Forecasting the Suitability of Online Mental Health Information for Effective Self-Care Developing Machine Learning Classifiers Using Natural Language Features
Abstract
1. Introduction
2. Methods
2.1. Data Collection
2.2. Feature Annotation
2.3. Classifier Training and Testing
2.4. Bayesian Machine Learning Classifiers (BMLC)
2.5. Bayesian Probabilistic Assessment of Personalised Mental Healthcare Quality
2.6. Classifier Optimisation
2.7. Feature Normalisation and Scaling
3. Results
4. Discussions
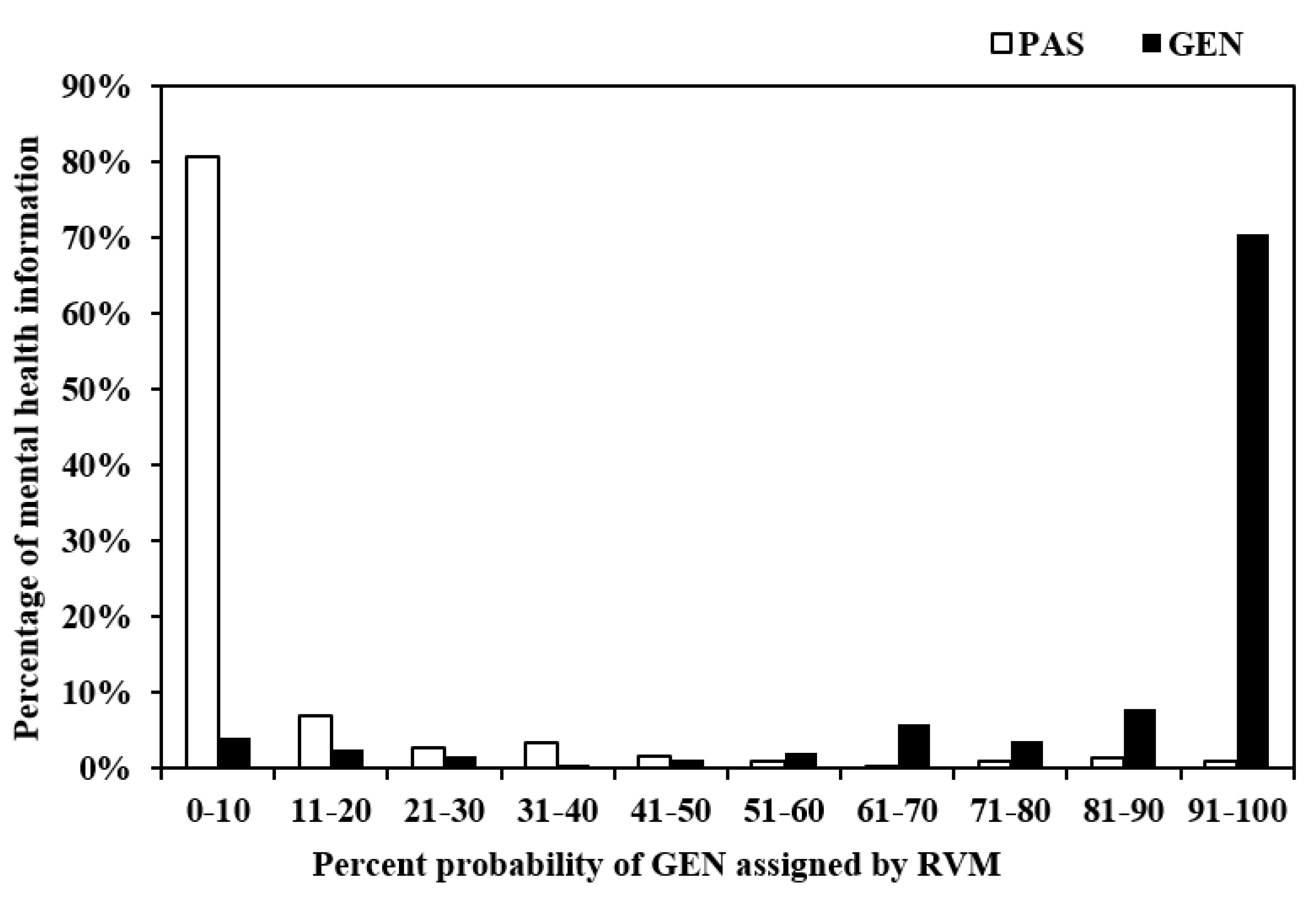
4.1. Probabilistic Outputs
4.2. Diagnostic Utility of Bayesian Classifiers to Predict Mental Healthcare Information Suitability for Young People
5. Conclusions
Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lund, C.; Brooke-Sumner, C.; Baingana, F.; Baron, E.C.; Breuer, E.; Chandra, P.; Haushofer, J.; Herrman, H.; Jordans, M.; Kieling, C.; et al. Social determinants of mental disorders and the Sustainable Development Goals: A systematic review of reviews. Lancet Psychiatry 2018, 5, 357–369. [Google Scholar] [CrossRef]
- Hamilton, M.P.; Hetrick, S.E.; Mihalopoulos, C.; Baker, D.; Browne, V.; Chanen, A.M.; Pennell, K.; Purcell, R.; Stavely, H.; McGorry, P.D. Identifying attributes of care that may improve cost-effectiveness in the youth mental health service system. Med. J. Aust. 2017, 207, S27–S37. [Google Scholar] [CrossRef]
- Moffitt, T.E. Childhood exposure to violence and lifelong health: Clinical intervention science and stress-biology research join forces. Dev. Psychopathol. 2013, 25 Pt 2, 1619–1634. [Google Scholar] [CrossRef]
- Ford, E.; Clark, C.; McManus, S.; Harris, J.; Jenkins, R.; Bebbington, P.; Brugha, T.; Meltzer, H.; Stansfeld, S. Common mental disorders, unemployment and welfare benefits in England. Public Health 2010, 124, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Meltzer, H.; Bebbington, P.; Dennis, M.S.; Jenkins, R.; McManus, S.; Brugha, T.S. Feelings of loneliness among adults with mental disorder. Soc. Psychiatry Psychiatr. Epidemiol. 2013, 48, 5–13. [Google Scholar] [CrossRef]
- Hosang, G.M.; Bhui, K. Gender discrimination, victimisation and women’s mental health. Br. J. Psychiatry 2018, 213, 682–684. [Google Scholar] [CrossRef]
- Lund, C.; Breen, A.; Flisher, A.J.; Kakuma, R.; Corrigall, J.; Joska, J.; Swartz, L.; Patel, V. Poverty and common mental disorders in low and middle income countries: A systematic review. Soc. Sci. Med. 2010, 71, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Bogic, M.; Ajdukovic, D.; Bremner, S.; Franciskovic, T.; Galeazzi, G.M.; Kucukalic, A.; Lecic-Tosevski, D.; Morina, N.; Popovski, M.; Schützwohl, M.; et al. Factors associated with mental disorders in long-settled war refugees: Refugees from the former Yugoslavia in Germany, Italy and the UK. Br. J. Psychiatry 2012, 200, 216–223. [Google Scholar] [CrossRef]
- Satcher, D.; Friel, S.; Bell, R. Natural and Manmade Disasters and Mental Health. JAMA 2007, 298, 2540–2542. [Google Scholar] [CrossRef] [PubMed]
- Carpinello, S.E.; Knight, E.L.; Markowitz, F.E.; Pease, E.A. The development of the Mental Health Confidence Scale: A measure of self-efficacy in individuals diagnosed with mental disorders. Psychiatr. Rehabil. J. 2000, 23, 236–243. [Google Scholar] [CrossRef]
- Falloon, I.R.H. Family interventions for mental disorders: Efficacy and effectiveness. World Psychiatry 2003, 2, 20–28. [Google Scholar]
- Clark, R.E. Family Support and Substance Use Outcomes for Persons with Mental Illness and Substance Use Disorders. Schizophr. Bull. 2001, 27, 93–101. [Google Scholar] [CrossRef]
- Hopkinson, J.B.; Brown, J.C.; Okamoto, I.; Addington-Hall, J.M. The Effectiveness of Patient-Family Carer (Couple) Inter-vention for the Management of Symptoms and Other Health-Related Problems in People Affected by Cancer: A Systematic Literature Search and Narrative Review. J. Pain Symptom Manag. 2012, 43, 111–142. [Google Scholar] [CrossRef] [PubMed]
- Musiat, P.; Hoffmann, L.; Schmidt, U. Personalised computerized feedback in E-mental health. J. Ment. Health 2012, 21, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Hollis, C.; Morriss, R.; Martin, J.; Amani, S.; Cotton, R.; Denis, M.; Lewis, S. Technological innovations in mental healthcare: Harnessing the digital revolution. Br. J. Psychiatry 2015, 206, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Hatton, C.M.; Paton, L.W.; McMillan, D.; Cussens, J.; Gilbody, S.; Tiffin, P.A. Predicting persistent depressive symptoms in older adults: A machine learning approach to personalized mental healthcare. J. Affect. Disord. 2019, 246, 857–860. [Google Scholar] [CrossRef]
- Chaturvedi, J. From Learning About Machines to Machine Learning: Applications for Mental Health Rehabilitation. J. Psy-chosoc. Rehabil. Ment. Health 2020, 7, 3–4. [Google Scholar] [CrossRef][Green Version]
- Aung, M.H.; Matthews, M.; Choudhury, T. Sensing behavioral symptoms of mental health and delivering personalized in-terventions using mobile technologies. Depress. Anxiety 2017, 34, 603–609. [Google Scholar] [CrossRef]
- Calvo, R.A.; Milne, D.N.; Hussain, M.S.; Christensen, H. Natural language processing in mental health applications using non-clinical texts. Nat. Lang. Eng. 2017, 23, 649–685. [Google Scholar] [CrossRef]
- Baclic, O.; Tunis, M.; Young, K.; Doan, C.; Swerdfeger, H.; Schonfeld, J. Challenges and opportunities for public health made possible by advances in natural language processing. Can. Commun. Dis. Rep. 2020, 46, 161–168. [Google Scholar] [CrossRef]
- Barak, A.; Miron, O. Writing Characteristics of Suicidal People on the Internet: A Psychological Investigation of Emerging Social Environments. Suicide Life-Threatening Behav. 2005, 35, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Percevic, R.; Okon, E.; Meermann, R.; Kordy, H. Use of text messaging in the aftercare of patients with bulimia nervosa. Eur. Eat. Disord. Rev. 2003, 11, 279–290. [Google Scholar] [CrossRef]
- Abbe, A.; Grouin, C.; Zweigenbaum, P.; Falissard, B. Text mining applications in psychiatry: A systematic literature review: Text Mining Applications in Psychiatry. Int. J. Methods Psychiatr. Res. 2016, 25, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, A.V.; Gupta, R.; Kim, C.; Kashkoush, A.; Hansberry, D.R.; Agarwal, N.; Koch, E. Patient Education Materials in Dermatology: Addressing the Health Literacy Needs of Patients. JAMA Dermatol. 2016, 152, 946–947. [Google Scholar] [CrossRef] [PubMed]
- Kasabwala, K.; Agarwal, N.; Hansberry, D.R.; Baredes, S.; Eloy, J.A. Readability Assessment of Patient Education Materials from the American Academy of Otolaryngology—Head and Neck Surgery Foundation. Otolaryngol.-Head Neck Surg. 2012, 147, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Rayson, P.; Archer, D.; Piao, S.; McEnery, A.M. (Eds.) The UCREL semantic analysis system. In Proceedings of the Beyond Named Entity Recognition Semantic Labeling for NLP Tasks Workshop, Lisbon, Portugal, 25–28 May 2004. [Google Scholar]
- McIntyre, D.; Archer, D. A corpus-based approach to mind style. J. Lit. Semant. 2010, 39, 167–182. [Google Scholar] [CrossRef]
- Piao, S.S.; Rayson, P.; Archer, D.; McEnery, T. Comparing and combining a semantic tagger and a statistical tool for MWE extraction. Comput. Speech Lang. 2005, 19, 378–397. [Google Scholar] [CrossRef]
- Tipping, M.E. (Ed.) The Relevance Vector Machine. In Advances in Neural Information Processing Systems; Mit Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Tipping, M.E. Sparse bayesian learning and the relevance vector machine. J. Mach. Learn. Res. 2001, 1, 211–244. [Google Scholar] [CrossRef]
- Caesarendra, W.; Widodo, A.; Yang, B.-S. Application of relevance vector machine and logistic regression for machine deg-radation assessment. Mech. Syst. Signal Process. 2010, 24, 1161–1171. [Google Scholar] [CrossRef]
- Matsumoto, M.; Hori, J. Classification of silent speech using support vector machine and relevance vector machine. Appl. Soft Comput. 2014, 20, 95–102. [Google Scholar] [CrossRef]
- Ranganathan, P.; Aggarwal, R. Understanding the properties of diagnostic tests—Part 2: Likelihood ratios. Perspect. Clin. Res. 2018, 9, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Deeks, J.J.; Altman, D.G. Diagnostic tests 4: Likelihood ratios. BMJ 2004, 329, 168–169. [Google Scholar] [CrossRef] [PubMed]
- Grimes, D.A.; Schulz, K.F. Refining clinical diagnosis with likelihood ratios. Lancet 2005, 365, 1500–1505. [Google Scholar] [CrossRef]
- Simel, D.L.; Samsa, G.P.; Matchar, D. Likelihood ratios with confidence: Sample size estimation for diagnostic test studies. J. Clin. Epidemiol. 1991, 44, 763–770. [Google Scholar] [CrossRef]
- Moayyedi, P.; Axon, A.T.R. The Usefulness of The Likelihood Ratio in The Diagnosis of Dyspepsia and Gastroesophageal Reflux Disease. Am. J. Gastroenterol. 1999, 94, 3122–3125. [Google Scholar] [CrossRef] [PubMed]
- Shreffler, J.; Huecker, M.R. Diagnostic Testing Accuracy: Sensitivity, Specificity, Predictive Values and Likelihood Ratios; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]

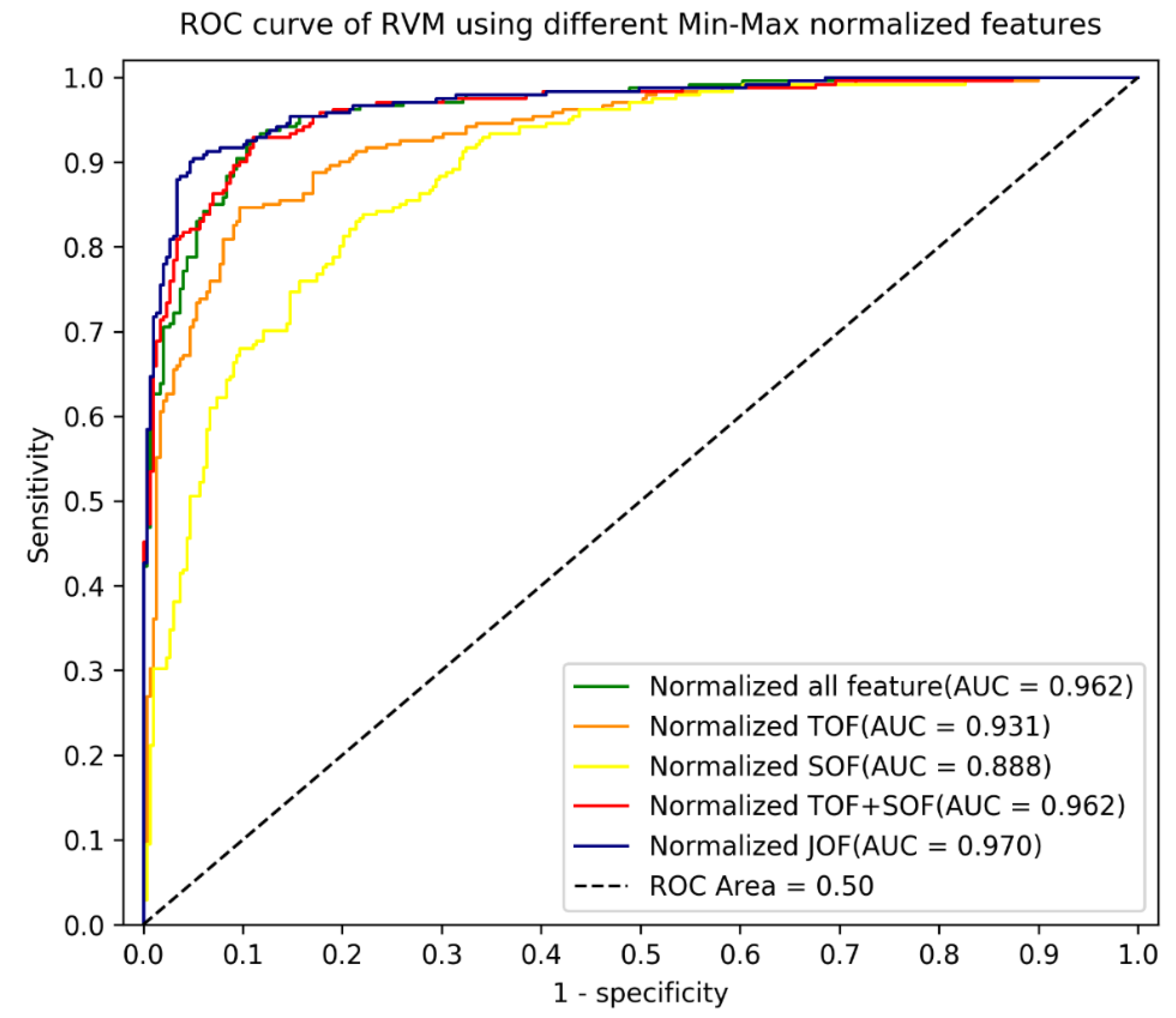
| Technique | Training/Testing (5-Fold Cross-Validation) | Validation | |||
|---|---|---|---|---|---|
| AUC Mean (SD) | AUC | Accuracy | Sensitivity | Specificity | |
| (i) Non-Optimised, Non-Normalised Feature Sets | |||||
| RVM_ SOF (22) | 0.858 (0.022) | 0.875 | 0.774 | 0.747 | 0.796 |
| RVM_ TOF (27) | 0.926 (0.018) | 0.927 | 0.848 | 0.805 | 0.883 |
| RVM_ SOF (22) + TOF (27) | 0.960 (0.009) | 0.965 | 0.9 | 0.8589 | 0.933 |
| (ii) Non-Optimised, Normalised Feature Sets | |||||
| RVM_ normalised SOF (22) | 0.861 (0.038) | 0.886 | 0.793 | 0.730 | 0.843 |
| RVM_ normalised TOF (27) | 0.953 (0.019) | 0.939 | 0.869 | 0.805 | 0.920 |
| RVM_ normalised SOF (22) + TOF (27) | 0.963 (0.013) | 0.962 | 0.889 | 0.851 | 0.920 |
| (iii) Separate Optimisation without Normalisation | |||||
| RVM_ SOF (17) | 0.858 (0.026) | 0.870 | 0.785 | 0.755 | 0.809 |
| RVM_ TOF (12) | 0.906 (0.026) | 0.887 | 0.802 | 0.739 | 0.853 |
| RVM_ SOF (17) + TOF (12) | 0.943 (0.013) | 0.940 | 0.867 | 0.830 | 0.896 |
| (iv) Separate Optimisation with Normalisation | |||||
| RVM_ normalised SOF (17) | 0.864 (0.029) | 0.888 | 0.8 | 0.734 | 0.853 |
| RVM_ normalised TOF (12) | 0.941 (0.010) | 0.931 | 0.874 | 0.838 | 0.903 |
| RVM_ normalised SOF (17) + TOF (12) | 0.951 (0.012) | 0.962 | 0.898 | 0.863 | 0.926 |
| (v) Joint Optimisation without Normalisation | |||||
| RVM_ JOF (39) | 0.960 (0.008) | 0.965 | 0.893 | 0.851 | 0.926 |
| (vi) Joint Optimisation with Normalisation | |||||
| RVM_ normalised JOF (39) | 0.966 (0.011) | 0.970 | 0.9296 | 0.9004 | 0.953 |
| Pairs | RVM Classifier Pair(s) | AUC Mean Difference | Asymptotic 95% Confidence Interval | ||
|---|---|---|---|---|---|
| Lower | Upper | p-Value | |||
| 1 | RVM_ normalised JOF (39) vs. RVM_ normalised SOF (22) | 0.105 | 0.037 | 0.172 | 0.0024 ** |
| 2 | RVM_ normalised JOF (39) vs. RVM_ normalised SOF (17) | 0.101 | 0.053 | 0.150 | 0.0008 ** |
| 3 | RVM_ normalised JOF (39) vs. RVM_ normalised TOF (27) | 0.040 | 0.009 | 0.070 | 0.0046 ** |
| 4 | RVM_ normalised JOF (39) vs. RVM_ normalised TOF (12) | 0.025 | 0.02 | 0.033 | 0.0002 ** |
| 5 | RVM_ normalised JOF (39) vs. RVM_ normalised SOF (12) + TOF (17) | 0.015 | 0.007 | 0.023 | 0.0014 ** |
| Pair | Mann–Whitney U Test | Sensitivity | Asymptotic 95% C.I. | Specificity | Asymptotic 95% C.I. | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean Difference | Lower | Upper | p-Value | Mean Difference | Lower | Upper | p-Value | ||
| 1 | RVM_ normalised JOF (39) vs. RVM_ TOF (27) | 0.073 | 0.009 | 0.137 | 0.022 ** | 0.083 | 0.034 | 0.132 | 0.022 ** |
| 2 | RVM_ normalised JOF (39) vs. RVM_ normalised TOF (27) | 0.079 | 0.006 | 0.152 | 0.022 ** | 0.037 | 0.019 | 0.056 | 0.047 ** |
| 3 | RVM_ normalised JOF (39) vs. RVM_ SOF (22) | 0.113 | −0.022 | 0.248 | 0.022 ** | 0.181 | 0.071 | 0.292 | 0.012 ** |
| 4 | RVM_ normalised JOF (39) vs. RVM_ normalised SOF (22) | 0.135 | 0.000 | 0.269 | 0.012 ** | 0.122 | 0.056 | 0.189 | 0.012 ** |
| 5 | RVM_ normalised JOF (39) vs. RVM_ TOF (12) | 0.148 | 0.083 | 0.213 | 0.012 ** | 0.109 | 0.057 | 0.162 | 0.012 ** |
| 6 | RVM_ normalised JOF (39) vs. RVM_ normalised TOF (12) | 0.066 | 0.029 | 0.104 | 0.012 ** | 0.062 | 0.031 | 0.092 | 0.028 ** |
| 7 | RVM_ normalised JOF (39) vs. RVM_SOF (17) | 0.114 | −0.036 | 0.263 | 0.021 ** | 0.189 | 0.055 | 0.322 | 0.012 ** |
| 8 | RVM_ normalised JOF (39) vs. RVM_ normalised SOF (17) | 0.128 | 0.016 | 0.240 | 0.012 ** | 0.126 | 0.033 | 0.220 | 0.012 ** |
| 9 | RVM_ normalised JOF (39) vs. RVM_ SOF (17) + TOF (12) | 0.068 | −0.001 | 0.137 | 0.022 ** | 0.063 | 0.025 | 0.100 | 0.022 ** |
| 10 | RVM_ normalised JOF (39) vs. RVM_ normalised SOF (17) + TOF (12) | 0.034 | 0.010 | 0.058 | 0.046 ** | 0.023 | 0.004 | 0.043 | 0.095 |
| 11 | RVM_ normalised JOF (39) vs. RVM_ SOF (22) + TOF (27) | 0.034 | 0.006 | 0.062 | 0.074 | 0.021 | 0.007 | 0.035 | 0.249 |
| 12 | RVM_ normalised JOF (39) vs. RVM_ normalised TOF (27) + SOF (22) | 0.025 | −0.024 | 0.074 | 0.346 | 0.026 | 0.005 | 0.048 | 0.144 |
| Probability Thresholds | Sensitivity (95% CI) | Specificity (95% CI) | Positive Likelihood Ratio (LR+) (95% CI) | Negative Likelihood Ratio (LR−) (95% CI) |
|---|---|---|---|---|
| 0.1 | 0.959 (0.933, 0.984) | 0.806 (0.761, 0.851) | 4.941 (3.916, 6.235) | 0.051 (0.028, 0.095) |
| 0.2 | 0.934 (0.902, 0.965) | 0.876 (0.839, 0.914) | 7.545 (5.570, 10.220) | 0.076 (0.047, 0.122) |
| 0.3 | 0.917 (0.882, 0.952) | 0.903 (0.869, 0.937) | 9.455 (6.676, 13.389) | 0.092 (0.060, 0.140) |
| 0.4 | 0.913 (0.877, 0.948) | 0.936 (0.909, 0.964) | 14.366 (9.281, 22.236) | 0.093 (0.062, 0.140) |
| 0.45 | 0.905 (0.867, 0.942) | 0.950 (0.925, 0.975) | 18.031 (10.992, 29.577) | 0.100 (0.068, 0.148) |
| 0.5 | 0.900 (0.863, 0.938) | 0.953 (0.929, 0.977) | 19.230 (11.512, 32.125) | 0.104 (0.071, 0.153) |
| 0.55 | 0.884 (0.843, 0.924) | 0.960 (0.938, 0.982) | 22.022 (12.627, 38.407) | 0.121 (0.085, 0.172) |
| 0.6 | 0.880 (0.839, 0.921) | 0.963 (0.942, 0.985) | 23.911 (13.369, 42.785) | 0.125 (0.089, 0.176) |
| 0.7 | 0.822 (0.773, 0.870) | 0.967 (0.947, 0.987) | 24.565 (13.318, 45.309) | 0.185 (0.141, 0.242) |
| 0.8 | 0.784 (0.732, 0.836) | 0.977 (0.959, 0.994) | 33.498 (16.061, 69.864) | 0.221 (0.174, 0.281) |
| 0.9 | 0.705 (0.648, 0.763) | 0.990 (0.979, 1.001) | 70.304 (22.737, 217.388) | 0.298 (0.245, 0.362) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, M.; Xie, W.; Huang, R.; Qian, X. Forecasting the Suitability of Online Mental Health Information for Effective Self-Care Developing Machine Learning Classifiers Using Natural Language Features. Int. J. Environ. Res. Public Health 2021, 18, 10048. https://doi.org/10.3390/ijerph181910048
Ji M, Xie W, Huang R, Qian X. Forecasting the Suitability of Online Mental Health Information for Effective Self-Care Developing Machine Learning Classifiers Using Natural Language Features. International Journal of Environmental Research and Public Health. 2021; 18(19):10048. https://doi.org/10.3390/ijerph181910048
Chicago/Turabian StyleJi, Meng, Wenxiu Xie, Riliu Huang, and Xiaobo Qian. 2021. "Forecasting the Suitability of Online Mental Health Information for Effective Self-Care Developing Machine Learning Classifiers Using Natural Language Features" International Journal of Environmental Research and Public Health 18, no. 19: 10048. https://doi.org/10.3390/ijerph181910048
APA StyleJi, M., Xie, W., Huang, R., & Qian, X. (2021). Forecasting the Suitability of Online Mental Health Information for Effective Self-Care Developing Machine Learning Classifiers Using Natural Language Features. International Journal of Environmental Research and Public Health, 18(19), 10048. https://doi.org/10.3390/ijerph181910048






