; Sansone, V.A.; Pizzocaro, A.; Fulceri, C.; Valaperta, R.; Borgato, S.; Costa, E.; Bandera, F.; Ambrosi, B.; et al. Gonadal failure is associated with visceral adiposity in myotonic dystrophies. Eur. J. Clin. Investig. 2015, 45, 702–710. [Google Scholar] [CrossRef]
- Ben Hamou, A.; Espiard, S.; Do Cao, C.; Ladsous, M.; Loyer, C.; Moerman, A.; Boury, S.; Kyheng, M.; Dhaenens, C.-M.; Tiffreau, V.; et al. Systematic thyroid screening in myotonic dystrophy: Link between thyroid volume and insulin resistance. Orphanet J. Rare Dis. 2019, 14, 42. [Google Scholar] [CrossRef]
- Rossi, S.; Della Marca, G.; Ricci, M.; Perna, A.; Nicoletti, T.F.; Brunetti, V.; Meleo, E.; Calvello, M.; Petrucci, A.; Antonini, G.; et al. Prevalence and predictor factors of respiratory impairment in a large cohort of patients with Myotonic Dystrophy type 1 (DM1): A retrospective, cross sectional study. J. Neurol. Sci. 2019, 399, 118–124. [Google Scholar] [CrossRef]
- Hawkins, A.M.; Hawkins, C.L.; Abdul Razak, K.; Khoo, T.K.; Tran, K.; Jackson, R.V. Respiratory dysfunction in myotonic dystrophy type 1: A systematic review. Neuromuscul. Disord. 2019, 29, 198–212. [Google Scholar] [CrossRef]
- Hermans, M.C.; Faber, C.G.; Bekkers, S.C.; de Die-Smulders, C.E.; Gerrits, M.M.; Merkies, I.S.; Snoep, G.; Pinto, Y.M.; Schalla, S. Structural and functional cardiac changes in myotonic dystrophy type 1: A cardiovascular magnetic resonance study. J. Cardiovasc. Magn. Reson. 2012, 14, 48. [Google Scholar] [CrossRef]
- Guedes, H.; Moreno, N.; dos Santos, R.P.; Marques, L.; Seabra, D.; Pereira, A.; Andrade, A.; Pinto, P. Importance of three-dimensional speckle tracking in the assessment of left atrial and ventricular dysfunction in patients with myotonic dystrophy type 1. Rev. Port. Cardiol. 2018, 37, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Chmielewski, L.; Bietenbeck, M.; Patrascu, A.; Rösch, S.; Sechtem, U.; Yilmaz, A.; Florian, A.-R. Non-invasive evaluation of the relationship between electrical and structural cardiac abnormalities in patients with myotonic dystrophy type 1. Clin. Res. Cardiol. 2019, 108, 857–867. [Google Scholar] [CrossRef]
- O’Cochlain, D.F.; Perez-Terzic, C.; Reyes, S.; Kane, G.C.; Behfar, A.; Hodgson, D.M.; Strommen, J.A.; Liu, X.-K.; van den Broek, W.; Wansink, D.G.; et al. Transgenic overexpression of human DMPK accumulates into hypertrophic cardiomyopathy, myotonic myopathy and hypotension traits of myotonic dystrophy. Hum. Mol. Genet. 2004, 13, 2505–2518. [Google Scholar] [CrossRef]
- Sarkar, P.S.; Han, J.; Reddy, S. In situ hybridization analysis of Dmpk mRNA in adult mouse tissues. Neuromuscul. Disord. 2004, 14, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Groenen, P.J.T.A. Constitutive and regulated modes of splicing produce six major myotonic dystrophy protein kinase (DMPK) isoforms with distinct properties. Hum. Mol. Genet. 2000, 9, 605–616. [Google Scholar] [CrossRef][Green Version]
- Harmon, E.B.; Harmon, M.L.; Larsen, T.D.; Yang, J.; Glasford, J.W.; Perryman, M.B. Myotonic Dystrophy Protein Kinase Is Critical for Nuclear Envelope Integrity. J. Biol. Chem. 2011, 286, 40296–40306. [Google Scholar] [CrossRef] [PubMed]
- Iyer, D.; Belaguli, N.; Flück, M.; Rowan, B.G.; Wei, L.; Weigel, N.L.; Booth, F.W.; Epstein, H.F.; Schwartz, R.J.; Balasubramanyam, A. Novel Phosphorylation Target in the Serum Response Factor MADS Box Regulates α-Actin Transcription †. Biochemistry 2003, 42, 7477–7486. [Google Scholar] [CrossRef]
- Murányi, A.; Zhang, R.; Liu, F.; Hirano, K.; Ito, M.; Epstein, H.F.; Hartshorne, D.J. Myotonic dystrophy protein kinase phosphorylates the myosin phosphatase targeting subunit and inhibits myosin phosphatase activity. FEBS Lett. 2001, 493, 80–84. [Google Scholar] [CrossRef]
- Sicot, G.; Gourdon, G.; Gomes-Pereira, M. Myotonic dystrophy, when simple repeats reveal complex pathogenic entities: New findings and future challenges. Hum. Mol. Genet. 2011, 20, R116–R123. [Google Scholar] [CrossRef]
- Perbellini, R.; Greco, S.; Sarra-Ferraris, G.; Cardani, R.; Capogrossi, M.C.; Meola, G.; Martelli, F. Dysregulation and cellular mislocalization of specific miRNAs in myotonic dystrophy type 1. Neuromuscul. Disord. 2011, 21, 81–88. [Google Scholar] [CrossRef]
- Rau, F.; Freyermuth, F.; Fugier, C.; Villemin, J.-P.; Fischer, M.-C.; Jost, B.; Dembele, D.; Gourdon, G.; Nicole, A.; Duboc, D.; et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011, 18, 840–845. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.M.; Krym, M.C.; Thornton, C.A. Ribonuclear foci at the neuromuscular junction in myotonic dystrophy type 1. Neuromuscul. Disord. 2007, 17, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Ravel-Chapuis, A.; Bélanger, G.; Yadava, R.S.; Mahadevan, M.S.; DesGroseillers, L.; Côté, J.; Jasmin, B.J. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J. Cell Biol. 2012, 196, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Cline, M.S.; Osborne, R.J.; Tuttle, D.L.; Clark, T.A.; Donohue, J.P.; Hall, M.P.; Shiue, L.; Swanson, M.S.; Thornton, C.A.; et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010, 17, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Ranum, L.P.W.; Day, J.W. Myotonic Dystrophy: RNA Pathogenesis Comes into Focus. Am. J. Hum. Genet. 2004, 74, 793–804. [Google Scholar] [CrossRef]
- Shin, J.-Y.; Hernandez-Ono, A.; Fedotova, T.; Östlund, C.; Lee, M.J.; Gibeley, S.B.; Liang, C.-C.; Dauer, W.T.; Ginsberg, H.N.; Worman, H.J. Nuclear envelope–localized torsinA-LAP1 complex regulates hepatic VLDL secretion and steatosis. J. Clin. Investig. 2019, 129, 4885–4900. [Google Scholar] [CrossRef] [PubMed]
- Hino, S.-I.; Kondo, S.; Sekiya, H.; Saito, A.; Kanemoto, S.; Murakami, T.; Chihara, K.; Aoki, Y.; Nakamori, M.; Takahashi, M.P.; et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum. Mol. Genet. 2007, 16, 2834–2843. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, S.; Vieira, S.I.; da Cruz e Silva, O.A.B.; Esselmann, H.; Wiltfang, J.; da Cruz e Silva, E.F. Tyr687 dependent APP endocytosis and abeta production. J. Mol. Neurosci. 2007, 32, 1–8. [Google Scholar] [CrossRef]
- Vieira, S.I.; Rebelo, S.; Esselmann, H.; Wiltfang, J.; Lah, J.; Lane, R.; Small, S.A.; Gandy, S.; da Cruz e Silva, E.F.; da Cruz e Silva, O.A. Retrieval of the Alzheimer’s amyloid precursor protein from the endosome to the TGN is S655 phosphorylation state-dependent and retromer-mediated. Mol. Neurodegener. 2010, 5, 40. [Google Scholar] [CrossRef]
- Rebelo, S.; Domingues, S.C.; Santos, M.; Fardilha, M.; Esteves, S.L.C.; Vieira, S.I.; Vintém, A.P.B.; Wu, W.; da Cruz e Silva, E.F.; da Cruz e Silva, O.A.B. Identification of a Novel Complex AβPP:Fe65:PP1 that Regulates AβPP Thr668 Phosphorylation Levels. J. Alzheimers Dis. 2013, 35, 761–775. [Google Scholar] [CrossRef]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Muzio, L. Lo The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef]
- Grundy, S.M. Metabolic syndrome update. Trends Cardiovasc. Med. 2016, 26, 364–373. [Google Scholar] [CrossRef]
- Rochlani, Y.; Pothineni, N.V.; Kovelamudi, S.; Mehta, J.L. Metabolic syndrome: Pathophysiology, management, and modulation by natural compounds. Ther. Adv. Cardiovasc. Dis. 2017, 11, 215–225. [Google Scholar] [CrossRef]
- Renna, L.V.; Bosè, F.; Iachettini, S.; Fossati, B.; Saraceno, L.; Milani, V.; Colombo, R.; Meola, G.; Cardani, R. Receptor and post-receptor abnormalities contribute to insulin resistance in myotonic dystrophy type 1 and type 2 skeletal muscle. PLoS ONE 2017, 12, e0184987. [Google Scholar] [CrossRef]
- Rakocevic Stojanovic, V.; Peric, S.; Lavrnic, D.; Popovic, S.; Ille, T.; Stevic, Z.; Basta, I.; Apostolski, S. Leptin and the metabolic syndrome in patients with myotonic dystrophy type 1. Acta Neurol. Scand. 2010, 121, 94–98. [Google Scholar] [CrossRef]
- Daniele, A.; De Rosa, A.; De Cristofaro, M.; Monaco, M.L.; Masullo, M.; Porcile, C.; Capasso, M.; Tedeschi, G.; Oriani, G.; Di Costanzo, A. Decreased concentration of adiponectin together with a selective reduction of its high molecular weight oligomers is involved in metabolic complications of myotonic dystrophy type 1. Eur. J. Endocrinol. 2011, 165, 969–975. [Google Scholar] [CrossRef]
- Shieh, K.; Gilchrist, J.M.; Promrat, K. Frequency and predictors of nonalcoholic fatty liver disease in myotonic dystrophy. Muscle Nerve 2010, 41, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Boman, K.; Cederquist, K.; Forsberg, H.; Olsson, T. Increased levels of tPA antigen and tPA/PAI-1 complex in myotonic dystrophy. J. Intern. Med. 2001, 249, 503–510. [Google Scholar] [CrossRef]
- Spaziani, M.; Semeraro, A.; Bucci, E.; Rossi, F.; Garibaldi, M.; Papassifachis, M.A.; Pozza, C.; Anzuini, A.; Lenzi, A.; Antonini, G.; et al. Hormonal and metabolic gender differences in a cohort of myotonic dystrophy type 1 subjects: A retrospective, case–control study. J. Endocrinol. Investig. 2020, 43, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Olsson, T.; Cederquist, K.; Forsberg, H.; Holst, J.; Ahren, B. Abnormal release of incretins and cortisol after oral glucose in subjects with insulin-resistant myotonic dystrophy. Eur. J. Endocrinol. 2002, 397–405. [Google Scholar] [CrossRef]
- Hudson, A.J.; Huff, M.W.; Wright, C.G.; Silver, M.M.; Lo, T.C.Y.; Banerjee, D. The role of insulin resistance in the pathogenesis of myotonic muscular dystrophy. Brain 1987, 110, 469–488. [Google Scholar] [CrossRef] [PubMed]
- Vujnic, M.; Peric, S.; Popovic, S.; Raseta, N.; Ralic, V.; Dobricic, V.; Novakovic, I.; Rakocevic-Stojanovic, V. Metabolic syndrome in patients with myotonic dystrophy type 1. Muscle Nerve 2015, 52, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Perseghin, G.; Comola, M.; Scifo, P.; Benedini, S.; De Cobelli, F.; Lanzi, R.; Costantino, F.; Lattuada, G.; Battezzati, A.; Del Maschio, A.; et al. Postabsorptive and insulin-stimulated energy and protein metabolism in patients with myotonic dystrophy type 1. Am. J. Clin. Nutr. 2004, 80, 357–364. [Google Scholar] [CrossRef][Green Version]
- Matsumura, T.; Iwahashi, H.; Funahashi, T.; Takahashi, M.P.; Saito, T.; Yasui, K.; Saito, T.; Iyama, A.; Toyooka, K.; Fujimura, H.; et al. A cross-sectional study for glucose intolerance of myotonic dystrophy. J. Neurol. Sci. 2009, 276, 60–65. [Google Scholar] [CrossRef]
- Heatwole, C.R.; Eichinger, K.J.; Friedman, D.I.; Hilbert, J.E.; Jackson, C.E.; Logigian, E.L.; Martens, W.B.; McDermott, M.P.; Pandya, S.K.; Quinn, C.; et al. Open-Label Trial of Recombinant Human Insulin-like Growth Factor 1/Recombinant Human Insulin-like Growth Factor Binding Protein 3 in Myotonic Dystrophy Type 1. Arch. Neurol. 2011, 68. [Google Scholar] [CrossRef] [PubMed]
- Perna, A.; Maccora, D.; Rossi, S.; Nicoletti, T.F.; Zocco, M.A.; Riso, V.; Modoni, A.; Petrucci, A.; Valenza, V.; Grieco, A.; et al. High Prevalence and Gender-Related Differences of Gastrointestinal Manifestations in a Cohort of DM1 Patients: A Perspective, Cross-Sectional Study. Front. Neurol. 2020, 11. [Google Scholar] [CrossRef]
- Moorjani, S.; Gaudet, D.; Laberge, C.; Thibault, M.C.; Mathieu, J.; Morissette, J.; Lupien, P.J.; Brun, D.; Gagné, C. Hypertriglyceridemia and Lower LDL Cholesterol Concentration in Relation to Apolipoprotein E Phenotypes in Myotonic Dystrophy. Can. J. Neurol. Sci. J. Can. Sci. Neurol. 1989, 16, 129–133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Takada, H.; Kon, S.; Oyama, Y.; Kimura, T.; Nagahata, F. Liver functional impairment in myotonic dystrophy type 1. Neuromuscul. Disord. 2016, 26, S195. [Google Scholar] [CrossRef]
- Renna, L.V.; Bosè, F.; Brigonzi, E.; Fossati, B.; Meola, G.; Cardani, R. Aberrant insulin receptor expression is associated with insulin resistance and skeletal muscle atrophy in myotonic dystrophies. PLoS ONE 2019, 14, e0214254. [Google Scholar] [CrossRef]
- Wang, E.T.; Treacy, D.; Eichinger, K.; Struck, A.; Estabrook, J.; Olafson, H.; Wang, T.T.; Bhatt, K.; Westbrook, T.; Sedehizadeh, S.; et al. Transcriptome alterations in myotonic dystrophy skeletal muscle and heart. Hum. Mol. Genet. 2019, 28, 1312–1321. [Google Scholar] [CrossRef] [PubMed]
- Han, G.-S.; Carman, G.M. Characterization of the Human LPIN1 -encoded Phosphatidate Phosphatase Isoforms. J. Biol. Chem. 2010, 285, 14628–14638. [Google Scholar] [CrossRef]
- Lutkewitte, A.J.; Finck, B.N. Regulation of Signaling and Metabolism by Lipin-mediated Phosphatidic Acid Phosphohydrolase Activity. Biomolecules 2020, 10, 1386. [Google Scholar] [CrossRef] [PubMed]
- Zhukovsky, M.A.; Filograna, A.; Luini, A.; Corda, D.; Valente, C. Phosphatidic acid in membrane rearrangements. FEBS Lett. 2019, 593, 2428–2451. [Google Scholar] [CrossRef] [PubMed]
- Péterfy, M.; Phan, J.; Xu, P.; Reue, K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 2001, 27, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Reue, K.; Brindley, D.N. Thematic Review Series: Glycerolipids. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 2008, 49, 2493–2503. [Google Scholar] [CrossRef]
- Nadra, K.; Charles, A.-S.d.P.; Medard, J.-J.; Hendriks, W.T.; Han, G.-S.; Gres, S.; Carman, G.M.; Saulnier-Blache, J.-S.; Verheijen, M.H.G.; Chrast, R. Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 2008, 22, 1647–1661. [Google Scholar] [CrossRef]
- Mitra, M.S.; Chen, Z.; Ren, H.; Harris, T.E.; Chambers, K.T.; Hall, A.M.; Nadra, K.; Klein, S.; Chrast, R.; Su, X.; et al. Mice with an adipocyte-specific lipin 1 separation-of-function allele reveal unexpected roles for phosphatidic acid in metabolic regulation. Proc. Natl. Acad. Sci. USA 2013, 110, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, J.; Qiu, W.; Han, G.-S.; Carman, G.M.; Adeli, K. Lipin-1γ isoform is a novel lipid droplet-associated protein highly expressed in the brain. FEBS Lett. 2011, 585, 1979–1984. [Google Scholar] [CrossRef]
- Reue, K.; Dwyer, J.R. Lipin proteins and metabolic homeostasis. J. Lipid Res. 2009, 50, S109–S114. [Google Scholar] [CrossRef]
- Reue, K.; Wang, H. Mammalian lipin phosphatidic acid phosphatases in lipid synthesis and beyond: Metabolic and inflammatory disorders. J. Lipid Res. 2019, 60, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Ter Horst, K.W.; Gilijamse, P.W.; Versteeg, R.I.; Ackermans, M.T.; Nederveen, A.J.; la Fleur, S.E.; Romijn, J.A.; Nieuwdorp, M.; Zhang, D.; Samuel, V.T.; et al. Hepatic Diacylglycerol-Associated Protein Kinase Cε Translocation Links Hepatic Steatosis to Hepatic Insulin Resistance in Humans. Cell Rep. 2017, 19, 1997–2004. [Google Scholar] [CrossRef]
- Ryu, D.; Oh, K.-J.; Jo, H.-Y.; Hedrick, S.; Kim, Y.-N.; Hwang, Y.-J.; Park, T.-S.; Han, J.-S.; Choi, C.S.; Montminy, M.; et al. TORC2 Regulates Hepatic Insulin Signaling via a Mammalian Phosphatidic Acid Phosphatase, LIPIN1. Cell Metab. 2009, 9, 240–251. [Google Scholar] [CrossRef]
- Chae, M.; Jung, J.-Y.; Bae, I.-H.; Kim, H.-J.; Lee, T.R.; Shin, D.W. Lipin-1 expression is critical for keratinocyte differentiation. J. Lipid Res. 2016, 57, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Meana, C.; Peña, L.; Lordén, G.; Esquinas, E.; Guijas, C.; Valdearcos, M.; Balsinde, J.; Balboa, M.A. Lipin-1 Integrates Lipid Synthesis with Proinflammatory Responses during TLR Activation in Macrophages. J. Immunol. 2014, 193, 4614–4622. [Google Scholar] [CrossRef]
- Grkovich, A.; Armando, A.; Quehenberger, O.; Dennis, E.A. TLR-4 mediated group IVA phospholipase A2 activation is phosphatidic acid phosphohydrolase 1 and protein kinase C dependent. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2009, 1791, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Grkovich, A.; Johnson, C.A.; Buczynski, M.W.; Dennis, E.A. Lipopolysaccharide-induced Cyclooxygenase-2 Expression in Human U937 Macrophages Is Phosphatidic Acid Phosphohydrolase-1-dependent. J. Biol. Chem. 2006, 281, 32978–32987. [Google Scholar] [CrossRef]
- Valdearcos, M.; Esquinas, E.; Meana, C.; Gil-de-Gómez, L.; Guijas, C.; Balsinde, J.; Balboa, M.A. Subcellular Localization and Role of Lipin-1 in Human Macrophages. J. Immunol. 2011, 186, 6004–6013. [Google Scholar] [CrossRef]
- Koh, Y.-K.; Lee, M.-Y.; Kim, J.-W.; Kim, M.; Moon, J.-S.; Lee, Y.-J.; Ahn, Y.-H.; Kim, K.-S. Lipin1 Is a Key Factor for the Maturation and Maintenance of Adipocytes in the Regulatory Network with CCAAT/Enhancer-binding Protein α and Peroxisome Proliferator-activated Receptor γ 2. J. Biol. Chem. 2008, 283, 34896–34906. [Google Scholar] [CrossRef]
- Chen, Z.; Gropler, M.C.; Mitra, M.S.; Finck, B.N. Complex Interplay between the Lipin 1 and the Hepatocyte Nuclear Factor 4 α (HNF4α) Pathways to Regulate Liver Lipid Metabolism. PLoS ONE 2012, 7, e51320. [Google Scholar] [CrossRef]
- Manmontri, B.; Sariahmetoglu, M.; Donkor, J.; Khalil, M.B.; Sundaram, M.; Yao, Z.; Reue, K.; Lehner, R.; Brindley, D.N. Glucocorticoids and cyclic AMP selectively increase hepatic lipin-1 expression, and insulin acts antagonistically. J. Lipid Res. 2008, 49, 1056–1067. [Google Scholar] [CrossRef]
- Kim, H.B.; Kumar, A.; Wang, L.; Liu, G.-H.; Keller, S.R.; Lawrence, J.C.; Finck, B.N.; Harris, T.E. Lipin 1 Represses NFATc4 Transcriptional Activity in Adipocytes To Inhibit Secretion of Inflammatory Factors. Mol. Cell. Biol. 2010, 30, 3126–3139. [Google Scholar] [CrossRef]
- Chandran, S.; Schilke, R.M.; Blackburn, C.M.R.; Yurochko, A.; Mirza, R.; Scott, R.S.; Finck, B.N.; Woolard, M.D. Lipin-1 Contributes to IL-4 Mediated Macrophage Polarization. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-H.; Gerace, L. Sumoylation Regulates Nuclear Localization of Lipin-1α in Neuronal Cells. PLoS ONE 2009, 4, e7031. [Google Scholar] [CrossRef][Green Version]
- Terracciano, C.; Rastelli, E.; Morello, M.; Celi, M.; Bucci, E.; Antonini, G.; Porzio, O.; Tarantino, U.; Zenobi, R.; Massa, R. Vitamin D deficiency in myotonic dystrophy type 1. J. Neurol. 2013, 260, 2330–2334. [Google Scholar] [CrossRef]
- Yao-Borengasser, A.; Rasouli, N.; Varma, V.; Miles, L.M.; Phanavanh, B.; Starks, T.N.; Phan, J.; Spencer, H.J.; McGehee, R.E.; Reue, K.; et al. Lipin Expression Is Attenuated in Adipose Tissue of Insulin-Resistant Human Subjects and Increases With Peroxisome Proliferator-Activated Receptor Activation. Diabetes 2006, 55, 2811–2818. [Google Scholar] [CrossRef][Green Version]
- Paran, C.W.; Zou, K.; Ferrara, P.J.; Song, H.; Turk, J.; Funai, K. Lipogenesis mitigates dysregulated sarcoplasmic reticulum calcium uptake in muscular dystrophy. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2015, 1851, 1530–1538. [Google Scholar] [CrossRef][Green Version]
- Lee, J.; Ridgway, N.D. Substrate channeling in the glycerol-3-phosphate pathway regulates the synthesis, storage and secretion of glycerolipids. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158438. [Google Scholar] [CrossRef]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR Complex 1 Regulates Lipin 1 Localization to Control the SREBP Pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Grigoraş, A.; Amalinei, C.; Balan, R.A.; Giuşcă, S.E.; Avădănei, E.R.; Lozneanu, L.; Căruntu, I.-D. Adipocytes spectrum—From homeostasia to obesity and its associated pathology. Ann. Anat. Anat. Anzeiger 2018, 219, 102–120. [Google Scholar] [CrossRef] [PubMed]
- Ferré, P.; Foufelle, F. Hepatic steatosis: A role for de novo lipogenesis and the transcription factor SREBP-1c. Diabetes, Obes. Metab. 2010, 12, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Ormazabal, V.; Nair, S.; Elfeky, O.; Aguayo, C.; Salomon, C.; Zuñiga, F.A. Association between insulin resistance and the development of cardiovascular disease. Cardiovasc. Diabetol. 2018, 17, 122. [Google Scholar] [CrossRef]
- Sesti, G. Pathophysiology of insulin resistance. Best Pract. Res. Clin. Endocrinol. Metab. 2006, 20, 665–679. [Google Scholar] [CrossRef]
- Samson, S.L.; Garber, A.J. Metabolic Syndrome. Endocrinol. Metab. Clin. North Am. 2014, 43, 1–23. [Google Scholar] [CrossRef]
- Taillandier, D.; Polge, C. Skeletal muscle atrogenes: From rodent models to human pathologies. Biochimie 2019, 166, 251–269. [Google Scholar] [CrossRef]
- Huffman, T.A.; Mothe-Satney, I.; Lawrence, J.C. Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc. Natl. Acad. Sci. USA 2002, 99, 1047–1052. [Google Scholar] [CrossRef]
- Saini-Chohan, H.K.; Mitchell, R.W.; Vaz, F.M.; Zelinski, T.; Hatch, G.M. Delineating the role of alterations in lipid metabolism to the pathogenesis of inherited skeletal and cardiac muscle disorders. J. Lipid Res. 2012, 53, 4–27. [Google Scholar] [CrossRef]
- Péterfy, M.; Harris, T.E.; Fujita, N.; Reue, K. Insulin-stimulated Interaction with 14-3-3 Promotes Cytoplasmic Localization of Lipin-1 in Adipocytes. J. Biol. Chem. 2010, 285, 3857–3864. [Google Scholar] [CrossRef]
- Song, K.-Y.; Guo, X.-M.; Wang, H.-Q.; Zhang, L.; Huang, S.-Y.; Huo, Y.-C.; Zhang, G.; Feng, J.-Z.; Zhang, R.-R.; Ma, Y.; et al. MBNL1 reverses the proliferation defect of skeletal muscle satellite cells in myotonic dystrophy type 1 by inhibiting autophagy via the mTOR pathway. Cell Death Dis. 2020, 11, 545. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weng, W.-C.; Stock, L.; Lindquist, D.; Martinez, A.; Gourdon, G.; Timchenko, N.; Snape, M.; Timchenko, L. Correction of Glycogen Synthase Kinase 3β in Myotonic Dystrophy 1 Reduces the Mutant RNA and Improves Postnatal Survival of DMSXL Mice. Mol. Cell. Biol. 2019, 39. [Google Scholar] [CrossRef]
- Wilcox, G. Insulin and insulin resistance. Clin. Biochem. Rev. 2005, 26, 19–39. [Google Scholar] [PubMed]
- van Harmelen, V.; Rydén, M.; Sjölin, E.; Hoffstedt, J. A role of lipin in human obesity and insulin resistance: Relation to adipocyte glucose transport and GLUT4 expression. J. Lipid Res. 2007, 48, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Reue, K.; Donkor, J. Lipin: A determinant of adiposity, insulin sensitivity and energy balance. Future Lipidol. 2006, 1, 91–101. [Google Scholar] [CrossRef]
- Rashid, T.; Nemazanyy, I.; Paolini, C.; Tatsuta, T.; Crespin, P.; Villeneuve, D.; Brodesser, S.; Benit, P.; Rustin, P.; Baraibar, M.A.; et al. Lipin1 deficiency causes sarcoplasmic reticulum stress and chaperone-responsive myopathy. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Chan, E.K.; Kornberg, A.J.; Ryan, M.M. A diagnostic approach to recurrent myalgia and rhabdomyolysis in children. Arch. Dis. Child. 2015, 100, 793–797. [Google Scholar] [CrossRef]
- Cheng, X.; Li, J.; Guo, D. SCAP/SREBPs are Central Players in Lipid Metabolism and Novel Metabolic Targets in Cancer Therapy. Curr. Top. Med. Chem. 2018, 18, 484–493. [Google Scholar] [CrossRef]
- Guo, D.; Bell, E.; Mischel, P.; Chakravarti, A. Targeting SREBP-1-driven Lipid Metabolism to Treat Cancer. Curr. Pharm. Des. 2014, 20, 2619–2626. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Todd, B.L.; Espenshade, P.J. SREBP Pathway Responds to Sterols and Functions as an Oxygen Sensor in Fission Yeast. Cell 2005, 120, 831–842. [Google Scholar] [CrossRef]
- Im, S.-S.; Yousef, L.; Blaschitz, C.; Liu, J.Z.; Edwards, R.A.; Young, S.G.; Raffatellu, M.; Osborne, T.F. Linking Lipid Metabolism to the Innate Immune Response in Macrophages through Sterol Regulatory Element Binding Protein-1a. Cell Metab. 2011, 13, 540–549. [Google Scholar] [CrossRef]
- Shimano, H.; Shimomura, I.; Hammer, R.E.; Herz, J.; Goldstein, J.L.; Brown, M.S.; Horton, J.D. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Investig. 1997, 100, 2115–2124. [Google Scholar] [CrossRef]
- García-Puga, M.; Saenz-Antoñanzas, A.; Fernández-Torrón, R.; de Munain, A.L.; Matheu, A. Myotonic Dystrophy type 1 cells display impaired metabolism and mitochondrial dysfunction that are reversed by metformin. Aging (Albany NY) 2020, 12, 6260–6275. [Google Scholar] [CrossRef] [PubMed]
- Gramegna, L.L.; Giannoccaro, M.P.; Manners, D.N.; Testa, C.; Zanigni, S.; Evangelisti, S.; Bianchini, C.; Oppi, F.; Poda, R.; Avoni, P.; et al. Mitochondrial dysfunction in myotonic dystrophy type 1. Neuromuscul. Disord. 2018, 28, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P. Skeletal muscle metabolism in myotonic dystrophy A 31P magnetic resonance spectroscopy study. Brain 1997, 120, 1699–1711. [Google Scholar] [CrossRef] [PubMed]
- Ihara, Y.; Mori, A.; Hayabara, T.; Namba, R.; Nobukuni, K.; Sato, K.; Miyata, S.; Edamatsu, R.; Liu, J.; Kawai, M. Free radicals, lipid peroxides and antioxidants in blood of patients with myotonic dystrophy. J. Neurol. 1995, 242, 119–122. [Google Scholar] [CrossRef]
- Zhang, P.; Reue, K. Lipin proteins and glycerolipid metabolism: Roles at the ER membrane and beyond. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1583–1595. [Google Scholar] [CrossRef]
- Higashida, K.; Higuchi, M.; Terada, S. Potential role of lipin-1 in exercise-induced mitochondrial biogenesis. Biochem. Biophys. Res. Commun. 2008, 374, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Phan, J.; Reue, K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005, 1, 73–83. [Google Scholar] [CrossRef]
- Phan, J.; Péterfy, M.; Reue, K. Lipin Expression Preceding Peroxisome Proliferator-activated Receptor-γ Is Critical for Adipogenesis in Vivo and in Vitro. J. Biol. Chem. 2004, 279, 29558–29564. [Google Scholar] [CrossRef] [PubMed]
- Guilherme, A.; Virbasius, J.V.; Puri, V.; Czech, M.P. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 2008, 9, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Rajala, M.W.; Scherer, P.E. Minireview: The Adipocyte—At the Crossroads of Energy Homeostasis, Inflammation, and Atherosclerosis. Endocrinology 2003, 144, 3765–3773. [Google Scholar] [CrossRef] [PubMed]
- Sartipy, P.; Loskutoff, D.J. Monocyte chemoattractant protein 1 in obesity and insulin resistance. Proc. Natl. Acad. Sci. USA 2003, 100, 7265–7270. [Google Scholar] [CrossRef]
- Hepler, C.; Gupta, R.K. The expanding problem of adipose depot remodeling and postnatal adipocyte progenitor recruitment. Mol. Cell. Endocrinol. 2017, 445, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, B.; Hedjazifar, S.; Gogg, S.; Hammarstedt, A.; Smith, U. Insulin resistance and impaired adipogenesis. Trends Endocrinol. Metab. 2015, 26, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Klöting, N.; Blüher, M. Adipocyte dysfunction, inflammation and metabolic syndrome. Rev. Endocr. Metab. Disord. 2014, 15, 277–287. [Google Scholar] [CrossRef]
- Valdearcos, M.; Esquinas, E.; Meana, C.; Peña, L.; Gil-de-Gómez, L.; Balsinde, J.; Balboa, M.A. Lipin-2 Reduces Proinflammatory Signaling Induced by Saturated Fatty Acids in Macrophages. J. Biol. Chem. 2012, 287, 10894–10904. [Google Scholar] [CrossRef]
- Balboa, M.A.; de Pablo, N.; Meana, C.; Balsinde, J. The role of lipins in innate immunity and inflammation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Cacciatore, S.; Loda, M. Innovation in metabolomics to improve personalized healthcare. Ann. N. Y. Acad. Sci. 2015, 1346, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Ellis, D.I.; Dunn, W.B.; Griffin, J.L.; Allwood, J.W.; Goodacre, R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics 2007, 8, 1243–1266. [Google Scholar] [CrossRef] [PubMed]
- Felgueiras, J.; Silva, J.V.; Nunes, A.; Fernandes, I.; Patrício, A.; Maia, N.; Pelech, S.; Fardilha, M. Investigation of spectroscopic and proteomic alterations underlying prostate carcinogenesis. J. Proteom. 2020, 226, 103888. [Google Scholar] [CrossRef]
- Santos, F.; Magalhaes, S.; Henriques, M.C.; Fardilha, M.; Nunes, A. Spectroscopic Features of Cancer Cells: FTIR Spectroscopy as a Tool for Early Diagnosis. Curr. Metab. 2018, 6, 103–111. [Google Scholar] [CrossRef]
- Santos, F.; Magalhães, S.; Henriques, M.C.; Silva, B.; Valença, I.; Ribeiro, D.; Fardilha, M.; Nunes, A. Understanding Prostate Cancer Cells Metabolome: A Spectroscopic Approach. Curr. Metab. 2019, 6, 218–224. [Google Scholar] [CrossRef]
- Schweitzer, G.G.; Collier, S.L.; Chen, Z.; Mccommis, K.S.; Pittman, S.K.; Yoshino, J.; Matkovich, S.J.; Hsu, F.-F.; Chrast, R.; Eaton, J.M.; et al. Loss of lipin 1-mediated phosphatidic acid phosphohydrolase activity in muscle leads to skeletal myopathy in mice. FASEB J. 2019, 33, 652–667. [Google Scholar] [CrossRef]
- Peric, S.; Stojanovic, V.R.; Nikolic, A.; Kacar, A.; Basta, I.; Pavlovic, S.; Lavrnic, D. Peripheral neuropathy in patients with myotonic dystrophy type 1. Neurol. Res. 2013, 35, 331–335. [Google Scholar] [CrossRef]
| Global Clinical Features | DM1 |
|---|---|
| Heredity | Autosomal dominant |
| Prevalence | 1 in 3000 and 8000 individuals |
| Cancer | Reproductive tract (endometrial, ovarian and testicular), thyroid and colorectal |
| High risk of tumour development | |
| Anticipation | Present |
| Life expectancy | Reduced |
| Mortality | 70%, Caused by cardiorespiratory complications |
| Appearance | Forehead balding |
| Myopathic face | |
| Temporal wasting | |
| Ptosis | |
| Nasal/slurred speech | |
| Dysphagia | |
| Age at onset | Childhood to adulthood |
| Congenital form | Present |
| Facial dysmorphism | Present in congenital form |
| Male hypogonadism | Present |
| Insulin Resistance | Present |
| Dyslipidemia | Present |
| Metabolic syndrome | Present |
| Thyroid deficiency | More common |
| Diabetes mellitus | High risk of development |
| Creatine kinase, liver enzymes and cholesterol | Elevated |
| Hypogammaglobulinemia | High incidence |
| Muscle weakness | Extreme at 50 years of age (distal) |
| Muscle Pain/myalgia | Less frequent |
| Clinical myotonia | Evident in early adulthood |
| Myotonia | Handgrip, tongue (distal) |
| Atrophy | Distal (early) |
| Type 1 fiber atrophy | Present |
| Calf hypertrophy | Absent |
| Cardiac arrhythmias | Present |
| Sudden death | More common |
| Sleep disorders | Present |
| Cognitive decline | Prominent in congenital form |
| Central nervous system problems | Present |
| Cataracts | Present |
| Respiratory failure | Present |
| Gastrointestinal problems | Present |
| Abdominal pain | Present |
| Constipation | Present |
| Gene | DMPK, chromosome 19q13.3, CTG expansion at 3’UTR |
| DMPK | Reduced |
| CNBP/ZNF-9 | Normal |
| CUGBP1 | Upregulated |
| MBNL1 | Down regulated/sequestered |
| Ribonuclear inclusion | Present |
| Spliceopathy | Present |
| Transcription dysregulation | Present |
| MicroRNA dysregulation | Present |
| RNA translation | Present |
| Patients Characteristics | DM1 (n = 623) [27,28,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70] | Controls (n = 428) [27,55,56,57,59,60,61,62,64,65,68,70] |
| Patients per study | 44 ± 33; (8–115) [27,28,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70] | 96.8 ± 203.3; (3–734) [27,55,56,57,59,60,61,62,64,65,68,70] |
| Age (years) | 41.7 ± 3.8; (34–47) [27,28,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70] | 42.35 ± 6.17; (33–54.6) [27,55,56,57,59,60,61,62,64,65,68,70] |
| Sex | 379 F, 417 M [27,28,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70] | 347 F, 815 M [27,55,56,57,59,60,61,62,64,65,68,70] |
| BMI (kg/m2) | 23.8 ± 1.75; (22.3–27.2); Median: 23.4 [27,28,55,57,58,59,61,63,64,65,66,67,70] | 23 ± 0.72; (21.7–23.7); Median: 23.2 [27,55,57,59,61,64,65,70] |
| Waist circumference (cm) | 95.8 ± 2.19; (94.3–97.4) Median: 93.43 [27,57,58,59] | 84.9; Median: 87.5 [27,57,59] |
| CTG repeat length | 555.22 ± 250.99; (355.9–973); Median: 413.75 [28,55,58,59,60,63,65,66,67,69,70] | ND |
| Central Obesity (%) | 13.6 [63] | ND |
| Metabolic syndrome (%) | 16.7–41.2 [58,63] | ND |
| Insulin metabolism (pmol/L) | 125.4 ± 63.5; (51.38–186.11); Median: 73.5 Male: 127.8 ± 33.4; (104.26–151.45) Female: 119.8 ± 22,1; (104.16–135.48) [27,28,55,56,57,58,59,60,61,62] | 69.9 ± 28.4; (45.8–86.0); Median: 44.29 Male: 56.25 Female: 63 [27,57,59,60,61,62] |
| HOMA-IR | 3.03 ± 1.7; (1.9–6.4); Median: 2.25 Male: 4.7 Female: 3.7 [27,55,56,57,58,61,65,70] | 1.43 ± 0.13; (1.3–1.6); Median: 1.42 [27,57,61,65] |
| Glucose metabolism (mg/dL) | 91.84 ± 7.5; (82.7–108.5); Median: 91.4 Male: 90.5 ± 6.3; (86–95) Female: 95.9 ± 24.1; (78.9–113.0) [27,28,55,56,57,58,60,61,62,63,64,65,66,67] | 89.65 ± 5.11; (81.6–95.6); Median: 90 Male: 83 Female: 88 [27,55,57,60,61,62,64,65] |
| Total cholesterol (mg/dL) | 200.34 ± 15.88; (176–228); Median: 200 Male: 216.37 ± 50.02; (181.0–251.7) Female: 200.8 ± 13.9; (191.0–210.7) [27,28,55,56,57,58,59,60,62,63,64,65,67] | 179.4 ± 24.5; (146.0–200.5); Median: 176.72 Male: 128 Female: 134 [27,55,57,59,60,62,64,65] |
| HDL (mg/dL) | 52.42 ± 3.64; (48.2–58.0); Median: 51.4 Male: 44 Female: 61 [27,28,58,59,60,62,63,64,65,66,68,69] | 51.4 ± 5.5; (45.7–56.7 51.82); Median: 51.82 Male: 67 Female: 55 [27,53,57,58,61,62,65,66] |
| LDL (mg/dL) | 118.38 ± 14.65; (106.34–143.08); Median: 122.6 Male: 104 Female: 101 [27,28,58,59,60,62,63,64,68,69] | 110.0 ± 17.7; (97.4–122.6); Median: 129.3 Male: 146 Female: 102 [27,59,60,62,64,68,69] |
| TAG (mg/dL) | 184.9 ± 52.5; (108–274); Median: 172.4 Male: 194.3 ± 11.7; (186.0–202.6) Female: 134.1 ± 18.2; (121–147) [27,28,56,57,58,59,60,62,63,64,65,66,67,68,69] | 115.9 ± 39.6; (77–168); Median: 95.2 Male: 75 Female: 105 [27,57,59,60,62,64,65,68,69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mateus, T.; Martins, F.; Nunes, A.; Herdeiro, M.T.; Rebelo, S. Metabolic Alterations in Myotonic Dystrophy Type 1 and Their Correlation with Lipin. Int. J. Environ. Res. Public Health 2021, 18, 1794. https://doi.org/10.3390/ijerph18041794
Mateus T, Martins F, Nunes A, Herdeiro MT, Rebelo S. Metabolic Alterations in Myotonic Dystrophy Type 1 and Their Correlation with Lipin. International Journal of Environmental Research and Public Health. 2021; 18(4):1794. https://doi.org/10.3390/ijerph18041794
Chicago/Turabian StyleMateus, Tiago, Filipa Martins, Alexandra Nunes, Maria Teresa Herdeiro, and Sandra Rebelo. 2021. "Metabolic Alterations in Myotonic Dystrophy Type 1 and Their Correlation with Lipin" International Journal of Environmental Research and Public Health 18, no. 4: 1794. https://doi.org/10.3390/ijerph18041794
APA StyleMateus, T., Martins, F., Nunes, A., Herdeiro, M. T., & Rebelo, S. (2021). Metabolic Alterations in Myotonic Dystrophy Type 1 and Their Correlation with Lipin. International Journal of Environmental Research and Public Health, 18(4), 1794. https://doi.org/10.3390/ijerph18041794









