Review of Deep Learning-Based Atrial Fibrillation Detection Studies
Abstract
:1. Introduction
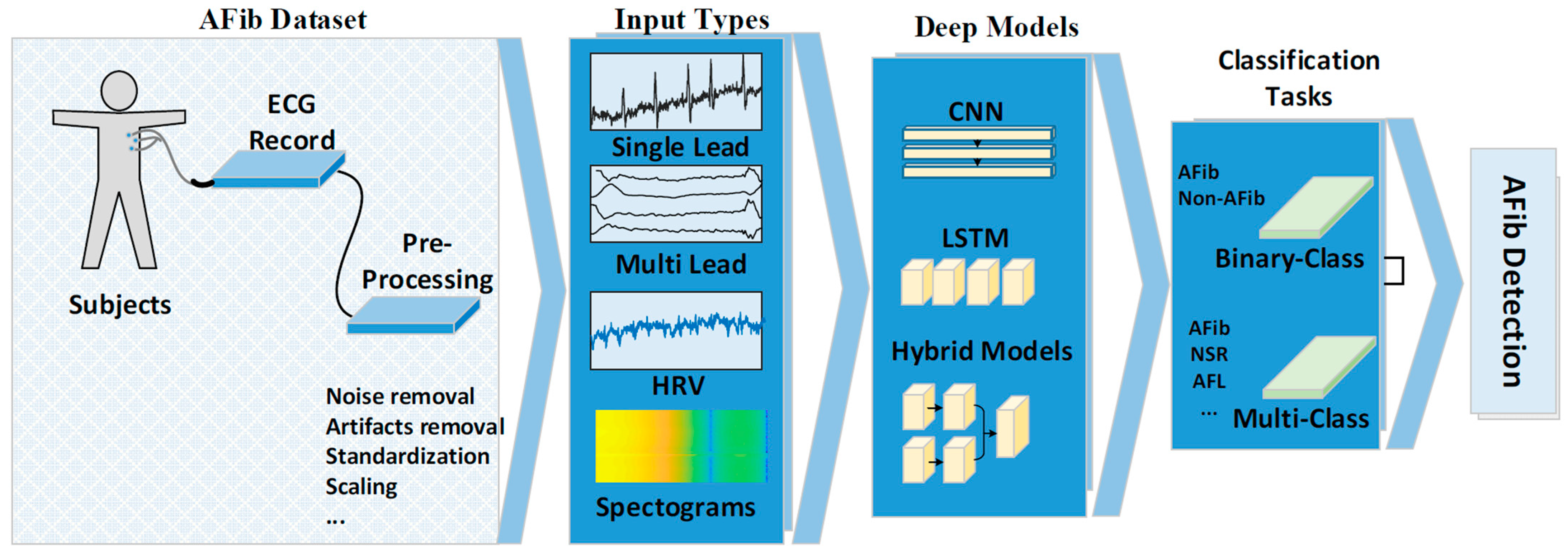
2. Materials and Methods
2.1. AF Datasets
Pre-Processing
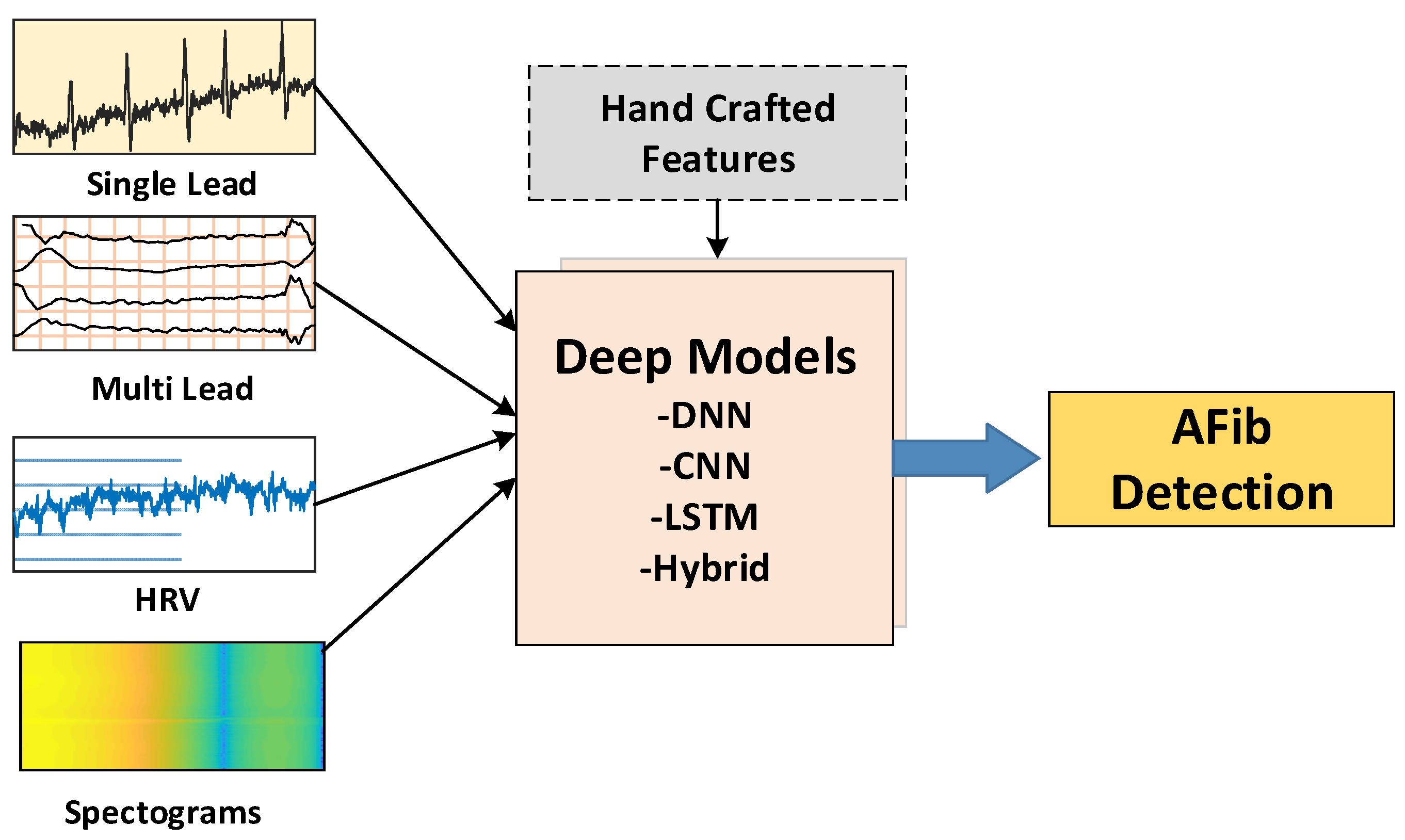
2.2. Model Input Types
2.3. Deep Models
2.3.1. Deep Neural Networks
2.3.2. Convolutional Neural Networks
2.3.3. Recurrent Neural Networks
2.3.4. Long Short-Term Memory
2.3.5. Hybrid Deep Models
2.4. Classification Task
3. Discussion and Comments
Cardiologist Comments
- Stroke and stroke-related complications can be prevented with early diagnosis of AF and initiation of oral anticoagulant therapy.
- AF-induced electrical and/or mechanical remodeling of the heart can be averted with rhythm and/or heart rate control.
- AF-associated heart failure can be prevented and/or ameliorated with specific heart failure drugs.
- AF-associated hospitalizations and healthcare expenditure can be reduced through optimal preventive management.
- Few public ECG databases are available for DL model training, which require a high volume of input data to develop accurate and robust models.
- Paroxysmal AF, which exacts similar stroke risk as persistent and permanent AF, may escape detection on 12-lead ECG and/or short-duration ECG monitoring.
- Related arrhythmia like AFL that are morphologically distinct from AF and yet also carries similar stroke risk as AF has only been included in selected studies.
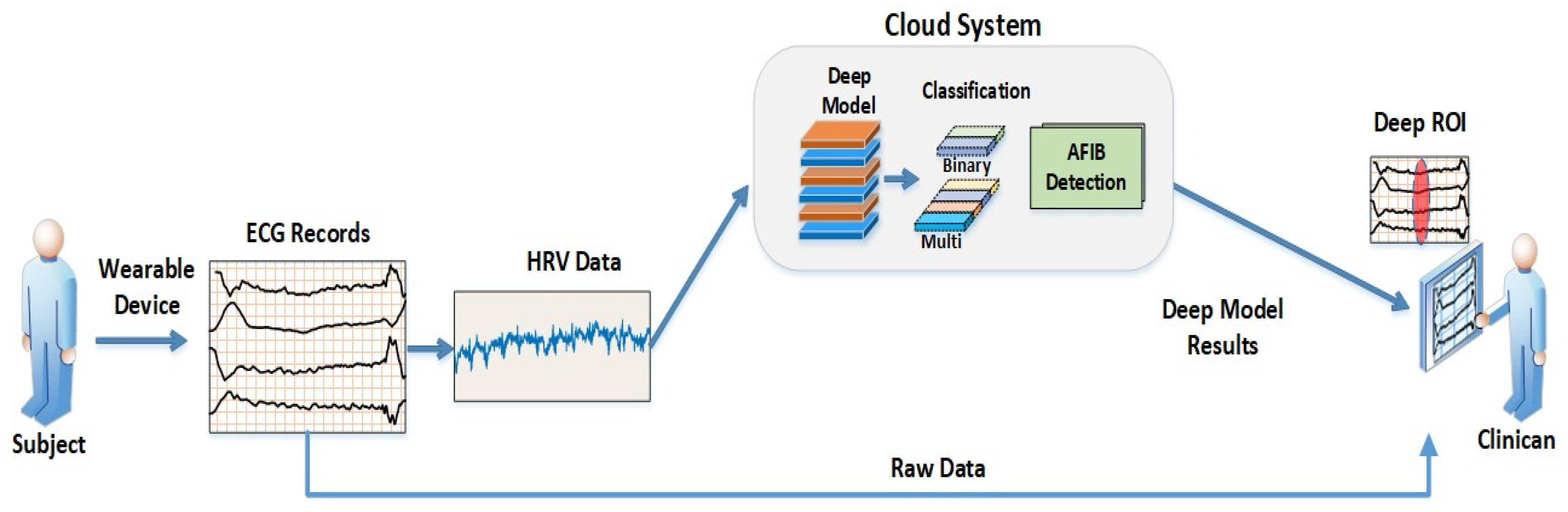
4. Future Work
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Furberg, C.D.; Psaty, B.M.; Manolio, T.A.; Gardin, J.M.; Smith, V.E.; Rautaharju, P.M. Prevalence of atrial fibrillation in elderly subjects (the Cardiovascular Health Study). Am. J. Cardiol. 1994, 74, 236–241. [Google Scholar] [CrossRef]
- Wolf, P.A.; Abbott, R.D.; Kannel, W.B. Atrial fibrillation as an independent risk factor for stroke: The Framingham Study. Stroke 1991, 22, 983–988. [Google Scholar] [CrossRef] [Green Version]
- Camm, A.J.; Lip, G.Y.; De Caterina, R.; Savelieva, I.; Atar, D.; Hohnloser, S.H.; Hindricks, G.; Kirchhof, P.; Bax, J.J.; Baumgartner, H.; et al. 2012 focused update of the ESC Guidelines for the management of atrial fibrillation: An update of the 2010 ESC Guidelines for the management of atrial fibrillation. Developed with the special contribution of the European Heart Rhythm Association. Eur. Heart J. 2012, 33, 2719–2747. [Google Scholar] [CrossRef] [Green Version]
- Hijazi, Z.; Oldgren, J.; Siegbahn, A.; Granger, C.B.; Wallentin, L. Biomarkers in atrial fibrillation: A clinical review. Eur. Heart J. 2013, 34, 1475–1480. [Google Scholar] [CrossRef]
- Gillis, A.M.; Krahn, A.D.; Skanes, A.C.; Nattel, S. Management of Atrial Fibrillation in the Year 2033: New Concepts, Tools, and Applications Leading to Personalized Medicine. Can. J. Cardiol. 2013, 29, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Markides, V.; Schilling, R.J. Atrial fibrillation: Classification, pathophysiology, mechanisms and drug treatment. Heart 2003, 89, 939–943. [Google Scholar] [CrossRef]
- Kareem, M.; Lei, N.; Ali, A.; Ciaccio, E.J.; Acharya, U.R.; Faust, O. A review of patient-led data acquisition for atrial fibrillation detection to prevent stroke. Biomed. Signal Process. Control 2021, 69, 102818. [Google Scholar] [CrossRef]
- Fuster, V.; Rydén, L.E.; Cannom, D.S.; Crijns, H.J.; Curtis, A.B.; Ellenbogen, K.A.; Halperin, J.L.; Kay, G.N.; Le Huezey, J.-Y.; Lowe, J.E.; et al. 2011 ACCF/AHA/HRS focused updates incorporated into the ACC/AHA/ESC 2006 guidelines for the management of patients with atrial fibrillation: A report of the American College of Cardiology Foundation/American Heart Association Task Force on practice guide. Circulation 2011, 123, e269–e367. [Google Scholar] [CrossRef]
- Acharya, U.R.; Faust, O.; Ciaccio, E.J.; Koh, J.E.W.; Oh, S.L.; Tan, R.S.; Garan, H. Application of nonlinear methods to discriminate fractionated electrograms in paroxysmal versus persistent atrial fibrillation. Comput. Methods Programs Biomed. 2019, 175, 163–178. [Google Scholar] [CrossRef] [PubMed]
- January, C.T.; Wann, L.S.; Alpert, J.S.; Calkins, H.; Cigarroa, J.E.; Cleveland, J.C.; Conti, J.B.; Ellinor, P.T.; Ezekowitz, M.D.; Field, M.E.; et al. 2014 AHA/ACC/HRS Guideline for the Management of Patients with Atrial Fibrillation: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. J. Am. Coll. Cardiol. 2014, 64, 2246–2280. [Google Scholar] [CrossRef] [Green Version]
- Fuster, V.; Rydén, L.E.; Cannom, D.S.; Crijns, H.J.; Curtis, A.B.; Ellenbogen, K.A.; Halperin, J.L.; Le Heuzey, J.-Y.; Kay, G.N.; Lowe, J.E.; et al. ACC/AHA/ESC 2006 Guidelines for the Management of Patients with Atrial Fibrillation: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the European Society of Cardiology Committee for Practice. Circulation 2006, 114, e257–e354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Zhang, C.; Liu, Y.; Yang, H.; Fu, D.; Wang, H.; Zhang, P. A global and updatable ECG beat classification system based on recurrent neural networks and active learning. Inf. Sci. 2019, 501, 523–542. [Google Scholar] [CrossRef]
- Larburu, N.; Lopetegi, T.; Romero, I. Comparative study of algorithms for Atrial Fibrillation detection. In 2011 Computing in Cardiology; IEEE: Manhattan, NY, USA, 2011; pp. 265–268. [Google Scholar]
- Slocum, J.; Sahakian, A.; Swiryn, S. Diagnosis of atrial fibrillation from surface electrocardiograms based on computer-detected atrial activity. J. Electrocardiol. 1992, 25, 1–8. [Google Scholar] [CrossRef]
- Ladavich, S.; Ghoraani, B. Rate-independent detection of atrial fibrillation by statistical modeling of atrial activity. Biomed. Signal Process. Control 2015, 18, 274–281. [Google Scholar] [CrossRef]
- Deng, M.; Wang, C.; Tang, M.; Zheng, T. Extracting cardiac dynamics within ECG signal for human identification and cardiovascular diseases classification. Neural Netw. 2018, 100, 70–83. [Google Scholar] [CrossRef]
- Parvaneh, S.; Rubin, J. Electrocardiogram Monitoring and Interpretation: From Traditional Machine Learning to Deep Learning, and Their Combination. In Proceedings of the 2018 Computing in Cardiology Conference (CinC), Maastricht, The Netherlands, 23–26 September 2018; Volume 45, pp. 1–4. [Google Scholar] [CrossRef]
- Henzel, N.; Wróbel, J.; Horoba, K. Atrial fibrillation episodes detection based on classification of heart rate derived features. In Proceedings of the 2017 MIXDES-24th International Conference “Mixed Design of Integrated Circuits and Systems”, Bydgoszcz, Poland, 22–24 June 2017; pp. 571–576. [Google Scholar] [CrossRef]
- Andersen, R.S.; Peimankar, A.; Puthusserypady, S. A deep learning approach for real-time detection of atrial fibrillation. Expert Syst. Appl. 2019, 115, 465–473. [Google Scholar] [CrossRef]
- Hagiwara, Y.; Fujita, H.; Oh, S.L.; Tan, J.H.; Tan, R.S.; Ciaccio, E.J.; Acharya, U.R. Computer-aided diagnosis of atrial fibrillation based on ECG Signals: A review. Inf. Sci. 2018, 467, 99–114. [Google Scholar] [CrossRef]
- Faust, O.; Ciaccio, E.J.; Majid, A.; Acharya, U.R. Improving the safety of atrial fibrillation monitoring systems through human verification. Saf. Sci. 2019, 118, 881–886. [Google Scholar] [CrossRef]
- Faust, O.; Ciaccio, E.J.; Acharya, U.R. A review of atrial fibrillation detection methods as a service. Int. J. Environ. Res. Public Health 2020, 17, 3093. [Google Scholar] [CrossRef]
- Mukami, B.; Dakshit, S.; Shamsuddin, R. CEFEs: A CNN Explainable Framework for ECG Signals. Artif. Intell. Med. 2021, 115, 102059. [Google Scholar] [CrossRef]
- Mashrur, F.R.; Islam, S.; Saha, D.K.; Moni, M.A. SCNN: Scalogram-based convolutional neural network to detect obstructive sleep apnea using single-lead electrocardiogram signals. Comput. Biol. Med. 2021, 134, 104532. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Z.; Deng, Y.; Zhang, X.; Zhang, Y. Biomedical Signal Processing and Control Human identification driven by deep CNN and transfer learning based on multiview feature representations of ECG. Biomed. Signal Process. Control 2021, 68, 102689. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X.; Yang, M.; Zhang, L. 12-Lead ECG arrhythmia classi fi cation using cascaded convolutional neural network and expert feature. J. Electrocardiol. 2021, 67, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Sepahvand, M.; Abdali-mohammadi, F. Biomedical Signal Processing and Control A novel multi-lead ECG personal recognition based on signals functional and structural dependencies using time-frequency representation and evolutionary morphological CNN. Biomed. Signal Process. Control 2021, 68, 102766. [Google Scholar] [CrossRef]
- Yao, Z.; Zhu, Z.; Chen, Y. Atrial Fibrillation Detection by Multi-scale Convolutional Neural Networks. In Proceedings of the 2017 20th International Conference on Information Fusion (Fusion), Xi’an, China, 10–13 July 2017. [Google Scholar]
- Huang, Y.; Lin, J.; Wang, G.; Ding, Z.; Sun, L. A Multi-dilation Convolution Neural Network for Atrial Fibrillation Detection. In Proceedings of the 2020 4th International Conference on Digital Signal Processing, Chengdu, China, 19–21 June 2020; pp. 136–140. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Lih, O.S.; Hagiwara, Y.; Tan, J.H.; Adam, M. Automated detection of arrhythmias using different intervals of tachycardia ECG segments with convolutional neural network. Inf. Sci. 2017, 405, 81–90. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Raghavendra, U.; Tan, J.H.; Adam, M.; Gertych, A.; Hagiwara, Y. Automated identification of shockable and non-shockable life-threatening ventricular arrhythmias using convolutional neural network. Future Gener. Comput. Syst. 2018, 79, 952–959. [Google Scholar] [CrossRef]
- Rahhal MMAl Bazi, Y.; AlHichri, H.; Alajlan, N.; Melgani, F.; Yager, R.R. Deep learning approach for active classification of electrocardiogram signals. Inf. Sci. 2016, 345, 340–354. [Google Scholar] [CrossRef]
- Zubair, M.; Kim, J.; Yoon, C. An Automated ECG Beat Classification System Using Convolutional Neural Networks. In Proceedings of the 2016 6th International Conference on IT Convergence and Security (ICITCS), Prague, Czech Republic, 26 September 2016; pp. 1–5. [Google Scholar] [CrossRef]
- Majumdar, A.; Ward, R. Robust greedy deep dictionary learning for ECG arrhythmia classification. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 4400–4407. [Google Scholar] [CrossRef]
- Wu, Z.; Ding, X.; Zhang, G.; Xu, X.; Wang, X.; Tao, Y.; Ju, C. A Novel Features Learning Method for ECG Arrhythmias Using Deep Belief Networks. In Proceedings of the 2016 6th International Conference on Digital Home (ICDH), Guangzhou, China, 2–4 December 2016; pp. 192–196. [Google Scholar] [CrossRef]
- Baek, Y.S.; Lee, S.C.; Choi, W.; Kim, D.H. OPEN A new deep learning algorithm of 12 lead electrocardiogram for identifying atrial fibrillation during sinus rhythm. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Shi, H.; Wang, H.; Qin, C.; Zhao, L.; Liu, C. Computer Methods and Programs in Biomedicine An incremental learning system for atrial fibrillation detection based on transfer learning and active learning. Comput. Methods Programs Biomed. 2020, 187, 105219. [Google Scholar] [CrossRef]
- Wang, J. An intelligent computer-aided approach for atrial fibrillation and atrial flutter signals classification using modified bidirectional LSTM network. Inf. Sci. 2021, 574, 320–332. [Google Scholar] [CrossRef]
- Radhakrishnan, T.; Karhade, J.; Ghosh, S.K.; Muduli, P.R.; Tripathy, R.K.; Acharya, U.R. AFCNNet: Automated detection of AF using chirplet transform and deep convolutional bidirectional long short term memory network with ECG signals. Comput. Biol. Med. 2021, 137, 104783. [Google Scholar] [CrossRef]
- Xia, Y.; Wulan, N.; Wang, K.; Zhang, H. Detecting atrial fibrillation by deep convolutional neural networks. Comput. Biol. Med. 2018, 93, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Shenfield, A.; Kareem, M.; San, T.R.; Fujita, H.; Acharya, U.R. Automated detection of atrial fibrillation using long short-term memory network with RR interval signals. Comput. Biol. Med. 2018, 102, 327–335. [Google Scholar] [CrossRef] [Green Version]
- Mousavi, S.; Afghah, F.; Acharya, U.R. HAN-ECG: An interpretable atrial fibrillation detection model using hierarchical attention networks. Comput. Biol. Med. 2020, 127, 104057. [Google Scholar] [CrossRef]
- Cai, W.; Chen, Y.; Guo, J.; Han, B.; Shi, Y.; Ji, L.; Wang, J.; Zhang, G.; Luo, J. Accurate detection of atrial fibrillation from 12-lead ECG using deep neural network. Comput. Biol. Med. 2020, 116, 103378. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.; Parvaneh, S.; Rahman, A.; Conroy, B.; Babaeizadeh, S. Densely connected convolutional networks for detection of atrial fibrillation from short single-lead ECG recordings. J. Electrocardiol. 2018, 51, S18–S21. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Yao, Q.; Cai, Y.; Miao, F.; Sun, F.; Li, Y. Multiscaled Fusion of Deep Convolutional Neural Networks for Screening Atrial Fibrillation from Single Lead Short ECG Recordings. IEEE J. Biomed. Health Inform. 2018, 22, 1744–1753. [Google Scholar] [CrossRef]
- Lai, D.; Bu, Y.; Su, Y.; Zhang, X.; Ma, C.S. Non-Standardized Patch-Based ECG Lead Together with Deep Learning Based Algorithm for Automatic Screening of Atrial Fibrillation. IEEE J. Biomed. Health Inform. 2020, 24, 1569–1578. [Google Scholar] [CrossRef]
- Attia, Z.I.; Noseworthy, P.A.; Lopez-Jimenez, F.; Asirvatham, S.J.; Deshmukh, A.J.; Gersh, B.J.; Carter, R.E.; Yao, X.; Rabinstein, A.A.; Erickson, B.J.; et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: A retrospective analysis of outcome prediction. Lancet 2019, 394, 861–867. [Google Scholar] [CrossRef]
- Baalman, S.W.; Schroevers, F.E.; Oakley, A.J.; Brouwer, T.F.; van der Stuijt, W.; Bleijendaal, H.; Ramos, L.A.; Lopes, R.R.; Marquering, H.; Knops, R.E.; et al. A morphology based deep learning model for atrial fibrillation detection using single cycle electrocardiographic samples. Int. J. Cardiol. 2020, 316, 130–136. [Google Scholar] [CrossRef]
- Jo, Y.-Y.; Cho, Y.; Lee, S.Y.; Kwon, J.-M.; Kim, K.-H.; Jeon, K.-H.; Cho, S.; Park, J.; Oh, B.-H. Explainable artificial intelligence to detect atrial fibrillation using electrocardiogram. Int. J. Cardiol. 2021, 328, 104–110. [Google Scholar] [CrossRef]
- Fan, X.; Hu, Z.; Wang, R.; Yin, L.; Li, Y.; Cai, Y. A novel hybrid network of fusing rhythmic and morphological features for atrial fibrillation detection on mobile ECG signals. Neural Comput. Appl. 2020, 32, 8101–8113. [Google Scholar] [CrossRef]
- Cao, P.; Li, X.; Mao, K.; Lu, F.; Ning, G.; Fang, L.; Pan, Q. A novel data augmentation method to enhance deep neural networks for detection of atrial fibrillation. Biomed. Signal Process. Control 2020, 56, 101675. [Google Scholar] [CrossRef]
- Petmezas, G.; Haris, K.; Stefanopoulos, L.; Kilintzis, V.; Tzavelis, A.; Rogers, J.A.; Katsaggelos, A.K.; Maglaveras, N. Automated Atrial Fibrillation Detection using a Hybrid CNN-LSTM Network on Imbalanced ECG Datasets. Biomed. Signal Process. Control 2021, 63, 102194. [Google Scholar] [CrossRef]
- Nguyen, Q.H.; Nguyen, B.P.; Nguyen, T.B.; Do, T.T.T.; Mbinta, J.F.; Simpson, C.R. Stacking segment-based CNN with SVM for recognition of atrial fibrillation from single-lead ECG recordings. Biomed. Signal Process. Control 2021, 68, 102672. [Google Scholar] [CrossRef]
- Chen, X.; Cheng, Z.; Wang, S.; Lu, G.; Xv, G.; Liu, Q.; Zhu, X. Atrial fibrillation detection based on multi-feature extraction and convolutional neural network for processing ECG signals. Comput. Methods Programs Biomed. 2021, 202, 106009. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, J.; Cai, Z.; Zhang, L.; Chen, Z.; Liu, C. Over-fitting suppression training strategies for deep learning-based atrial fibrillation detection. Med. Biol. Eng. Comput. 2021, 59, 165–173. [Google Scholar] [CrossRef]
- Fujita, H.; Cimr, D. Computer Aided detection for fibrillations and flutters using deep convolutional neural network. Inf. Sci. 2019, 486, 231–239. [Google Scholar] [CrossRef]
- Zhao, Z.; Särkkä, S.; Rad, A.B. Kalman-based Spectro-Temporal ECG Analysis using Deep Convolutional Networks for Atrial Fibrillation Detection. J. Signal Process. Syst. 2020, 92, 621–636. [Google Scholar] [CrossRef]
- Tran, L.; Li, Y.; Nocera, L.; Shahabi, C.; Xiong, L. MultiFusionNet: Atrial Fibrillation Detection With Deep Neural Networks. AMIA Jt. Summits Transl. Sci. Proc. 2020, 2020, 654–663. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7233068/ (accessed on 26 October 2021).
- Jin, Y.; Qin, C.; Huang, Y.; Zhao, W.; Liu, C. Multi-domain modeling of atrial fibrillation detection with twin attentional convolutional long short-term memory neural networks. Knowl.-Based Syst. 2020, 193, 105460. [Google Scholar] [CrossRef]
- Wang, J. Automated detection of atrial fibrillation and atrial flutter in ECG signals based on convolutional and improved Elman neural network. Knowl.-Based Syst. 2020, 193, 105446. [Google Scholar] [CrossRef]
- Nurmaini, S.; Tondas, A.E.; Darmawahyuni, A.; Rachmatullah, M.N.; Partan, R.U.; Firdaus, F.; Tutuko, B.; Pratiwi, F.; Juliano, A.H.; Khoirani, R. Robust detection of atrial fibrillation from short-term electrocardiogram using convolutional neural networks. Future Gener. Comput. 2020, 113, 304–317. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.H.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhyisoNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [Green Version]
- PhysioNet, PhysioNet MIT-BIH Atrial Fibrillation Database. Available online: https://physionet.org/content/afdb/1.0.0/ (accessed on 26 October 2021).
- PhysioNet, AF Classification from a Short Single Lead ECG Recording—The PhysioNet Computing in Cardiology Challenge 2017. Available online: https://physionet.org/physiobank/database/mitdb/ (accessed on 26 October 2021).
- CUDB Website. Available online: http://www.physionet.org/physiobank/database/cudb/ (accessed on 26 October 2021).
- Wu, X.; Zheng, Y.; Chu, C.H.; He, Z. Extracting deep features from short ECG signals for early atrial fibrillation detection. Artif. Intell. Med. 2020, 109, 101896. [Google Scholar] [CrossRef]
- Wang, J.; Wang, P.; Wang, S. Automated detection of atrial fibrillation in ECG signals based on wavelet packet transform and correlation function of random process. Biomed. Signal Process. Control 2020, 55, 101662. [Google Scholar] [CrossRef]
- Luz, E.; Schwartz, W.R.; Chavez, G.C.; Menotti, D. ECG-based heartbeat classification for arrhythmia detection: A survey. Comput. Methods Programs Biomed. 2016, 127, 144–164. [Google Scholar] [CrossRef]
- Acharya, U.R.; Joseph, K.P.; Kannathal, N.; Lim, C.M.; Suri, J.S. Heart rate variability: A review. Med Biol. Eng. Comput. 2006, 44, 1031–1051. [Google Scholar] [CrossRef] [PubMed]
- Ishaque, S.; Khan, N.; Krishnan, S. Trends in Heart-Rate Variability Signal Analysis. Front. Digit. Health 2021, 3, 1–18. [Google Scholar] [CrossRef]
- Faust, O.; Kareem, M.; Shenfield, A.; Ali, A.; Acharya, U.R. Validating the robustness of an internet of things based atrial fibrillation detection system. Pattern Recognit. Lett. 2020, 133, 55–61. [Google Scholar] [CrossRef]
- Mei, Z.; Gu, X.; Chen, H.; Chen, W. Automatic atrial fibrillation detection based on heart rate variability and spectral features. IEEE Access 2018, 6, 53566–53575. [Google Scholar] [CrossRef]
- Mohebbi, M.; Ghassemian, H. Prediction of paroxysmal atrial fibrillation based on non-linear analysis and spectrum and bispectrum features of the heart rate variability signal. Comput. Methods Programs Biomed. 2012, 105, 40–49. [Google Scholar] [CrossRef]
- Narin, A.; Isler, Y.; Ozer, M.; Perc, M. Early prediction of paroxysmal atrial fibrillation based on short-term heart rate variability. Phys. A Stat. Mech. Its Appl. 2018, 509, 56–65. [Google Scholar] [CrossRef]
- Chesnokov, Y.V. Complexity and spectral analysis of the heart rate variability dynamics for distant prediction of paroxysmal atrial fibrillation with artificial intelligence methods. Artif. Intell. Med. 2008, 43, 151–165. [Google Scholar] [CrossRef]
- Hirsch, G.; Jensen, S.H.; Poulsen, E.S.; Puthusserypady, S. Atrial fibrillation detection using heart rate variability and atrial activity: A hybrid approach. Expert Syst. Appl. 2021, 169, 114452. [Google Scholar] [CrossRef]
- Ebrahimzadeh, E.; Kalantari, M.; Joulani, M.; Shahraki, R.S.; Fayaz, F.; Ahmadi, F. Prediction of paroxysmal Atrial Fibrillation: A machine learning based approach using combined feature vector and mixture of expert classification on HRV signal. Comput. Methods Programs Biomed. 2018, 165, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Boon, K.H.; Khalil-Hani, M.; Malarvili, M.B.; Sia, C.W. Paroxysmal atrial fibrillation prediction method with shorter HRV sequences. Comput. Methods Programs Biomed. 2016, 134, 187–196. [Google Scholar] [CrossRef]
- Marinucci, D.; Sbrollini, A.; Marcantoni, I.; Morettini, M.; Swenne, C.A.; Burattini, L. Artificial neural network for atrial fibrillation identification in portable devices. Sensors 2020, 20, 3570. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, B.; Yao, B.; He, W. ECG Arrhythmia Classification Using STFT-Based Spectrogram and Convolutional Neural Network. IEEE Access 2019, 7, 92871–92880. [Google Scholar] [CrossRef]
- Murat, F.; Yildirim, O.; Talo, M.; Baloglu, U.B.; Demir, Y.; Acharya, U.R. Application of deep learning techniques for heartbeats detection using ECG signals-analysis and review. Comput. Biol. Med. 2020, 120, 103726. [Google Scholar] [CrossRef] [PubMed]
- Izci, E.; Ozdemir, M.A.; Degirmenci, M.; Akan, A. Cardiac arrhythmia detection from 2d ecg images by using deep learning technique. In TIPTEKNO 2019—Tip Teknolojileri Kongresi; IEEE: Manhattan, NY, USA, 2019. [Google Scholar] [CrossRef]
- Abel Latif, A.K.; Peng, X.; Messinger-Rapport, B.J. Predictors of anticoagulation prescription in nursing home residents with atrial fibrillation. J. Am. Med Dir. Assoc. 2005, 6, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Afdala, A.; Nuryani, N. Detection of atrial fibrillation using coherency of power spectrum in electrocardiogram. In AIP Conference Proceedings; AIP: College Park, MD, USA, 2016. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Adv. Neural. Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015-Conference Track Proceedings), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef] [Green Version]
- Robinson, A.J.; Fallside, F. The Utility Driven Dynamic Error Propagation Network; IEEE: Manhattan, NY, USA, 1987. [Google Scholar]
- Werbos, P.J. Generalization of backpropagation with application to a recurrent gas market model. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [Green Version]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Gertsch, M. The ECG: A Two-Step Approach to Diagnosis; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2004. [Google Scholar]

| Database | Records | Papers |
|---|---|---|
| MIT-BIH DB | 0.5 h duration, 48 records from 47 subjects, 360 Hz sampling rate | [19,30,46,50,54,56,60] |
| MIT-BIH AFDB | 10 h duration, 25 records, 250 Hz sampling rate | [19,30,40,41,42,52,55,56,59,60,61] |
| PhysioNet/CinC 2017 | 8528 single-lead ECG, 300 Hz | [42,44,45,49,50,51,53,55,57,58] |
| MIT-BIH VFDB | 0.5 h duration, 22 records | [56] |
| CU VTDB | 8 min, 35 records, 250 Hz sampling rate | [30] |
| Others | Details in individual papers | [43,46,47,48,49,54,55,61] |
| Author, Year | Purpose | Classifier | Input | Performance (%) | ||
|---|---|---|---|---|---|---|
| Spec. | Sen. | Acc. | ||||
| Faust et al., 2018 [41] | AF detection | Bidirectional LSTM | 23 subjects | 99.61 | 99.87 | 99.77 |
| Mei et al., 2018 [72] | AF detection | SVM + BT | 8528 single-lead ECG | 98.6 | 83.2 | 96.6 |
| Mohebbi et al., 2012 [73] | PAF prediction | SVM | 30-min ECG | 93.10 | 96.30 | - |
| Narin et al., 2018 [74] | PAF prediction | KNN | 5-min ECG | 88 | 92 | 90 |
| Chesnokov, 2008 [75] | PAF prediction | SVM | 30-min segments | 93 | 76 | - |
| Hirsch et al., 2021 [76] | AF detection | BoT, RF, LDA | 30-beat window | 96.1 | 95.9 | 97.4 |
| Ebrahimzadeh et al., 2018 [77] | PAF prediction | MLP, KNN, SVM | 5-min ECG | 95.55 | 100 | 98.21 |
| Boon et al., 2016 [78] | PAF prediction | SVM | 30-min ECG | 79.3 | 81.1 | 80.2 |
| Marinnucci et al., 2020 [79] | AF identification | ANN | 8244 ECG | 75.0 | 88.7 | - |
| Deep Models | Related Publications | Advantage/Disadvantage |
|---|---|---|
| DNN | [43] | In terms of speed, it is more advantageous. |
| CNN | [30,40,44,45,46,47,49,50,53,54,56,57,60,61] | Strong in obtaining representative properties, but lacking in design difficulties and parameter tuning. |
| RNN | [42,48] | Although it is used because of its memory structure, it is poor at representing sequences. |
| LSTM | [41,51] | Although useful for sequence representations, it is slow and consumes a lot of resources. |
| Hybrid (CNN+LSTM) | [19,52,55,58,59] | The use of both representation and sequence features together is advantageous, but it takes more time and cost. |
| Authors, Year | Number of Subjects | Leads | Classes | Database | Method | Performance (%) | ||
|---|---|---|---|---|---|---|---|---|
| Spec. | Sen. | Acc. | ||||||
| Acharya et al., 2017 [30] | 21,709 2 s ECG segments 8683 5 s ECG segments | Lead II | SR, AF, AFL and VF | MIT-BIH DB, MIT-BIH AFDB, CU VTDB | 11-layer CNN | 93.13 81.44 | 98.09 99.13 | 92.50 94.90 |
| Xia et al., 2018 [40] | 162,536 5 s ECG segments | 2 Lead | AF and non-AF | MIT-BIH AFDB | STFT (RGB) + CNN STFT (grayscale) + CNN SWT + CNN | 98.24 97.17 97.87 | 98.34 98.60 98.79 | 98.29 97.74 98.63 |
| Faust et al., 2018 [41] | CV: 20 subjects BV: 3 subjects | - | SR and AF | MIT-BIH AFDB | HRV + bidirectional LSTM | 98.67 99.61 | 98.32 99.87 | 98.51 99.77 |
| Fan et al., 2018 [45] | 5154 SR recordings 7713 AF recordings | Single Lead | SR and AF, AF and O | PhysioNet/CinC 2017 | MS-CNN | 98.77 98.84 | 93.77 80.26 | 98.13 97.19 |
| Andersen et al., 2019 [19] | 23 long-term recordings 48 short-term recordings 18 long-term recordings | Single Lead | SR and AF | MIT-BIH AFDB, MIT-BIH DB, MIT-BIH SRDB | CNN + LSTM | 96.95 86.04 95.01 | 98.98 98.96 - | 97.80 87.40 - |
| Fujita et al., 2019 [56] | 25,287 2 s ECG segments | Single Lead | SR, AF, AFL and VF | MIT-BIH DB, MIT-BIH AFDB, MIT-BIH VFDB | 8-layer CNN | 96.07 | 99.43 | 98.61 |
| Attia et al., 2019 [47] | 649,931 10 s ECG recordings | 12 Lead | SR and AF (includes AFL) | Mayo Clinic ECG Laboratory | CNN | 83.4 | 82.3 | 83.3 |
| Baalman et al., 2020 [48] | 1499 10 s ECG recordings | Lead II, 8 Lead | SR and AF | AFACT | R-centered SC-ECG + RNN R-to-R-wave SC-ECG + RNN | - | - | 94.00 96.00 |
| Cai et al., 2020 [43] | 16,557 10 s ECG recordings | 12 Lead | SR and AF AF and non-AF SR, AF and O | Chinese PLA General Hospital Wearable 12-Lead, The China Physiological Signal 2018 | DDNN | 99.19 97.04 95.85 | 99.44 98.63 98.38 | 99.35 98.21 97.74 |
| Lai et al., 2020 [46] | 510,472 10 s ECG recordings | Multi Lead | AF and non-AF | Hexin Patch Lead II, MIT-BIH DB | 8-layer CNN | 93.4 | 93.1 | 93.1 |
| Jin et al., 2020 [59] | 150,060 5 s ECG recordings | - | AF and non-AF | MIT-BIH AFDB | Multi-domain feature + TAC-LSTM | 98.76 | 98.14 | 98.51 |
| Wang et al., 2020 [60] | 22,174 ECG segments 1265 ECG segments | Single Lead | SR, AF and AFL | MIT-BIH AFDB, MIT-BIH DB | CNN + MLP CNN + ENN CNN + IENN | 99.3 99.6 | 97.1 99.3 | 98.3 99.4 |
| Nurmaini et al., 2020 [61] | 6114 samples (9 s) | Single Lead | SR and AF SR, AF and non-AF | PhysioNet AFDB, MIT-BIH AFDB, MIT-BIH Malignant Ventricular Entropy, An Indonesian Hospital | 13-layer one-dimensional CNN | 99.91 99.17 | 99.91 98.90 | 99.98 99.17 |
| Mousavi et al., 2020 [42] | 167,422 5 s ECG recordings 8528 ECG recordings | Single Lead | AF and non-AF SR and AF | MIT-BIH AFDB, PhysioNet/CinC 2017 | BiRNN (HAN-ECG) | 98.54 | 99.08 | 98.81 |
| Chen et al., 2021 [54] | - | 2 Lead 12 Lead | SR and AF | MIT-BIH DB, AHA DB, QT DB, CSE DB | Multiple feature extraction + CNN | - | - | 98.92 |
| Petmezas et al., 2021 [52] | 970,009 beats | 2 Lead | SR, AF, AFL and J | MIT-BIH AFDB | CNN + LSTM + FL | 99.29 | 97.87 | - |
| Jo et al., 2021 [49] | - | 12 lead, 6 Lead, Single Lead | AF and non-AF | Sejong ECG DB, PTB-XL ECG DB, Charman et al. ECG DB, PhysioNet DB | CNN | 99.5 | 99.9 | 99.6 |
| Zhang et al., 2021 [55] | 80,000 ECG segments 83,464 ECG segments 19,220 ECG segments | Lead I | AF and non-AF | Wearable Lead I-II, MIT-BIH AFDB, PhysioNet/CinC 2017 | LSTM + CNN | 95.19 94.49 96.66 | 97.73 96.46 92.09 | 95.44 95.28 96.23 |
| Authors, Year | Classes | Method | F1N | F1A | F1O | F1 |
|---|---|---|---|---|---|---|
| Rubin et al., 2018 [44] | SR, AF, O and N | SQA + DCNN | 0.91 | 0.83 | 0.72 | 0.82 |
| Fan et al., 2020 [50] | SR, AF and O | FRM-CNN | 0.93 | 0.88 | 0.74 | 0.85 |
| Zhao et al., 2020 [57] | SR, AF, O and N | Kalman filter + DCNN | 0.89 | 0.79 | 0.72 | 0.80 |
| Tran et al., 2020 [58] | SR, AF, O and N | CNN + LSTM | 0.90 | 0.83 | 0.75 | 0.80 |
| Cao et al., 2020 [51] | SR, AF, O and N | 2-layer LSTM | 0.91 | 0.84 | 0.70 | 0.82 |
| Nguyen et al., 2021 [53] | SR, AF, O and N | Stacking CNN + SVM | 0.93 | 0.78 | 0.79 | 0.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murat, F.; Sadak, F.; Yildirim, O.; Talo, M.; Murat, E.; Karabatak, M.; Demir, Y.; Tan, R.-S.; Acharya, U.R. Review of Deep Learning-Based Atrial Fibrillation Detection Studies. Int. J. Environ. Res. Public Health 2021, 18, 11302. https://doi.org/10.3390/ijerph182111302
Murat F, Sadak F, Yildirim O, Talo M, Murat E, Karabatak M, Demir Y, Tan R-S, Acharya UR. Review of Deep Learning-Based Atrial Fibrillation Detection Studies. International Journal of Environmental Research and Public Health. 2021; 18(21):11302. https://doi.org/10.3390/ijerph182111302
Chicago/Turabian StyleMurat, Fatma, Ferhat Sadak, Ozal Yildirim, Muhammed Talo, Ender Murat, Murat Karabatak, Yakup Demir, Ru-San Tan, and U. Rajendra Acharya. 2021. "Review of Deep Learning-Based Atrial Fibrillation Detection Studies" International Journal of Environmental Research and Public Health 18, no. 21: 11302. https://doi.org/10.3390/ijerph182111302
APA StyleMurat, F., Sadak, F., Yildirim, O., Talo, M., Murat, E., Karabatak, M., Demir, Y., Tan, R.-S., & Acharya, U. R. (2021). Review of Deep Learning-Based Atrial Fibrillation Detection Studies. International Journal of Environmental Research and Public Health, 18(21), 11302. https://doi.org/10.3390/ijerph182111302








