Contamination of Hospital Surfaces with Bacterial Pathogens under the Current COVID-19 Outbreak
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Transportation
2.2. Nucleic Acid Extraction
2.3. qPCR for Bacterial Pathogen Identification
2.4. 16S rRNA Gene Amplicon Sequencing
2.5. Sequence Analysis
2.6. Ethical Consideration
3. Results
3.1. Detection of Bacterial Pathogens on Various Surfaces
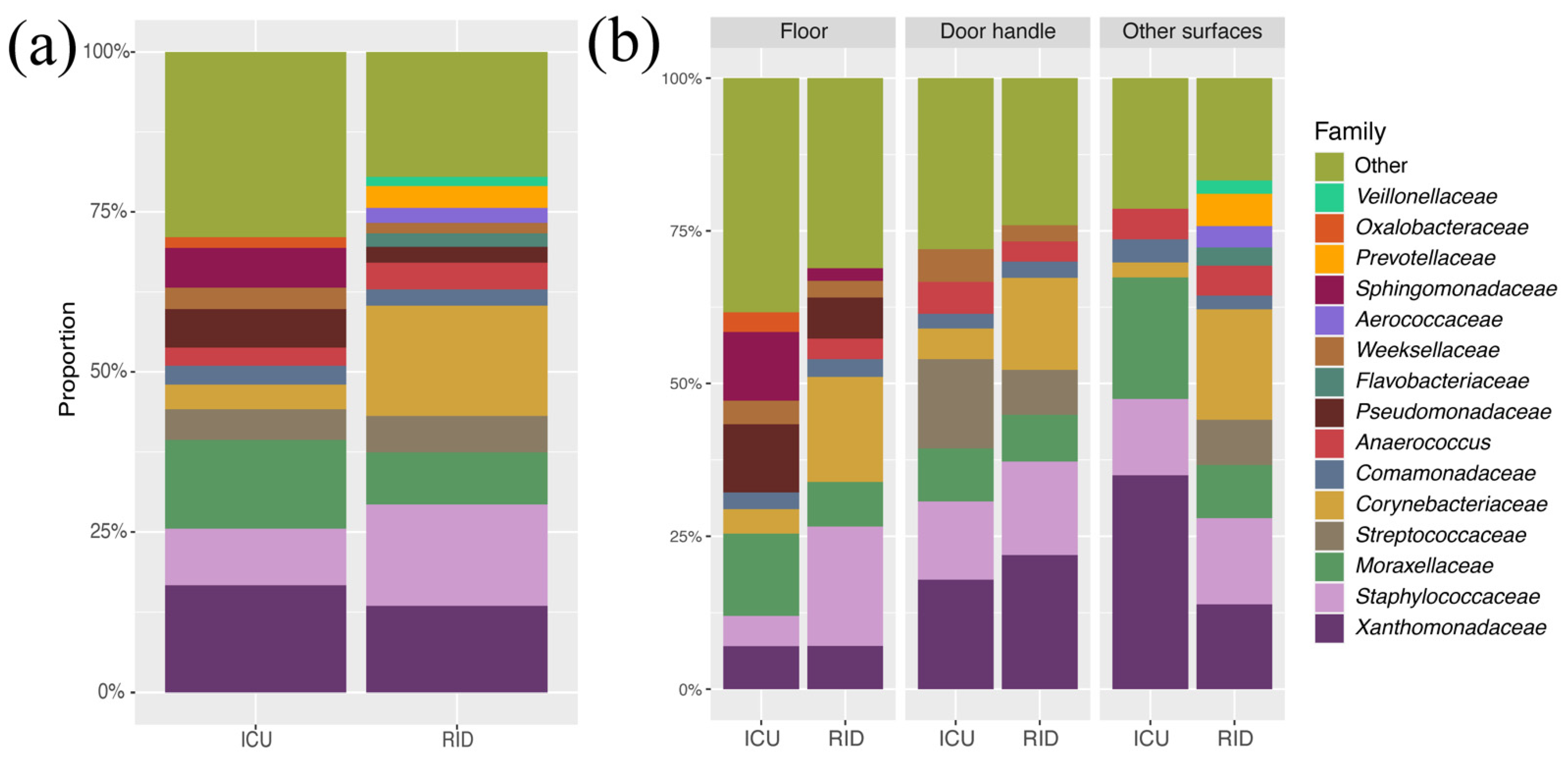
3.2. Taxonomy Composition of Departments and Surface Types
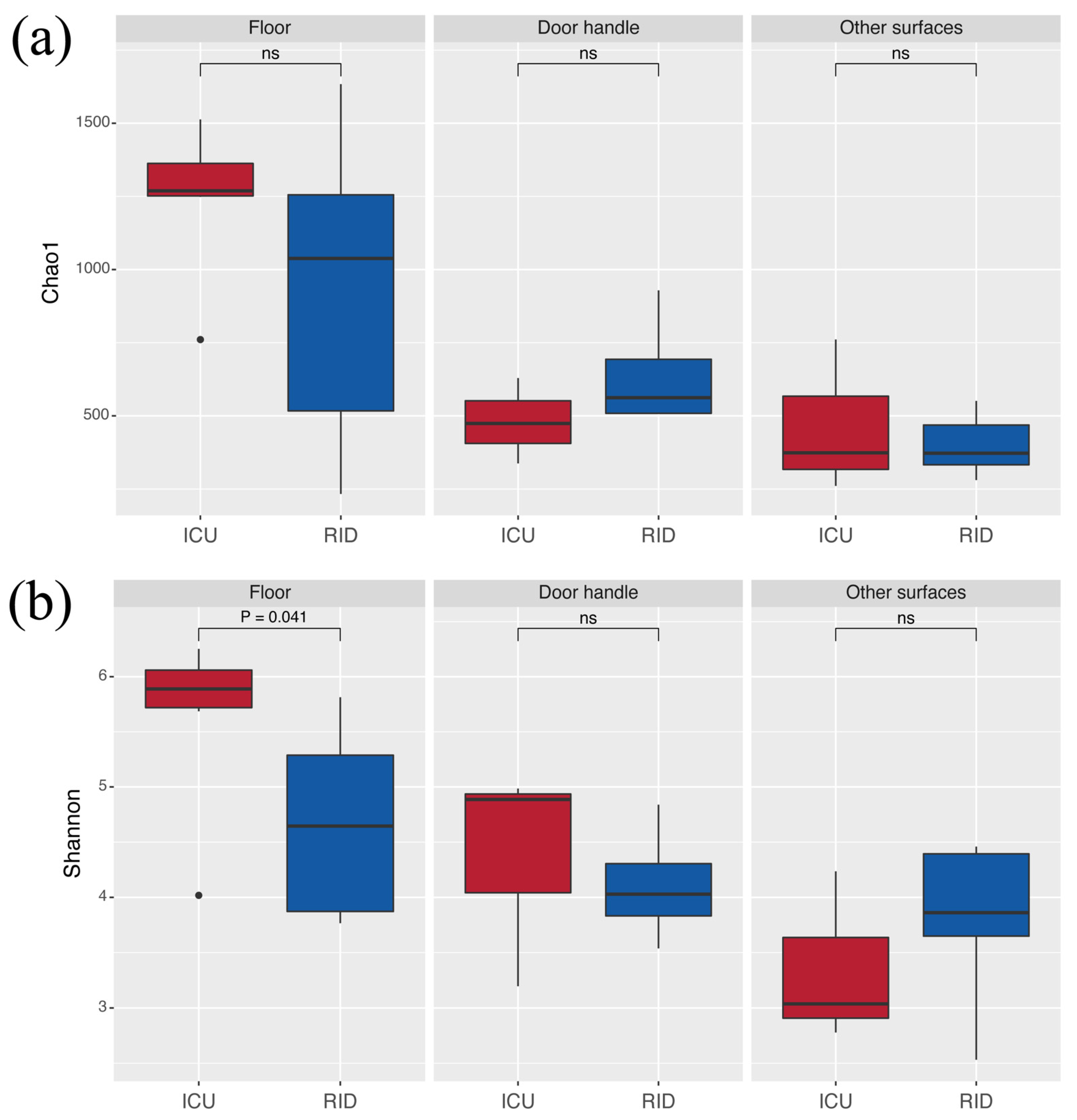
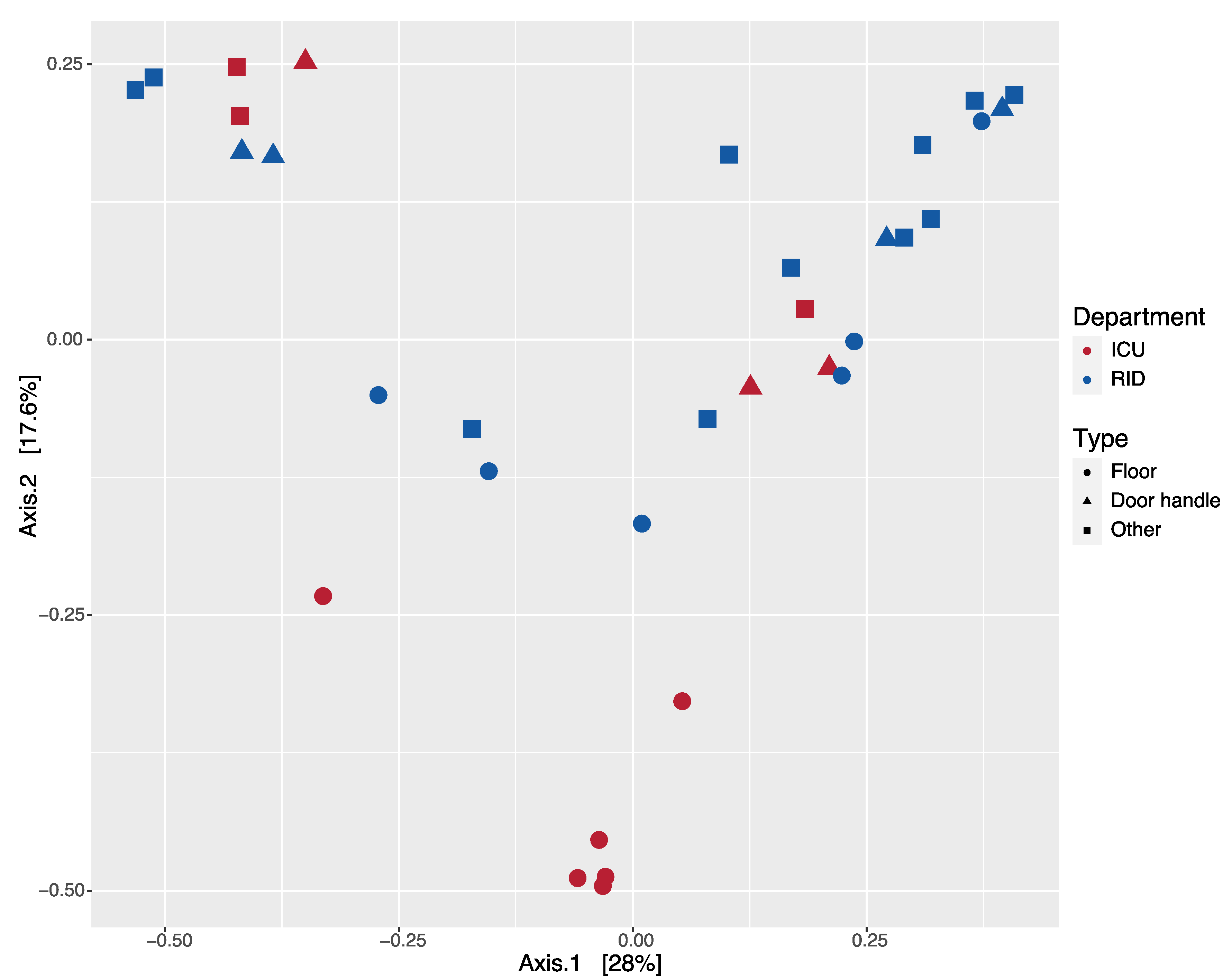
3.3. Diversity
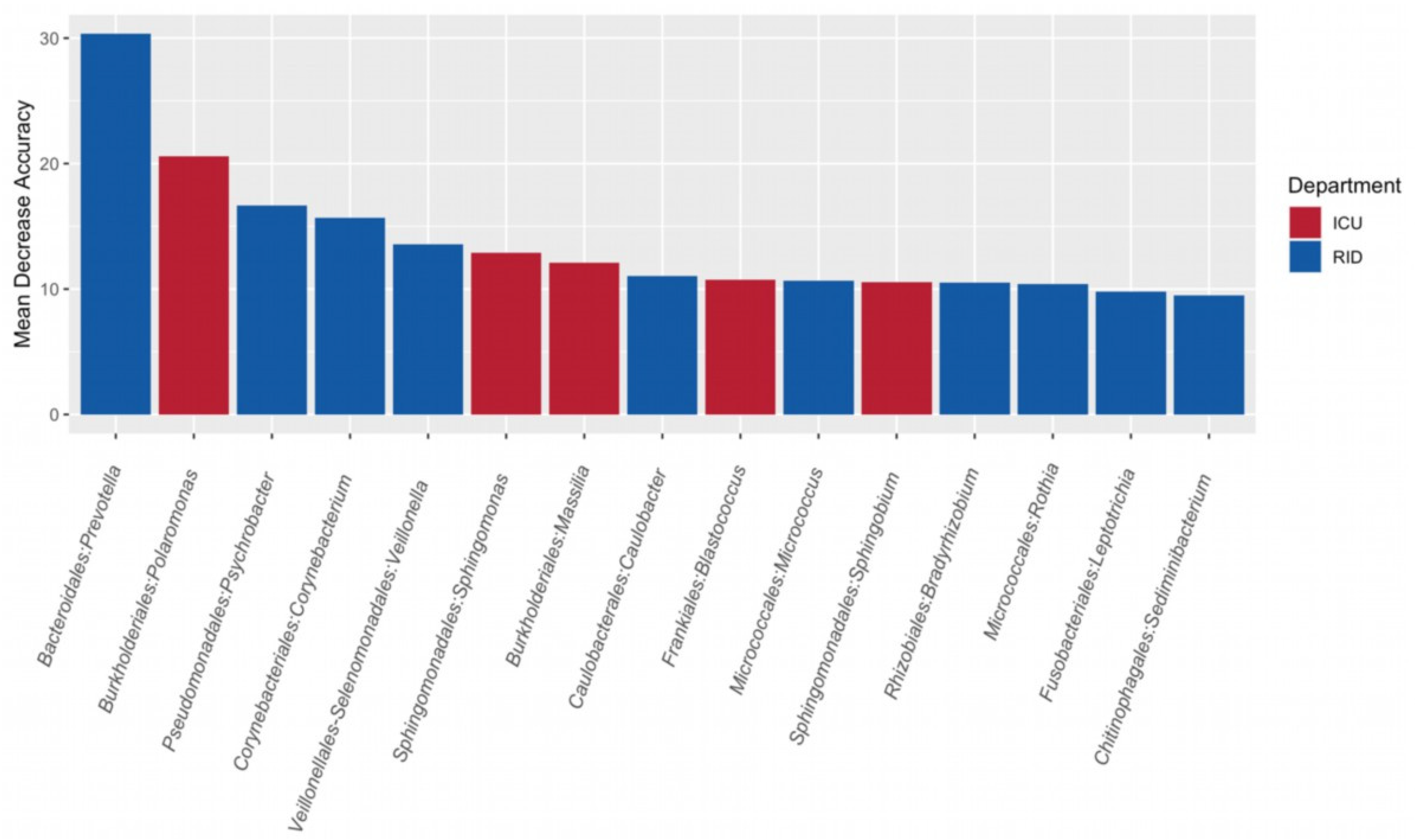
3.4. Search for Microbial Indicators of Departments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, X.; Chen, Z.; Azman, A.S.; Sun, R.; Lu, W.; Zheng, N.; Zhou, J.; Wu, Q.; Deng, X.; Zhao, Z.; et al. Neutralizing antibodies against SARS-CoV-2 variants induced by natural infection or vaccination: A systematic review and pooled meta-analysis. Clin. Infect. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Alenquer, M.; Ferreira, F.; Lousa, D.; Valério, M.; Medina-Lopes, M.; Bergman, M.-L.; Gonçalves, J.; Demengeot, J.; Leite, R.B.; Lilue, J.; et al. Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies. PLoS Pathog. 2021, 17, e1009772. [Google Scholar] [CrossRef]
- Ghinai, I.; McPherson, T.D.; Hunter, J.C.; Kirking, H.L.; Christiansen, D.; Joshi, K.; Rubin, R.; Morales-Estrada, S.; Black, S.R.; Pacilli, M.; et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet 2020, 395, 1137–1144. [Google Scholar] [CrossRef]
- Pastorino, B.; Touret, F.; Gilles, M.; de Lamballerie, X.; Charrel, R.N. Prolonged Infectivity of SARS-CoV-2 in Fomites. Emerg. Infect. Dis. 2020, 26, 2256–2257. [Google Scholar] [CrossRef]
- Peng, X.; Xu, X.; Li, Y.; Cheng, L.; Zhou, X.; Ren, B. Transmission routes of 2019-nCoV and controls in dental practice. Int. J. Oral Sci. 2020, 12, 1–6. [Google Scholar] [CrossRef]
- Richard, M.; Kok, A.; de Meulder, D.; Bestebroer, T.M.; Lamers, M.M.; Okba, N.M.A.; Fentener van Vlissingen, M.; Rockx, B.; Haagmans, B.L.; Koopmans, M.P.G.; et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020, 11, 3496. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.W.X.; Tan, Y.K.; Chia, P.Y.; Lee, T.H.; Ng, O.T.; Wong, M.S.Y.; Marimuthu, K. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) from a Symptomatic Patient. J. Am. Med. Assoc. 2020, 323, 1610–1612. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Qiao, F.; Zhou, F.; Yuan, Y. Aerosol and Surface Distribution of Severe Acute Respiratory Syndrome Coronavirus 2 in Hospital Wards, Wuhan, China, 2020. Indoor Built Environ. 2020, 26, 1586–1591. [Google Scholar] [CrossRef]
- Nissen, K.; Krambrich, J.; Akaberi, D.; Hoffman, T.; Ling, J.; Lundkvist, Å.; Svensson, L.; Salaneck, E. Long-distance airborne dispersal of SARS-CoV-2 in COVID-19 wards. Sci. Rep. 2020, 10, 19589. [Google Scholar] [CrossRef]
- Santarpia, J.L.; Rivera, D.N.; Herrera, V.L.; Morwitzer, M.J.; Creager, H.M.; Santarpia, G.W.; Crown, K.K.; Brett-Major, D.M.; Schnaubelt, E.R.; Broadhurst, M.J.; et al. Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Sci. Rep. 2020, 10, 12732. [Google Scholar] [CrossRef]
- Liu, Y.; Ning, Z.; Chen, Y.; Guo, M.; Liu, Y.; Gali, N.K.; Sun, L.; Duan, Y.; Cai, J.; Westerdahl, D.; et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature 2020, 582, 557–560. [Google Scholar] [CrossRef]
- Bardi, T.; Pintado, V.; Gomez-rojo, M.; Escudero-sanchez, R.; Lopez, A.A. Nosocomial infections associated to COVID-19 in the intensive care unit: Clinical characteristics and outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 495–502. [Google Scholar] [CrossRef]
- Li, Z.; Chen, Z.M.; Chen, L.D.; Zhan, Y.Q.; Li, S.Q.; Cheng, J.; Zhu, A.R.; Chen, L.Y.; Zhong, N.S.; Li, S.Y.; et al. Coinfection with SARS-CoV-2 and other respiratory pathogens in patients with COVID-19 in Guangzhou, China. J. Med. Virol. 2020, 92, 2381–2383. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Liu, W.; Jiang, M.; Huang, P.; Xiang, Z.; Deng, D.; Chen, P.; Xie, L. Clinical characteristics of COVID-19 patients with clinically diagnosed bacterial co-infection: A multi-center study. PLoS ONE 2021, 16, e0249668. [Google Scholar] [CrossRef] [PubMed]
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Westwood, D.; MacFadden, D.R.; Soucy, J.P.R.; Daneman, N. Bacterial co-infection and secondary infection in patients with COVID-19: A living rapid review and meta-analysis. Clin. Microbiol. Infect. 2020, 26, 1622–1629. [Google Scholar] [CrossRef]
- Lai, C.C.; Wang, C.Y.; Hsueh, P.R. Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-CoV-2 agents? J. Microbiol. Immunol. Infect. 2020, 53, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Arcari, G.; Raponi, G.; Sacco, F.; Bibbolino, G.; Di Lella, F.M.; Alessandri, F.; Coletti, M.; Trancassini, M.; Deales, A.; Pugliese, F.; et al. Klebsiella pneumoniae infections in COVID-19 patients: A 2-month retrospective analysis in an Italian hospital. Int. J. Antimicrob. Agents 2021, 57, 106245. [Google Scholar] [CrossRef]
- Cusumano, J.A.; Dupper, A.C.; Malik, Y.; Gavioli, E.M.; Banga, J.; Berbel Caban, A.; Nadkarni, D.; Obla, A.; Vasa, C.V.; Mazo, D.; et al. Staphylococcus aureus Bacteremia in Patients Infected with COVID-19: A Case Series. Open Forum Infect. Dis. 2020, 7, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Rusic, D.; Vilovic, M.; Bukic, J.; Leskur, D.; Seselja Perisin, A.; Kumric, M.; Martinovic, D.; Petric, A.; Modun, D.; Bozic, J. Implications of COVID-19 pandemic on the emergence of antimicrobial resistance: Adjusting the response to future outbreaks. Life 2021, 11, 220. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Schwebke, I.; Kampf, G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 2006, 6, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatta, D.R.; Hamal, D.; Shrestha, R.; Hosuru Subramanya, S.; Baral, N.; Singh, R.K.; Nayak, N.; Gokhale, S. Bacterial contamination of frequently touched objects in a tertiary care hospital of Pokhara, Nepal: How safe are our hands? Antimicrob. Resist. Infect. Control 2018, 7, 4–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Beek, J.; de Graaf, M.; Al-Hello, H.; Allen, D.J.; Ambert-Balay, K.; Botteldoorn, N.; Brytting, M.; Buesa, J.; Cabrerizo, M.; Chan, M.; et al. Molecular surveillance of norovirus, 2005–2016: An epidemiological analysis of data collected from the NoroNet network. Lancet Infect. Dis. 2018, 18, 545–553. [Google Scholar] [CrossRef]
- Desdouits, M.; de Graaf, M.; Strubbia, S.; Oude Munnink, B.B.; Kroneman, A.; Le Guyader, F.S.; Koopmans, M.P.G. Novel opportunities for NGS-based one health surveillance of foodborne viruses. One Health Outlook 2020, 2. [Google Scholar] [CrossRef]
- Rampelotto, P.H.; Sereia, A.F.R.; De Oliveira, L.F.V.; Margis, R. Exploring the hospital microbiome by high-resolution 16s rRNA profiling. Int. J. Mol. Sci. 2019, 20, 3099. [Google Scholar] [CrossRef] [Green Version]
- Pochtovyi, A.A.; Bacalin, V.V.; Kuznetsova, N.A.; Nikiforova, M.A.; Shidlovskaya, E.V.; Verdiev, B.I.; Milashenko, E.N.; Shchetinin, A.M.; Burgasova, O.A.; Kolobukhina, L.V.; et al. SARS-CoV-2 Aerosol and Surface Contamination in Health Care Settings: The Moscow Pilot Study. Aerosol Air Qual. Res. 2021, 21, 200604. [Google Scholar] [CrossRef]
- Chia, P.Y.; Coleman, K.K.; Tan, Y.K.; Ong, S.W.X.; Gum, M.; Lau, S.K.; Lim, X.F.; Lim, A.S.; Sutjipto, S.; Lee, P.H.; et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020, 11, 2800. [Google Scholar] [CrossRef]
- Hemati, S.; Mobini, G.R.; Heidari, M.; Rahmani, F.; Soleymani Babadi, A.; Farhadkhani, M.; Nourmoradi, H.; Raeisi, A.; Ahmadi, A.; Khodabakhshi, A.; et al. Simultaneous monitoring of SARS-CoV-2, bacteria, and fungi in indoor air of hospital: A study on Hajar Hospital in Shahrekord, Iran. Environ. Sci. Pollut. Res. 2021. [Google Scholar] [CrossRef]
- Hassan, M.Z.; Sturm-Ramirez, K.; Rahman, M.Z.; Hossain, K.; Aleem, M.A.; Bhuiyan, M.U.; Islam, M.M.; Rahman, M.; Gurley, E.S. Contamination of hospital surfaces with respiratory pathogens in Bangladesh. PLoS ONE 2019, 14, e0224065. [Google Scholar] [CrossRef] [Green Version]
- Price, E.P.; Arango, V.S.; Kidd, T.J.; Fraser, T.A.; Nguyen, T.K.; Bell, S.C.; Sarovich, D.S. Duplex real-time PCR assay for the simultaneous detection of Achromobacter xylosoxidans and Achromobacter spp. Microb. Genom. 2020, 6, 1–11. [Google Scholar] [CrossRef]
- Deschaght, P.; De Baere, T.; Van Simaey, L.; Van Daele, S.; De Baets, F.; De Vos, D.; Pirnay, J.P.; Vaneechoutte, M. Comparison of the sensitivity of culture, PCR and quantitative real-time PCR for the detection of Pseudomonas aeruginosa in sputum of cystic fibrosis patients. BMC Microbiol. 2009, 9, 244. [Google Scholar] [CrossRef] [Green Version]
- Martinucci, M.; Roscetto, E.; Iula, V.D.; Votsi, A.; Catania, M.R.; De Gregorio, E. Accurate identification of members of the Burkholderia cepacia complex in cystic fibrosis sputum. Lett. Appl. Microbiol. 2016, 62, 221–229. [Google Scholar] [CrossRef] [Green Version]
- Hartman, L.J.; Selby, E.B.; Whitehouse, C.A.; Coyne, S.R.; Jaissle, J.G.; Twenhafel, N.A.; Burke, R.L.; Kulesh, D.A. Rapid real-time PCR assays for detection of Klebsiella pneumoniae with the rmpA or magA genes associated with the hypermucoviscosity phenotype: Screening of nonhuman primates. J. Mol. Diagn. 2009, 11, 464–471. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 1–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Lucien, M.A.B.; Canarie, M.F.; Kilgore, P.E.; Jean-Denis, G.; Fénélon, N.; Pierre, M.; Cerpa, M.; Joseph, G.A.; Maki, G.; Zervos, M.J.; et al. Antibiotics and antimicrobial resistance in the COVID-19 era: Perspective from resource-limited settings. Int. J. Infect. Dis. 2021, 104, 250–254. [Google Scholar] [CrossRef]
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Soucy, J.P.R.; Westwood, D.; Daneman, N.; MacFadden, D.R. Antibiotic prescribing in patients with COVID-19: Rapid review and meta-analysis. Clin. Microbiol. Infect. 2021, 27, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peña, C.; Pujol, M.; Ardanuy, C.; Ricart, A.; Pallarés, R.; Liñares, J.; Ariza, J.; Gudiol, F. An outbreak of hospital-acquired Klebsiella pneumoniae bacteraemia, including strains producing extended-spectrum β-lactamase. J. Hosp. Infect. 2001, 47, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, T.; Harada, S.; Okamoto, K.; Ishino, S.; Kaneko, M.; Suzuki, M.; Ito, R.; Mizoguchi, M. COVID-19 and fatal sepsis caused by hypervirulent Klebsiella pneumoniae, Japan, 2020. Emerg. Infect. Dis. 2021, 27, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.; Papineni, S.; Ramachandruni, S.; Molina, J.; Surani, S. Achromobacter xylosoxidans/denitrificans Bacteremia in a Patient with Good’s Syndrome. Cureus 2021, 13, 11–15. [Google Scholar] [CrossRef]
- Awadh, H.; Mansour, M.; Aqtash, O.; Shweihat, Y. Pneumonia due to a Rare Pathogen: Achromobacter xylosoxidans, Subspecies denitrificans. Case Rep. Infect. Dis. 2017, 2017, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osman, H.; Nguyen, P. First Case of COVID-19 Complicated with Burkolderia Cepacia Pneumonia and Bacteremia. Chest 2020, 158, A544. [Google Scholar] [CrossRef]
- Zhong, H.; Wang, Y.; Shi, Z.; Zhang, L.; Ren, H.; He, W.; Zhang, Z.; Zhu, A.; Zhao, J.; Xiao, F.; et al. Characterization of respiratory microbial dysbiosis in hospitalized COVID-19 patients. Cell Discov. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Sirivongrangson, P.; Kulvichit, W.; Payungporn, S.; Pisitkun, T.; Chindamporn, A.; Peerapornratana, S.; Pisitkun, P.; Chitcharoen, S.; Sawaswong, V.; Worasilchai, N.; et al. Endotoxemia and circulating bacteriome in severe COVID-19 patients. Intensive Care Med. Exp. 2020, 8. [Google Scholar] [CrossRef]
- Rosas-Salazar, C.; Kimura, K.S.; Shilts, M.H.; Strickland, B.A.; Freeman, M.H.; Wessinger, B.C.; Gupta, V.; Brown, H.M.; Rajagopala, S.V.; Turner, J.H.; et al. SARS-CoV-2 infection and viral load are associated with the upper respiratory tract microbiome. J. Allergy Clin. Immunol. 2021, 147, 1226–1233.e2. [Google Scholar] [CrossRef]
- Ma, S.; Zhang, F.; Zhou, F.; Li, H.; Ge, W.; Gan, R.; Nie, H.; Li, B.; Wang, Y.; Wu, M.; et al. Metagenomic analysis reveals oropharyngeal microbiota alterations in patients with COVID-19. Signal Transduct. Target. Ther. 2021, 6. [Google Scholar] [CrossRef]
- Oie, S.; Hosokawa, I.; Kamiya, A. Contamination of room door handles by methicillin-sensitive/methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 2002, 51, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Caneiras, C.; Lito, L.; Melo-Cristino, J.; Duarte, A. Community-and hospital-acquired Klebsiella pneumoniae urinary tract infections in Portugal: Virulence and antibiotic resistance. Microorganisms 2019, 7, 138. [Google Scholar] [CrossRef] [Green Version]
- Engelhart, S.T.; Krizek, L.; Glasmacher, A.; Fischnaller, E.; Marklein, G.; Exner, M. Pseudomonas aeruginosa outbreak in a haematology-oncology unit associated with contaminated surface cleaning equipment. J. Hosp. Infect. 2002, 52, 93–98. [Google Scholar] [CrossRef]
- Nkuwi, E.J.; Kabanangi, F.; Joachim, A.; Rugarabamu, S.; Majigo, M. Methicillin-resistant Staphylococcus aureus contamination and distribution in patient’s care environment at Muhimbili National Hospital, Dar es Salaam-Tanzania. BMC Res. Notes 2018, 11, 4–9. [Google Scholar] [CrossRef]
- De Abreu, P.M.; Farias, P.G.; Paiva, G.S.; Almeida, A.M.; Morais, P.V. Persistence of microbial communities including Pseudomonas aeruginosa in a hospital environment: A potential health hazard. BMC Microbiol. 2014, 14, 118. [Google Scholar] [CrossRef] [Green Version]
| Bacterial Pathogen | Intensive Care Unit | Respiratory Infections Department | ||
|---|---|---|---|---|
| Surface (n = 13) | Patient (n = 5) | Surface (n = 22) | Patient (n = 6) | |
| Klebsiella pneumoniae | 13 (100%) | 0 (0%) | 14 (63.64%) | 0 (0%) |
| Pseudomonas aeruginosa | 1 (7.69%) | 0 (0%) | 6 (27.27%) | 0 (0%) |
| Staphylococcus aureus | 2 (15.38%) | 0 (0%) | 0 (0%) | 0 (0%) |
| CoNS | 13 (100%) | 0 (0%) | 22 (100%) | 0 (0%) |
| Achromobacter spp. | 3 (23.08%) | 0 (0%) | 3 (13.64%) | 0 (0%) |
| Burkholderia cepacia complex | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pochtovyi, A.A.; Vasina, D.V.; Kustova, D.D.; Divisenko, E.V.; Kuznetsova, N.A.; Burgasova, O.A.; Kolobukhina, L.V.; Tkachuk, A.P.; Gushchin, V.A.; Gintsburg, A.L. Contamination of Hospital Surfaces with Bacterial Pathogens under the Current COVID-19 Outbreak. Int. J. Environ. Res. Public Health 2021, 18, 9042. https://doi.org/10.3390/ijerph18179042
Pochtovyi AA, Vasina DV, Kustova DD, Divisenko EV, Kuznetsova NA, Burgasova OA, Kolobukhina LV, Tkachuk AP, Gushchin VA, Gintsburg AL. Contamination of Hospital Surfaces with Bacterial Pathogens under the Current COVID-19 Outbreak. International Journal of Environmental Research and Public Health. 2021; 18(17):9042. https://doi.org/10.3390/ijerph18179042
Chicago/Turabian StylePochtovyi, Andrei A., Daria V. Vasina, Daria D. Kustova, Elizaveta V. Divisenko, Nadezhda A. Kuznetsova, Olga A. Burgasova, Ludmila V. Kolobukhina, Artem P. Tkachuk, Vladimir A. Gushchin, and Alexander L. Gintsburg. 2021. "Contamination of Hospital Surfaces with Bacterial Pathogens under the Current COVID-19 Outbreak" International Journal of Environmental Research and Public Health 18, no. 17: 9042. https://doi.org/10.3390/ijerph18179042
APA StylePochtovyi, A. A., Vasina, D. V., Kustova, D. D., Divisenko, E. V., Kuznetsova, N. A., Burgasova, O. A., Kolobukhina, L. V., Tkachuk, A. P., Gushchin, V. A., & Gintsburg, A. L. (2021). Contamination of Hospital Surfaces with Bacterial Pathogens under the Current COVID-19 Outbreak. International Journal of Environmental Research and Public Health, 18(17), 9042. https://doi.org/10.3390/ijerph18179042








