Artificial Intelligence and Computer Vision in Low Back Pain: A Systematic Review
Abstract
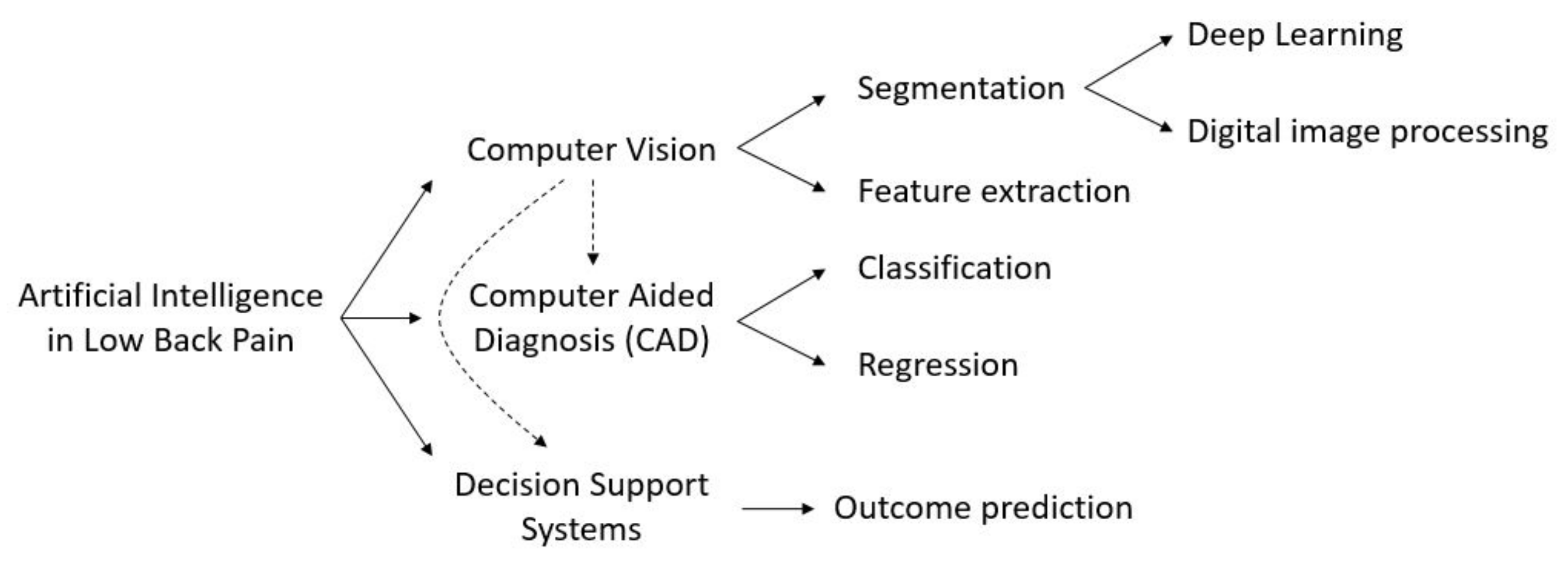
:1. Introduction
2. Materials and Methods
2.1. Inclusion and Exclusion Criteria
- •
- Chronic LBP or lumbar diseases must have been among the main topics of the article. We included works on the prevention, diagnosis or treatment of chronic LBP and treating at least one of the structures involved in LBP (i.e., vertebrae, discs, muscles);
- •
- AI must have been used in the work with application to clinical images. We included articles exploiting AI methods falling in the areas of computer vision, machine learning and artificial Neural Networks (NNs);
- •
- Subjects of the study: all the articles must have been based on studies of human low back and related pathology, regardless of the age or employment of the subjects included in the study;
- •
- Language: all articles must have been written in English.
- •
- A different medical problem was considered: we excluded articles which did not consider chronic LBP and its related physical structures and medical data. For example, we excluded studies that considered only cervical or thoracic vertebrae, or that focused on osteoporosis, metastases, traumatic LBP, and other causes of non-discogenic LBP;
- •
- AI was not considered: some articles in the search results proposed definitions and practice for LBP based only on medical observation without utilization of AI;
- •
- Computer vision and clinical images were not considered in the study, regardless of whether AI was utilized for developing diagnosis or support systems;
- •
- Animal studies: we excluded studies based on vertebral structures of animals;
- •
- Embryonal studies: we excluded studies performed on embryos and concerning the embryogenesis of spinal structures.
2.2. Evaluation Metrics
3. Quality of Evidence
4. Results
4.1. Feature Extraction
- •
- six articles on MRI (1 of which considers 3D MRI);
- •
- one article on 3D images of the back surface;
- •
- one article on X-ray imaging.
4.2. Segmentation
4.2.1. Digital Image Processing
- •
- 15 articles on MRI (2 of which considered 3D MRI);
- •
- 15 articles on CT images;
- •
- 1 articles on both MRI and CT images;
- •
- 3 articles on fluoroscopic images;
- •
- 2 articles on ultrasound images;
- •
- 2 articles on X-ray images.
4.2.2. Deep Learning
- •
- 13 articles on MRI (2 of which considered 3D MRI and 1 with the addition of clinical notes);
- •
- 5 articles on CT images;
- •
- 4 articles on X-ray images (1 of which in combination with Moire images);
- •
- 1 article on ultrasound images.
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Galbusera, F.; Casaroli, G.; Bassani, T. Artificial intelligence and machine learning in spine research. JOR Spine 2019, 2, e1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, H.S.; Shan, H.; Dahoun, T.; Vogel, H.; Yuan, S. Advancing drug discovery via artificial intelligence. Trends Pharmacol. Sci. 2019, 40, 592–604. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.B.M.A.; Chow, E.K.H. Artificial intelligence-driven designer drug combinations: From drug development to personalized medicine. SLAS Technol. Transl. Life Sci. Innov. 2019, 24, 124–125. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Bur, A.M.; Shew, M.; New, J. Artificial intelligence for the otolaryngologist: A state of the art review. Otolaryngol.-Neck Surg. 2019, 160, 603–611. [Google Scholar] [CrossRef]
- Boon, I.S.; Au Yong, T.; Boon, C.S. Assessing the role of artificial intelligence (AI) in clinical oncology: Utility of machine learning in radiotherapy target volume delineation. Medicines 2018, 5, 131. [Google Scholar] [CrossRef] [Green Version]
- Loftus, T.J.; Tighe, P.J.; Filiberto, A.C.; Efron, P.A.; Brakenridge, S.C.; Mohr, A.M.; Rashidi, P.; Upchurch, G.R.; Bihorac, A. Artificial intelligence and surgical decision-making. JAMA Surg. 2020, 155, 148–158. [Google Scholar] [CrossRef]
- Yang, T.; Li, R.; Liang, N.; Li, J.; Yang, Y.; Huang, Q.; Li, Y.; Cao, W.; Wang, Q.; Zhang, H. The application of key feature extraction algorithm based on Gabor wavelet transformation in the diagnosis of lumbar intervertebral disc degenerative changes. PLoS ONE 2020, 15, e0227894. [Google Scholar] [CrossRef] [Green Version]
- Thong, W.; Parent, S.; Wu, J.; Aubin, C.E.; Labelle, H.; Kadoury, S. Three-dimensional morphology study of surgical adolescent idiopathic scoliosis patient from encoded geometric models. Eur. Spine J. 2016, 25, 3104–3113. [Google Scholar] [CrossRef]
- Garcia-Cano, E.; Cosío, F.A.; Duong, L.; Bellefleur, C.; Roy-Beaudry, M.; Joncas, J.; Parent, S.; Labelle, H. Prediction of spinal curve progression in adolescent idiopathic scoliosis using random forest regression. Comput. Biol. Med. 2018, 103, 34–43. [Google Scholar] [CrossRef]
- Franklin, G.M.; Wickizer, T.M.; Coe, N.B.; Fulton-Kehoe, D. Workers’ compensation: Poor quality health care and the growing disability problem in the United States. Am. J. Ind. Med. 2015, 58, 245–251. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Tagliaferri, S.D.; Angelova, M.; Zhao, X.; Owen, P.J.; Miller, C.T.; Wilkin, T.; Belavy, D.L. Artificial intelligence to improve back pain outcomes and lessons learnt from clinical classification approaches: Three systematic reviews. NPJ Digit. Med. 2020, 3, 1–16. [Google Scholar] [CrossRef]
- Tack, C. Artificial intelligence and machine learning| applications in musculoskeletal physiotherapy. Musculoskelet. Sci. Pract. 2019, 39, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Azimi, P.; Yazdanian, T.; Benzel, E.C.; Aghaei, H.N.; Azhari, S.; Sadeghi, S.; Montazeri, A. A Review on the Use of Artificial Intelligence in Spinal Diseases. Asian Spine J. 2020, 14, 543. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Burns, J.E.; Forsberg, D.; Seitel, A.; Rasoulian, A.; Abolmaesumi, P.; Hammernik, K.; Urschler, M.; Ibragimov, B.; Korez, R.; et al. A multi-center milestone study of clinical vertebral CT segmentation. Comput. Med. Imaging Graph. 2016, 49, 16–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belasso, C.J.; Behboodi, B.; Benali, H.; Boily, M.; Rivaz, H.; Fortin, M. LUMINOUS database: Lumbar multifidus muscle segmentation from ultrasound images. BMC Musculoskelet. Disord. 2020, 21, 1–11. [Google Scholar] [CrossRef]
- Burian, E.; Rohrmeier, A.; Schlaeger, S.; Dieckmeyer, M.; Diefenbach, M.N.; Syväri, J.; Klupp, E.; Weidlich, D.; Zimmer, C.; Rummeny, E.J.; et al. Lumbar muscle and vertebral bodies segmentation of chemical shift encoding-based water-fat MRI: The reference database myosegmentum spine. BMC Musculoskelet. Disord. 2019, 20, 1–7. [Google Scholar] [CrossRef]
- Adankon, M.M.; Dansereau, J.; Labelle, H.; Cheriet, F. Non invasive classification system of scoliosis curve types using least-squares support vector machines. Artif. Intell. Med. 2012, 56, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Castro-Mateos, I.; Pozo, J.M.; Eltes, P.E.; Del Rio, L.; Lazary, A.; Frangi, A.F. 3D segmentation of annulus fibrosus and nucleus pulposus from T2-weighted magnetic resonance images. Phys. Med. Biol. 2014, 59, 7847. [Google Scholar] [CrossRef]
- Raudner, M.; Schreiner, M.M.; Hilbert, T.; Kober, T.; Weber, M.; Szelényi, A.; Windhager, R.; Juras, V.; Trattnig, S. Clinical implementation of accelerated T 2 mapping: Quantitative magnetic resonance imaging as a biomarker for annular tear and lumbar disc herniation. Eur. Radiol. 2020, 31, 3590–3599. [Google Scholar] [CrossRef] [PubMed]
- Abdollah, V.; Parent, E.C.; Dolatabadi, S.; Marr, E.; Croutze, R.; Wachowicz, K.; Kawchuk, G. Texture Analysis in the Classification of T2 Weighted Magnetic Resonance Images in Persons with and without Low Back Pain. J. Orthop. Res.® 2020, 39, 2187–2196. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-España, S.; Arana, E.; Moratal, D. Semiautomatic computer-aided classification of degenerative lumbar spine disease in magnetic resonance imaging. Comput. Biol. Med. 2015, 62, 196–205. [Google Scholar] [CrossRef]
- Ketola, J.H.; Inkinen, S.I.; Karppinen, J.; Niinimäki, J.; Tervonen, O.; Nieminen, M.T. T 2-weighted magnetic resonance imaging texture as predictor of low back pain: A texture analysis-based classification pipeline to symptomatic and asymptomatic cases. J. Orthop. Res.® 2020. [Google Scholar] [CrossRef]
- Haq, R.; Schmid, J.; Borgie, R.; Cates, J.; Audette, M.A. Deformable multisurface segmentation of the spine for orthopedic surgery planning and simulation. J. Med. Imaging 2020, 7, 015002. [Google Scholar] [CrossRef]
- Haq, R.; Aras, R.; Besachio, D.A.; Borgie, R.C.; Audette, M.A. 3D lumbar spine intervertebral disc segmentation and compression simulation from MRI using shape-aware models. Int. J. Comput. Assist. Radiol. Surg. 2015, 10, 45–54. [Google Scholar] [CrossRef]
- Neubert, A.; Fripp, J.; Engstrom, C.; Schwarz, R.; Lauer, L.; Salvado, O.; Crozier, S. Automated detection, 3D segmentation and analysis of high resolution spine MR images using statistical shape models. Phys. Med. Biol. 2012, 57, 8357. [Google Scholar] [CrossRef]
- Li, Y.; Liang, W.; Zhang, Y.; Tan, J. Automatic global level set approach for lumbar vertebrae CT image segmentation. BioMed Res. Int. 2018, 2018, 6319879. [Google Scholar] [CrossRef]
- Ibragimov, B.; Korez, R.; Likar, B.; Pernuš, F.; Xing, L.; Vrtovec, T. Segmentation of pathological structures by landmark-assisted deformable models. IEEE Trans. Med. Imaging 2017, 36, 1457–1469. [Google Scholar] [CrossRef]
- Yu, W.; Liu, W.; Tan, L.; Zhang, S.; Zheng, G. Multi-object Model-Based Multi-atlas Segmentation Constrained Grid Cut for Automatic Segmentation of Lumbar Vertebrae from CT Images. In Intelligent Orthopaedics; Springer: Singapore, 2018; pp. 65–71. [Google Scholar] [CrossRef]
- Korez, R.; Ibragimov, B.; Likar, B.; Pernuš, F.; Vrtovec, T. A framework for automated spine and vertebrae interpolation-based detection and model-based segmentation. IEEE Trans. Med. Imaging 2015, 34, 1649–1662. [Google Scholar] [CrossRef] [PubMed]
- Al-Helo, S.; Raja’S, A.; Chaudhary, V.; Al-Zoubi, M. Segmentation of lumbar vertebrae from clinical CT using active shape models and GVF-snake. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 8033–8036. [Google Scholar] [CrossRef]
- Ruiz-España, S.; Díaz-Parra, A.; Arana, E.; Moratal, D. A fully automated level-set based segmentation method of thoracic and lumbar vertebral bodies in Computed Tomography images. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 3049–3052. [Google Scholar] [CrossRef]
- Huang, J.; Jian, F.; Wu, H.; Li, H. An improved level set method for vertebra CT image segmentation. Biomed. Eng. Online 2013, 12, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Mahdy, L.N.; Ezzat, K.A.; Hassanien, A.E. Automatic detection System for Degenerative Disk and simulation for artificial disc replacement surgery in the spine. ISA Trans. 2018, 81, 244–258. [Google Scholar] [CrossRef]
- Courbot, J.B.; Rust, E.; Monfrini, E.; Collet, C. Vertebra segmentation based on two-step refinement. J. Comput. Surg. 2016, 4, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Rasoulian, A.; Rohling, R.; Abolmaesumi, P. Lumbar spine segmentation using a statistical multi-vertebrae anatomical shape+ pose model. IEEE Trans. Med. Imaging 2013, 32, 1890–1900. [Google Scholar] [CrossRef]
- Mastmeyer, A.; Engelke, K.; Fuchs, C.; Kalender, W.A. A hierarchical 3D segmentation method and the definition of vertebral body coordinate systems for QCT of the lumbar spine. Med. Image Anal. 2006, 10, 560–577. [Google Scholar] [CrossRef]
- Jimenez-Pastor, A.; Alberich-Bayarri, A.; Fos-Guarinos, B.; Garcia-Castro, F.; Garcia-Juan, D.; Glocker, B.; Marti-Bonmati, L. Automated vertebrae localization and identification by decision forests and image-based refinement on real-world CT data. La Radiol. Medica 2020, 125, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, S.; Kim, Y.S.; Chung, W.K. Automated segmentation of the lumbar pedicle in CT images for spinal fusion surgery. IEEE Trans. Biomed. Eng. 2011, 58, 2051–2063. [Google Scholar] [CrossRef]
- Klinder, T.; Ostermann, J.; Ehm, M.; Franz, A.; Kneser, R.; Lorenz, C. Automated model-based vertebra detection, identification, and segmentation in CT images. Med. Image Anal. 2009, 13, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Štern, D.; Likar, B.; Pernuš, F.; Vrtovec, T. Automated detection of spinal centrelines, vertebral bodies and intervertebral discs in CT and MR images of lumbar spine. Phys. Med. Biol. 2009, 55, 247. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Mishra, A.; Fieguth, P.; Clausi, D.; Dunk, N.M.; Callaghan, J.P. Shape-guided active contour based segmentation and tracking of lumbar vertebrae in video fluoroscopy using complex wavelets. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; pp. 863–866. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.; Nolte, L.P.; Ferguson, S.J. Scaled, patient-specific 3D vertebral model reconstruction based on 2D lateral fluoroscopy. Int. J. Comput. Assist. Radiol. Surg. 2011, 6, 351–366. [Google Scholar] [CrossRef]
- Michopoulou, S.K.; Costaridou, L.; Panagiotopoulos, E.; Speller, R.; Panayiotakis, G.; Todd-Pokropek, A. Atlas-based segmentation of degenerated lumbar intervertebral discs from MR images of the spine. IEEE Trans. Biomed. Eng. 2009, 56, 2225–2231. [Google Scholar] [CrossRef] [PubMed]
- Fallah, F.; Walter, S.S.; Bamberg, F.; Yang, B. Simultaneous volumetric segmentation of vertebral bodies and intervertebral discs on fat-water MR images. IEEE J. Biomed. Health Inform. 2018, 23, 1692–1701. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Chaudhary, V. Supervised methods for detection and segmentation of tissues in clinical lumbar MRI. Comput. Med. Imaging Graph. 2014, 38, 639–649. [Google Scholar] [CrossRef]
- Kim, S.; Bae, W.C.; Masuda, K.; Chung, C.B.; Hwang, D. Semi-automatic segmentation of vertebral bodies in MR images of human lumbar spines. Appl. Sci. 2018, 8, 1586. [Google Scholar] [CrossRef] [Green Version]
- Gaonkar, B.; Xia, Y.; Villaroman, D.S.; Ko, A.; Attiah, M.; Beckett, J.S.; Macyszyn, L. Multi-parameter ensemble learning for automated vertebral body segmentation in heterogeneously acquired clinical MR images. IEEE J. Transl. Eng. Health Med. 2017, 5, 1–12. [Google Scholar] [CrossRef]
- Gaweł, D.; Główka, P.; Kotwicki, T.; Nowak, M. Automatic spine tissue segmentation from MRI data based on cascade of boosted classifiers and active appearance model. BioMed Res. Int. 2018, 2018, 7952946. [Google Scholar] [CrossRef]
- Engstrom, C.M.; Fripp, J.; Jurcak, V.; Walker, D.G.; Salvado, O.; Crozier, S. Segmentation of the quadratus lumborum muscle using statistical shape modeling. J. Magn. Reson. Imaging 2011, 33, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Baum, T.; Lorenz, C.; Buerger, C.; Freitag, F.; Dieckmeyer, M.; Eggers, H.; Zimmer, C.; Karampinos, D.C.; Kirschke, J.S. Automated assessment of paraspinal muscle fat composition based on the segmentation of chemical shift encoding-based water/fat-separated images. Eur. Radiol. Exp. 2018, 2, 1–5. [Google Scholar] [CrossRef]
- Zheng, Y.; Nixon, M.S.; Allen, R. Automated segmentation of lumbar vertebrae in digital videofluoroscopic images. IEEE Trans. Med. Imaging 2004, 23, 45–52. [Google Scholar] [CrossRef]
- Jurcak, V.; Fripp, J.; Engstrom, C.; Walker, D.; Salvado, O.; Ourselin, S.; Crozier, S. Automated segmentation of the quadratus lumborum muscle from magnetic resonance images using a hybrid atlas based-geodesic active contour scheme. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; pp. 867–870. [Google Scholar] [CrossRef]
- Fortin, M.; Omidyeganeh, M.; Battié, M.C.; Ahmad, O.; Rivaz, H. Evaluation of an automated thresholding algorithm for the quantification of paraspinal muscle composition from MRI images. Biomed. Eng. Online 2017, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Neubert, A.; Fripp, J.; Engstrom, C.; Walker, D.; Weber, M.; Schwarz, R.; Crozier, S. Three-dimensional morphological and signal intensity features for detection of intervertebral disc degeneration from magnetic resonance images. J. Am. Med. Inform. Assoc. 2013, 20, 1082–1090. [Google Scholar] [CrossRef] [Green Version]
- Oktay, A.B.; Akgul, Y.S. Localization of the Lumbar Discs Using Machine Learning and Exact Probabilistic Inference. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2011; Fichtinger, G., Martel, A., Peters, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 158–165. [Google Scholar] [CrossRef] [Green Version]
- Castro-Mateos, I.; Hua, R.; Pozo, J.M.; Lazary, A.; Frangi, A.F. Intervertebral disc classification by its degree of degeneration from T2-weighted magnetic resonance images. Eur. Spine J. 2016, 25, 2721–2727. [Google Scholar] [CrossRef]
- Kim, K.B.; Park, H.J.; Song, D.H. Automatic Characterizations of Lumbar Multifidus Muscle and Intramuscular Fat with Fuzzy C-means based Quantization from Ultrasound Images. Curr. Med. Imaging 2020, 16, 592–600. [Google Scholar] [CrossRef]
- Lui, D.; Scharfenberger, C.; De Carvalho, D.E.; Callaghan, J.P.; Wong, A. Semi-automatic Fisher-Tippett guided active contour for lumbar multifidus muscle segmentation. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 5530–5533. [Google Scholar] [CrossRef]
- Ribeiro, E.A.; Nogueira-Barbosa, M.H.; Rangayyan, R.M.; Azevedo-Marques, P.M. Detection of vertebral plateaus in lateral lumbar spinal X-ray images with Gabor filters. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 4052–4055. [Google Scholar] [CrossRef]
- Sa, R.; Owens, W.; Wiegand, R.; Chaudhary, V. Fast scale-invariant lateral lumbar vertebrae detection and segmentation in X-ray images. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 1054–1057. [Google Scholar] [CrossRef]
- Iriondo, C.; Pedoia, V.; Majumdar, S. Lumbar intervertebral disc characterization through quantitative MRI analysis: An automatic voxel-based relaxometry approach. Magn. Reson. Med. 2020, 84, 1376–1390. [Google Scholar] [CrossRef] [PubMed]
- Staartjes, V.E.; Seevinck, P.R.; Vandertop, W.P.; van Stralen, M.; Schröder, M.L. Magnetic resonance imaging–based synthetic computed tomography of the lumbar spine for surgical planning: A clinical proof-of-concept. Neurosurg. Focus 2021, 50, E13. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Han, I.H.; Kim, D.H.; Yu, S.; Lee, I.S.; Song, Y.S.; Joo, S.; Jin, C.B.; Kim, H. Spine computed tomography to magnetic resonance image synthesis using generative adversarial networks: A preliminary study. J. Korean Neurosurg. Soc. 2020, 63, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, G.; Liu, H.; Wang, D.; Feng, C.; Li, Y.; Yin, B.; Zhou, Z.; Gu, X.; Zhang, H.; Lu, Y.; et al. Deep learning-based lumbosacral reconstruction for difficulty prediction of percutaneous endoscopic transforaminal discectomy at L5/S1 level: A retrospective cohort study. Int. J. Surg. 2020, 82, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Malinda, V.; Lee, D. Lumbar Vertebrae Synthetic Segmentation in Computed Tomography Images Using Hybrid Deep Generative Adversarial Networks. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1327–1330. [Google Scholar] [CrossRef]
- Siemionow, K.; Luciano, C.; Forsthoefel, C.; Aydogmus, S. Autonomous image segmentation and identification of anatomical landmarks from lumbar spine intraoperative computed tomography scans using machine learning: A validation study. J. Craniovertebral Junction Spine 2020, 11, 99. [Google Scholar] [CrossRef] [PubMed]
- Netherton, T.J.; Rhee, D.J.; Cardenas, C.E.; Chung, C.; Klopp, A.H.; Peterson, C.B.; Howell, R.M.; Balter, P.A.; Court, L.E. Evaluation of a multiview architecture for automatic vertebral labeling of palliative radiotherapy simulation CT images. Med. Phys. 2020, 47, 5592. [Google Scholar] [CrossRef]
- Watanabe, K.; Aoki, Y.; Matsumoto, M. An application of artificial intelligence to diagnostic imaging of spine disease: Estimating spinal alignment from moire images. Neurospine 2019, 16, 697. [Google Scholar] [CrossRef]
- Kim, S.; Bae, W.C.; Masuda, K.; Chung, C.B.; Hwang, D. Fine-grain segmentation of the intervertebral discs from MR spine images using deep convolutional neural networks: BSU-Net. Appl. Sci. 2018, 8, 1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Huang, J.; Zheng, Q.; Zhu, Z.; Lv, X.; Liu, Y.; Wang, Y. A Deep-Learning–Based, Fully Automated Program to Segment and Quantify Major Spinal Components on Axial Lumbar Spine Magnetic Resonance Imaging. Phys. Ther. 2021, 101, pzab041. [Google Scholar] [CrossRef]
- Gaonkar, B.; Villaroman, D.; Beckett, J.; Ahn, C.; Attiah, M.; Babayan, D.; Villablanca, J.; Salamon, N.; Bui, A.; Macyszyn, L. Quantitative Analysis of Spinal Canal Areas in the Lumbar Spine: An Imaging Informatics and Machine Learning Study. Am. J. Neuroradiol. 2019, 40, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Shen, H.; Wu, J.; Hu, X.; Zhu, Z.; Lv, X.; Liu, Y.; Wang, Y. Spine Explorer: A deep learning based fully automated program for efficient and reliable quantifications of the vertebrae and discs on sagittal lumbar spine MR images. Spine J. 2020, 20, 590–599. [Google Scholar] [CrossRef]
- Li, H.; Luo, H.; Huan, W.; Shi, Z.; Yan, C.; Wang, L.; Mu, Y.; Liu, Y. Automatic lumbar spinal MRI image segmentation with a multi-scale attention network. Neural Comput. Appl. 2021, 1–14. [Google Scholar] [CrossRef]
- Li, H.; Luo, H.; Liu, Y. Paraspinal muscle segmentation based on deep neural network. Sensors 2019, 19, 2650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Damasceno, P.F.; Chachad, R.; Cheung, J.R.; Ballatori, A.; Lotz, J.C.; Lazar, A.A.; Link, T.M.; Fields, A.J.; Krug, R. Automatic Vertebral Body Segmentation Based on Deep Learning of Dixon Images for Bone Marrow Fat Fraction Quantification. Front. Endocrinol. 2020, 11, 612. [Google Scholar] [CrossRef]
- Jamaludin, A.; Lootus, M.; Kadir, T.; Zisserman, A.; Urban, J.; Battié, M.C.; Fairbank, J.; McCall, I. ISSLS PRIZE IN BIOENGINEERING SCIENCE 2017: Automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur. Spine J. 2017, 26, 1374–1383. [Google Scholar] [CrossRef] [Green Version]
- Natalia, F.; Meidia, H.; Afriliana, N.; Young, J.C.; Yunus, R.E.; Al-Jumaily, M.; Al-Kafri, A.; Sudirman, S. Automated measurement of anteroposterior diameter and foraminal widths in MRI images for lumbar spinal stenosis diagnosis. PLoS ONE 2020, 15, e0241309. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Y.; Chen, Q.; Gu, G.; Sui, X. Automatic lumbar MRI detection and identification based on deep learning. J. Digit. Imaging 2019, 32, 513–520. [Google Scholar] [CrossRef]
- Forsberg, D.; Sjöblom, E.; Sunshine, J.L. Detection and labeling of vertebrae in MR images using deep learning with clinical annotations as training data. J. Digit. Imaging 2017, 30, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Baka, N.; Leenstra, S.; van Walsum, T. Ultrasound aided vertebral level localization for lumbar surgery. IEEE Trans. Med. Imaging 2017, 36, 2138–2147. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.H.; Kaji, D.; Cheung, Z.B.; Ye, I.B.; Tang, R.; Ahn, A.; Carrillo, O.; Schwartz, J.T.; Valliani, A.A.; Oermann, E.K.; et al. Automated measurement of lumbar lordosis on radiographs using machine learning and computer vision. Glob. Spine J. 2020, 10, 611–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Liang, W.; Zhang, Y.; An, H.; Tan, J. Automatic lumbar vertebrae detection based on feature fusion deep learning for partial occluded C-arm X-ray images. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 647–650. [Google Scholar] [CrossRef]
- Sa, R.; Owens, W.; Wiegand, R.; Studin, M.; Capoferri, D.; Barooha, K.; Greaux, A.; Rattray, R.; Hutton, A.; Cintineo, J.; et al. Intervertebral disc detection in X-ray images using faster R-CNN. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 564–567. [Google Scholar] [CrossRef]
- Liu, X.; Deng, Z.; Yang, Y. Recent progress in semantic image segmentation. Artif. Intell. Rev. 2019, 52, 1089–1106. [Google Scholar] [CrossRef] [Green Version]



| Author/Year | Main Task | Data Type | # Patients | Structures Involved | Results | Model |
|---|---|---|---|---|---|---|
| Adankon, 2012 [19] | Feature Extraction and Classification | 3D image of the back surface | 165 | Vertebrae | Acc = 95% | Local Geometric Descriptors and SVM |
| Castro-Mateos, 2014 [20] | Feature Extraction and Segmentation | 3D MRI | 59 | Discs | DICE = 88.4% | Statistical shape model space and B-Spline space |
| Raudner, 2020 [21] | Feature Extraction | MRI | 58 | Discs | / | GRAPPATINI |
| Abdollah, 2020 [22] | Feature Extraction | MRI | 28 | Discs, Vertebrae | / | Random Forest and texture analysis |
| Yang 2020 [8] | Feature Extraction and Classification | MRI | 109 | Discs | Acc = 88.3% | Gabor wavelet transformation and KLT feature tracker |
| Ruiz-España, 2015 [23] | Feature Extraction and Classification | MRI | 67 | Discs | Acc > 90% | Gradient Vector Flow, several ML models |
| Ketola, 2020 [24] | Feature Extraction and Classification | MRI | 518 | LBP | Acc = 83% | Texture feature extraction and Logistic Regression |
| Garcia-Cano, 2018 [10] | Feature Extraction and Regression | X-rays | 150 | Vertebrae | Cobb angle MAE = 4.79° | Independent component analysis and Random Forest |
| Author/Year | Main Task | Data Type | # Patients | Structures Involved | Results | Model |
|---|---|---|---|---|---|---|
| Haq, 2015 [26] | Segmentation | 3D MRI | 21 | Discs | DICE = 91.7% | Shape-aware models |
| Neubert, 2012 [27] | Segmentation and Identification | 3D MRI | 28 | Discs and Vertebrae | DICE = 89 and 91%, Sen = 100%, Spec = 98% | Statistical shape model |
| Haq, 2020 [25] | Segmentation | CT images | 18 SpineWeb | Discs | DICE = from 91,7 to 95,4% | Shape statistics deformable model |
| Li, 2018 [28] | Segmentation | CT images | 115 (Microsoft R.+ SpineWeb) | Vertebrae | DICE = 92.1% | Gaussian Mixture Model + threshold |
| Ibragimov, 2017 [29] | Segmentation | CT images | 30 vertebrae | Vertebrae | DICE = 84.7% | Landmark detection and deformable models |
| Yu, 2018 [30] | Segmentation | CT images | 21 images | Vertebrae | DICE = 93.9% | Bone-sheetness assisted grid cut |
| Korez, 2015 [31] | Segmentation | CT images | 220 | Vertebrae | DICE = 94.6% | Shape-constrained deformable model |
| Al-Helo, 2011 [32] | Segmentation | CT images | 50 | Vertebrae | Visual evaluation | Active shape models and GVF-snake |
| Ruiz-España, 2015 [33] | Segmentation | CT images | 10 | Vertebrae | DICE = 95% | Selective Binary Gaussian Filtering Regularized Level Set |
| Huang, 2013 [34] | Segmentation | CT images | 56 | Vertebrae | DICE = 94% | Otsu thresholding, edge- and region-based level set |
| Mahdy, 2018 [35] | Segmentation and Localization | CT images | 10 | Vertebrae | Visual evaluation | Threshold and adaptive K-Means |
| Courbot, 2016 [36] | Localization | CT images | 15 | Vertebrae | Visual evaluation, Acc = 89.4% | Hidden Markov Chain segmentation |
| Rasoulian, 2013 [37] | Localization | CT images | 32 | Vertebrae | Visual evaluation, Center of mass MAE = 2mm | Multi-object shape model |
| Mastmeyer, 2006 [38] | Segmentation | CT images | 41 | Vertebrae | DICE > 98.6% | Volume growing and morphological operations |
| Jimenez-Pastor, 2020 [39] | Localization and Identification | CT images | 272 images | Vertebrae | Localization error = 13.7mm, Acc = 74,8% | Decision forest + morphological image processing |
| Lee, 2011 [40] | Localization and Identification | CT images | 19 | Vertebrae | Localization error = 0.14mm, Acc = 93.2% | Threshold and thinning-based integrated cost |
| Klinder, 2009 [41] | Localization and Identification | CT images | 64 | Vertebrae | Localization error = 1.1mm, Acc = 92% | Triangulated shape models |
| Štern, 2009 [42] | Localization | MRI and CT images | 13 and 29 images | Discs and Vertebrae | Localization error = 2.8 and 1.8 mm | Analysis of the geometry of spinal structures |
| Wong, 2008 [43] | Segmentation and Tracking | Fluoroscopic images | 2 videos | Vertebrae | Visual evaluation | Wavelet and shape-active contour based |
| Zheng, 2011 [44] | Segmentation and 3D reconstruction | Fluoroscopic images | 4 | Vertebrae | Mean reconstruction error<1.6mm | Statistical shape models |
| Michopoulou, 2009 [45] | Segmentation | MRI | 34 | Discs | DICE = 90% | Atlas-robust-fuzzy C-Means |
| Fallah, 2018 [46] | Segmentation | MRI | 50 | Discs and Vertebrae | DICE = 92.5 and 91.4% | Hierarchical conditional random field and Random Forest |
| Ghosh, 2014 [47] | Segmentation | MRI | 212 | Discs and Vertebrae | DICE = 87 and 84% | Random Forest and context features |
| Kim, 2018 [48] | Segmentation | MRI | 19 | Vertebrae | DICE = 90% | Graph-based and line-based segmentation algorithms |
| Gaonkar, 2017 [49] | Segmentation | MRI | 63 | Vertebrae | DICE = 83% | Multi-parametric ensemble |
| Gawel, 2018 [50] | Segmentation | MRI | 50 | Vertebrae | DICE = 91.4% | Cascade classifier and Active Appearance Model |
| Engstrom, 2011 [51] | Segmentation | MRI | 20 | Muscles | DICE = 87% | Statistical shape model |
| Baum, 2018 [52] | Segmentation | MRI | 10 | Muscles | DICE = 83% | Average shape model and dual feature model |
| Zheng, 2004 [53] | Segmentation | Fluoroscopic images | 1 | Vertebrae | Visual evaluation | Hough transform and Fourier descriptors |
| Jurcak, 2008 [54] | Segmentation | MRI | 20 | Muscles | DICE = 77% | Probabilistic atlases and geodesic active contours |
| Fortin, 2017 [55] | Segmentation and Regression | MRI | 30 | Muscles | Reliability coefficient = 97-99% | Threshold |
| Neubert, 2013 [56] | Segmentation and Localization | MRI | 44 | Discs | DICE = 92.3%, AUC = 0.98 | Active shape model, Linear Discriminant Analysis, SVM |
| Oktay, 2011 [57] | Localization and Identification | MRI | 40 | Discs | Localization rate = 95.4%, Acc = 97% | Probabilistic model and SVM |
| Castro-Mateos, 2016 [58] | Identification | MRI | 48 | Discs | Sensitivity = 87% | Active contour model and Feedforward NN |
| Kim, 2020 [59] | Localization | Ultrasound | 50 | Muscles | 2mm discrepancy | Fuzzy C-Means Clustering |
| Lui, 2014 [60] | Localization | Ultrasound | 10 | Muscles | F1-Score = 90.9% | Decoupled Active Contour |
| Ribeiro, 2010 [61] | Segmentation | X-rays | 41 | Vertebrae | DICE = 91.7% | Gabor Filters and NN |
| Sa, 2016 [62] | Localization | X-rays | 30 | Vertebrae | True Positive Rate = 75% | GVF-snake and SVM |
| Author/Year | Main Task | Data Type | # Patients | Structures Involved | Results | Model |
|---|---|---|---|---|---|---|
| Iriondo, 2020 [63] | Segmentation | 3D MRI | 31 | Discs | DICE > 85% | Coarse-to-fine context memory NN |
| Staartjes, 2021 [64] | Segmentation and Reconstruction | 3D MRI | 3 | All structures | Visual evaluation | CNN |
| Lee, 2020 [65] | Segmentation and Reconstruction | CT images | 280 images | All structures | MAE = 21 pixels | Generative Adversarial Networks |
| Fan, 2020 [66] | Segmentation and Reconstruction | CT images | 108 | All structures | Kambin triangle = 161 mm | U-net |
| Malinda, 2020[67] | Segmentation | CT images | 120 | Vertebrae | DICE = 94.2% | Generative Adversarial Networks |
| Siemionow, 2020 [68] | Identification | CT images | 45 | Vertebrae | Acc = 96 to 99% | CNN |
| Netherton, 2020 [69] | Localization and Identification | CT images | 330 images | Vertebrae | Localization error = 2.2 mm, Acc = 94% | X-net ensemble |
| Watanabe 2019 [70] | Regression | Moire images + X-rays | 1996 | Vertebrae | Cobb angle MAE = 3.42° | CNN |
| Kim, 2018 [71] | Segmentation | MRI | SpineWeb 20 | Discs | DICE = 89.4% | CNN (BSU-net) |
| Shen, 2021 [72] | Segmentation | MRI | 120 | Discs, Spinal canal and Muscles | Jaccard: 87, 82 and 85% | Feedforward NN |
| Gaonkar, 2019 [73] | Segmentation | MRI | 39295 | Discs and Spinal canal | DICE = 88 and 87% | Discs: U-net, Canal: SVM and RT |
| Huang, 2020 [74] | Segmentation | MRI | 100 | Discs and Vertebrae | Jaccard = 92.6 and 94.7% | U-net |
| Li, 2021 [75] | Segmentation | MRI | 120 | Vertebrae and Spinal canal | DICE = 92.5% | CNN |
| Li, 2019 [76] | Segmentation | MRI | 120 | Muscles | DICE > 91.3% | Deformed U-net |
| Zhou, 2020 [77] | Segmentation | MRI | 57 | Vertebrae | DICE = 84.9% | U-net |
| Jamaludin, 2017 [78] | Classification | MRI | 2009 | Discs and Vertebrae | Acc = 95.6% | CNN |
| Natalia, 2020 [79] | Regression | MRI | 515 | Discs and Spinal canal | Mean error: 0.9 mm | SegNet and Contour Evolution Algorithm |
| Zhou, 2019 [80] | Identification | MRI | 1318 | Vertebrae | Acc = 98.9% | CNN |
| Forsberg, 2017 [81] | Identification | MRI with clinical notes | 475 | Vertebrae | Acc = 97% | CNN and parts-based graphical models |
| Baka, 2017 [82] | Identification | Ultrasound | 19 data sets | Vertebrae | Acc = 92% | CNN and matching strategy |
| Cho, 2020 [83] | Segmentation and Regression | X-rays | 629 | Vertebrae | DICE = 82.1%, MAE = 8,055° | U-net |
| Li, 2016 [84] | Identification | X-rays | 110 | Vertebrae | Acc = 80.4% | CNN |
| Sa, 2017 [85] | Localization | X-rays | 1081 images | Discs | Precision = 90.5% | Faster R-CNN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Antoni, F.; Russo, F.; Ambrosio, L.; Vollero, L.; Vadalà, G.; Merone, M.; Papalia, R.; Denaro, V. Artificial Intelligence and Computer Vision in Low Back Pain: A Systematic Review. Int. J. Environ. Res. Public Health 2021, 18, 10909. https://doi.org/10.3390/ijerph182010909
D’Antoni F, Russo F, Ambrosio L, Vollero L, Vadalà G, Merone M, Papalia R, Denaro V. Artificial Intelligence and Computer Vision in Low Back Pain: A Systematic Review. International Journal of Environmental Research and Public Health. 2021; 18(20):10909. https://doi.org/10.3390/ijerph182010909
Chicago/Turabian StyleD’Antoni, Federico, Fabrizio Russo, Luca Ambrosio, Luca Vollero, Gianluca Vadalà, Mario Merone, Rocco Papalia, and Vincenzo Denaro. 2021. "Artificial Intelligence and Computer Vision in Low Back Pain: A Systematic Review" International Journal of Environmental Research and Public Health 18, no. 20: 10909. https://doi.org/10.3390/ijerph182010909
APA StyleD’Antoni, F., Russo, F., Ambrosio, L., Vollero, L., Vadalà, G., Merone, M., Papalia, R., & Denaro, V. (2021). Artificial Intelligence and Computer Vision in Low Back Pain: A Systematic Review. International Journal of Environmental Research and Public Health, 18(20), 10909. https://doi.org/10.3390/ijerph182010909












