Rapid Test Ag 2019-nCoV (PROGNOSIS, BIOTECH, Larissa, Greece); Performance Evaluation in Hospital Setting with Real Time RT-PCR
Abstract
:1. Introduction
- To estimate the sensitivity and specificity of a local produced and recently approved RAT, Rapid Test Ag 2019-nCoV (PROGNOSIS, BIOTECH, Larissa, Greece), in a hospital setting.
- To evaluate its accuracy for viral load determination in comparison to the reference method RT-qPCR.
2. Material and Methods
2.1. Ethics Statement
2.2. Principal of the Assay
2.3. Limit of Detection (LOD) Estimation
2.4. Determination of Clinical Sensitivity-Specificity
2.5. Reader Description and Standard Curve
2.6. Statistical Analysis
3. Results
3.1. Determination of the Lower Limit of Detection (LOD)
3.2. Clinical Evaluation
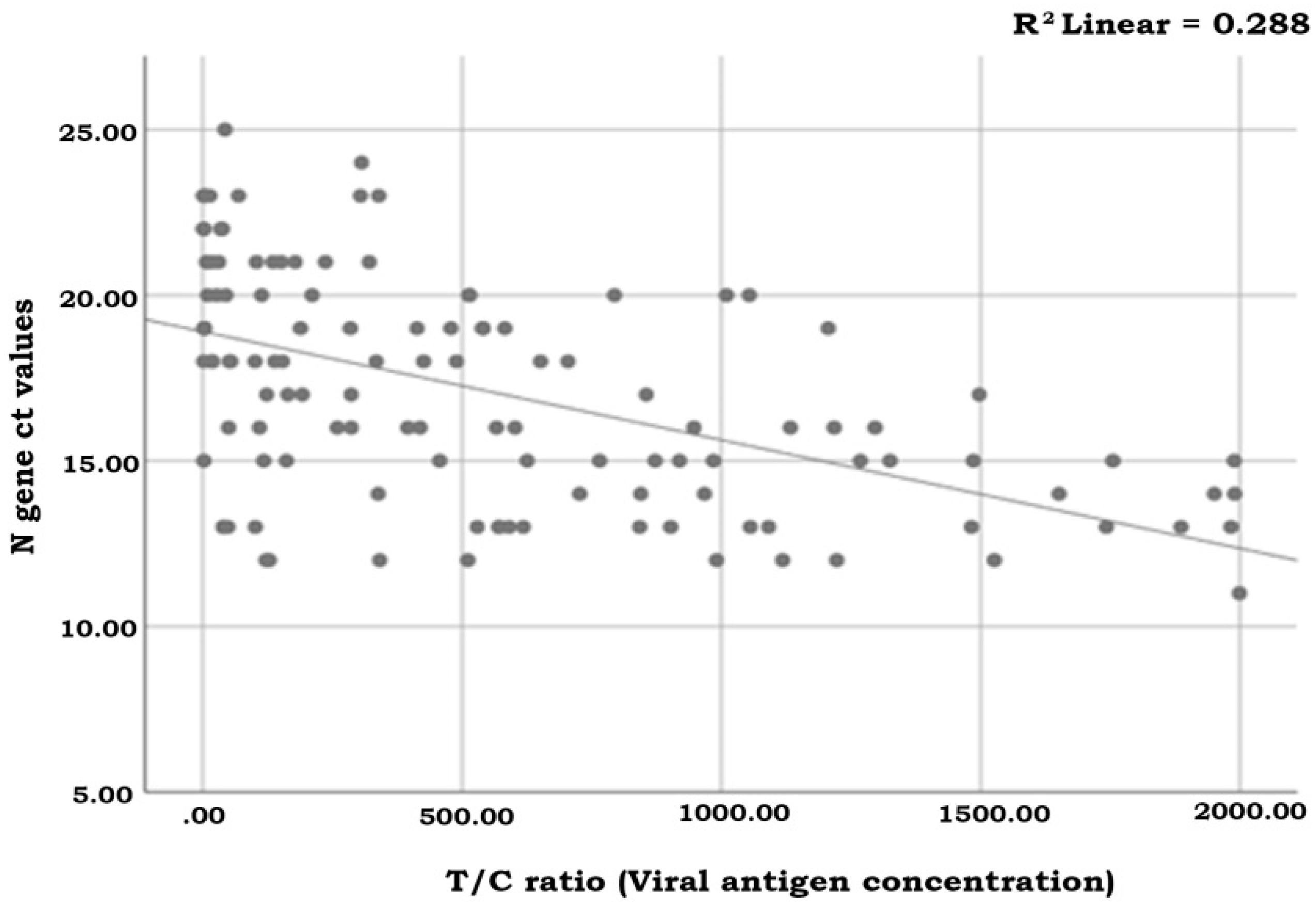
3.3. Semi-Quantification Using the Standard Curve
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 8 June 2021).
- Jiang, S.; Du, L.; Shi, Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: Calling for developingtherapeutic and prophylactic strategies. Emerg. Microbes Infect. 2020, 9, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, W.; Liu, H.; Qi, B.; Liang, K.; Xu, H.; Peng, Z.; Xiao, S.Y. Clinical outcomes of 402 patients with COVID-2019 from a single center in Wuhan, China. J. Med. Virol. 2020, 92, 2751–2757. [Google Scholar] [CrossRef]
- Anderson, E.L.; Turnham, P.; Griffin, J.R.; Clarke, C.C. Consideration of the Aerosol Transmission for COVID-19 and Public Health. Risk Anal. 2020, 40, 902–907. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Duong Bang, D.; Wolff, A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. In Vitro Diagnostics Detecting Sars-CoV-2 Nucleic Acid and Rapid Diagnostics Tests Detecting Sars-CoV-2 Antigen. Published 9 June 2020. Available online: https://www.who.int/diagnostics_laboratory/EUL/en/ (accessed on 8 June 2021).
- Ye, Q.; West, A.M.V.; Silletti, S.; Corbett, K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020, 29, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Clopper, C.J.; Pearson, E.S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 1934, 26, 404–413. [Google Scholar] [CrossRef]
- Cohen, J. Weighted kappa: Nominal scale agreement with provision for scaled disagreement or partial credit. Psychol. Bull. 1968, 70, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Arevalo-Rodriguez, I.; Buitrago-Garcia, D.; Simancas-Racines, D.; Zambrano-Achig, P.; Del Campo, R.; Ciapponi, A.; Sued, O.; Martinez-García, L.; Rutjes, A.W.; Low, N.; et al. False negative results of initial RT-PCR assays for COVID-19: A systematic review. PLoS ONE 2020, 15, e0242958. [Google Scholar] [CrossRef] [PubMed]
- Zaman, F.A.; Aggarwal, S.; Pal, R.; Chatterjee, P.K.; Kiran, K.A.; Panda, S.; Bhattacharya, T. Exploratory study on the operational issues faced in collection, transportation, and laboratory testing related to COVID-19 in remote areas of selected EAG states of North East and East India. J. Fam. Med. Prim. Care 2021, 10, 1443. [Google Scholar]
- Linares, M.; P’erez-Tanoira, R.; Carrero, A.; Romanyk, J.; P’erez-García, F.; G’omez-Herruz, P.; Arroyo, T.; Cuadros, J. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J. Clin. Virol. 2020, 133, 104659. [Google Scholar] [CrossRef] [PubMed]
- Mak, G.C.; Lau, S.S.; Wong, K.K.; Chow, N.L.; Lau, C.S.; Lam, E.T.; Chan, R.C.; Tsang, D.N. Analytical sensitivity and clinical sensitivity of the three rapid antigen detection kits for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 133, 104684. [Google Scholar] [CrossRef] [PubMed]
- Scohy, A.; Anantharajah, A.; Bod’eus, M.; Kabamba-Mukadi, B.; Verroken, A.; Rodriguez-Villalobos, H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020, 129, 104455. [Google Scholar] [CrossRef] [PubMed]
- Mertens, P.; De Vos, N.; Martiny, D.; Jassoy, C.; Mirazimi, A.; Cuypers, L.; Van den Wijngaert, S.; Monteil, V.; Melin, P.; Stoffels, K.; et al. Development and potential usefulness of the COVID-19 Ag respi-strip diagnostic assay in a pandemic context. Front. Med. 2020, 7, 225. [Google Scholar] [CrossRef] [PubMed]
- Kohmer, N.; Toptan, T.; Pallas, C.; Karaca, O.; Pfeiffer, A.; Westhaus, S. The comparative clinical performance of four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. J. Clin. Med. 2021, 10, 328. [Google Scholar] [CrossRef] [PubMed]
- Mak, G.C.K.; Lau, S.S.Y.; Wong, K.K.Y.; Chow, N.L.S.; Lau, C.S.; Lam, E.T.K.; Chan, R.C.; Tsang, D.N. Evaluation of rapid antigen detection kit from the WHO emergency use list for detecting SARS-CoV-2. J. Clin. Virol. 2021, 134, 104712. [Google Scholar] [CrossRef] [PubMed]
- Fourati, S.; Audureau, E.; Chevaliez, S.; Pawlotsky, J.M. Evaluation de la Performance Diagnostique des Tests Rapides D’orientation Diagnostique Antigeniques COVID-19. Laboratoire de Virologie et le Service de Santé Publique des Hôpitaux Universitaires Henri-Mondor (AP-HP, Université Paris-est-Créteil, et INSERM). Available online: https://www.sfm-microbiologie.org/covid-19-reactifs-evaluations/ (accessed on 8 June 2021).
- Kim, D.; Lee, J.; Bal, J.; Seo, S.K.; Chong, C.K.; Lee, J.H.; Park, H. Development and Clinical Evaluation of an Immunochromatography-Based Rapid Antigen Test (GenBody™ COVAG025) for COVID-19 Diagnosis. Viruses 2021, 13, 796. [Google Scholar] [CrossRef] [PubMed]


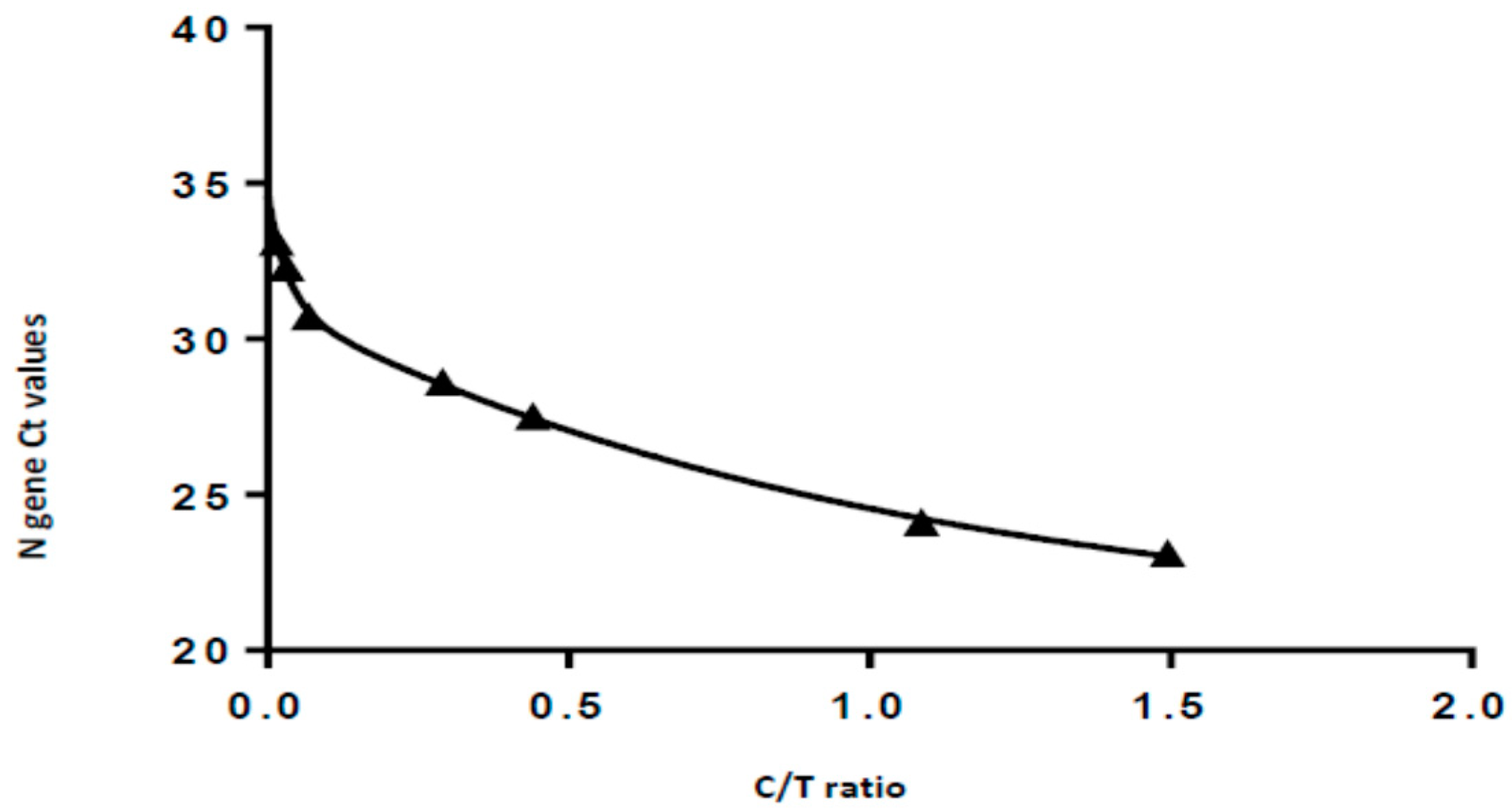
| Viral Load (gc/mL) | N Gene Ct Value | Test Line Density (T) | Control Line Density (C) | Ratio T/C | |
|---|---|---|---|---|---|
| 1 | 187 500 | 22.6 | 387.817 | 266.465 | 1.452 |
| 2 | 18 750 | 27.5 | 111.206 | 252.339 | 0.441 |
| 3 | 1 875 | 30.7 | 16.153 | 237.341 | 0.068 |
| 4 | 937.5 | 31.6 | 10.191 | 226.687 | 0.045 |
| 5 | 468.8 | 32.5 | 7.152 | 217.968 | 0.033 |
| 6 | 0 (negative control) | undetermined | 0.255 | 203.568 | 0.001 |
| Sensitivity (%) | Specificity (%) | Inter-Rater Agreement (Kappa) | PPV (%) | NPV (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive Samples (n) | 95% CI | Negative Samples (n) | 95% CI | 95% CI | Prevalence | Prevalence | |||||||||||
| 1‰ | 1% | 5% | 1‰ | 1% | 5% | ||||||||||||
| Total | 141/165 | 85.5 | 79.1 | 90.5 | 458/459 | 99.8 | 98.8 | 100.0 | 0.89 | 85.1 | 93.3 | 28.2 | 79.9 | 95.4 | 99.99 | 99.9 | 99.2 |
| Ct < 30 | 141/156 | 90.4 | 84.6 | 94.5 | 0.93 | 89.5 | 96.3 | 28.6 | 80.7 | 95.6 | 99.99 | 99.9 | 99.5 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kyritsi, M.; Vontas, A.; Voulgaridi, I.; Matziri, A.; Komnos, A.; Babalis, D.; Papadogoulas, A.; Oikonomou, A.; Mouchtouri, V.A.; Speletas, M.; et al. Rapid Test Ag 2019-nCoV (PROGNOSIS, BIOTECH, Larissa, Greece); Performance Evaluation in Hospital Setting with Real Time RT-PCR. Int. J. Environ. Res. Public Health 2021, 18, 9151. https://doi.org/10.3390/ijerph18179151
Kyritsi M, Vontas A, Voulgaridi I, Matziri A, Komnos A, Babalis D, Papadogoulas A, Oikonomou A, Mouchtouri VA, Speletas M, et al. Rapid Test Ag 2019-nCoV (PROGNOSIS, BIOTECH, Larissa, Greece); Performance Evaluation in Hospital Setting with Real Time RT-PCR. International Journal of Environmental Research and Public Health. 2021; 18(17):9151. https://doi.org/10.3390/ijerph18179151
Chicago/Turabian StyleKyritsi, Maria, Alexandros Vontas, Ioanna Voulgaridi, Alexia Matziri, Apostolos Komnos, Dimitris Babalis, Antonios Papadogoulas, Aikaterini Oikonomou, Varvara A. Mouchtouri, Matthaios Speletas, and et al. 2021. "Rapid Test Ag 2019-nCoV (PROGNOSIS, BIOTECH, Larissa, Greece); Performance Evaluation in Hospital Setting with Real Time RT-PCR" International Journal of Environmental Research and Public Health 18, no. 17: 9151. https://doi.org/10.3390/ijerph18179151
APA StyleKyritsi, M., Vontas, A., Voulgaridi, I., Matziri, A., Komnos, A., Babalis, D., Papadogoulas, A., Oikonomou, A., Mouchtouri, V. A., Speletas, M., & Hadjichristodoulou, C. (2021). Rapid Test Ag 2019-nCoV (PROGNOSIS, BIOTECH, Larissa, Greece); Performance Evaluation in Hospital Setting with Real Time RT-PCR. International Journal of Environmental Research and Public Health, 18(17), 9151. https://doi.org/10.3390/ijerph18179151









