Abstract
This paper analyzed the degradation pathways of petroleum hydrocarbon degradation bacteria, screened the main degradation pathways, and found the petroleum hydrocarbon degradation enzymes corresponding to each step of the degradation pathway. Through the Copeland method, the best inoculation program of petroleum hydrocarbon degradation bacteria in a polluted site was selected as follows: single oxygenation path was dominated by Streptomyces avermitilis, hydroxylation path was dominated by Methylosinus trichosporium OB3b, secondary oxygenation path was dominated by Pseudomonas aeruginosa, secondary hydroxylation path was dominated by Methylococcus capsulatus, double oxygenation path was dominated by Acinetobacter baylyi ADP1, hydrolysis path was dominated by Rhodococcus erythropolis, and CoA path was dominated by Geobacter metallireducens GS-15 to repair petroleum hydrocarbon contaminated sites. The Copeland method score for this solution is 22, which is the highest among the 375 solutions designed in this paper, indicating that it has the best degradation effect. Meanwhile, we verified its effect by the Cdocker method, and the Cdocker energy of this solution is −285.811 kcal/mol, which has the highest absolute value. Among the inoculation programs of the top 13 petroleum hydrocarbon degradation bacteria, the effect of the best inoculation program of petroleum hydrocarbon degradation bacteria was 18% higher than that of the 13th group, verifying that this solution has the best overall degradation effect. The inoculation program of petroleum hydrocarbon degradation bacteria designed in this paper considered the main pathways of petroleum hydrocarbon pollutant degradation, especially highlighting the degradability of petroleum hydrocarbon intermediate degradation products, and enriching the theoretical program of microbial remediation of petroleum hydrocarbon contaminated sites.
1. Introduction
The rapid development of the petroleum industry and its products, and the increase in the use of petroleum fossil fuels have led to serious environmental pollution. The pollution can be largely attributed to petroleum hydrocarbon pollutants [1]. Petroleum hydrocarbon pollutants are classified as priority environmental pollutants [2], which primarily exist in the form of n-alkanes, branched-chain alkanes, cycloalkanes, and aromatic hydrocarbons in the environment [3,4]. Most of them can exist stably in the environment for a long time and are not easily degraded. Crude oil leakage is one of the fundamental causes of petroleum hydrocarbon pollution, which has caused great harm to the soil ecosystem [5,6,7]. The petroleum hydrocarbon pollutants can affect the physical and chemical properties (such as soil texture, compactness, osmotic resistance, and the concentration and content of heavy metals) of soil [8]. Petroleum hydrocarbon pollutant-enriched soil adversely affects the growth of plants and animals, and in some cases can even cause death, resulting in extensive ecosystem damage [9,10,11,12]. At the same time, petroleum hydrocarbon pollutants exhibit carcinogenic, neurotoxic, genus toxic, and other toxicological properties, which pose a serious threat to human health [13,14]. Therefore, the repair of petroleum-contaminated sites has become a focus of attention.
Petroleum hydrocarbon pollution has always been a concern in various countries, and standards have been set in various countries for petroleum hydrocarbon pollution. The US government has established regulations or guidelines for some petroleum hydrocarbon fractions and their components to protect the public from the potentially harmful health effects of petroleum compounds. In turn, all states have their own standards and methods for cleaning up petroleum hydrocarbons or their components [15]. For example, the Washington State Department of Ecology has set limits for soils of 120–6000 mg/kg (gasoline series organics), 240–6000 mg/kg (diesel series organics) and 3600–4400 mg/kg (residual petroleum hydrocarbons) [16]. Compared to developed countries such as the USA, UK, and Canada, Asia’s petroleum hydrocarbon contaminated land management systems lag behind in their effectiveness in identifying petroleum hydrocarbon contaminated sites, conducting appropriate detailed risk assessments and initiating remediation activities. Asian countries generally lack specific regulations for the management of petroleum hydrocarbon contamination of soil and groundwater. In those countries or regions that do not have any cleanup standards, or legal and regulatory frameworks in place to assess and manage petroleum hydrocarbon, contaminated sites still use the US EPA standards [17]. The latest Soil Environmental Quality Soil Contamination Risk Control Standard for Construction Land (Trial) (GB 36600-2018), promulgated by China in 2018, sets out risk screening values for soil contamination based on human health, but this standard only sets limit values for extractable petroleum hydrocarbons (C10 to C40). Under China’s current environmental management framework, there are still many gaps in the control of petroleum-based pollutants in soil, and the few contaminated sites that have been evaluated and remediated have had to draw on standards already in place in other countries. Therefore, further improving the delineation and establishing supporting analytical methods for petroleum hydrocarbon pollutants is one of the priorities for future environmental management in China.
To achieve effective remediation of petroleum contaminated sites, suitable remediation methods need to be selected based on the nature of the land type and extent of contamination of the petroleum contaminated sites [18,19,20]. Chemical drenching, electrokinetic remediation, solvent extraction, microwave radiation, and bioremediation are currently the commonly used methods to realize the remediation of petroleum-contaminated sites [21,22,23]. Among them, bioremediation is the most important method followed for the remediation of oil-contaminated sites. This method does not cause secondary pollution and is economical and effective in realizing remediation. Therefore, it has been used as a common remediation method to repair petroleum-contaminated sites [24]. The microbial remediation method, another of the commonly used bioremediation methods, can be used to remove petroleum hydrocarbon pollutants present. In this method, the microbial population in the environment is biodegraded [25]. This method is more economical and environmentally friendly than other remediation methods [26].
Petroleum hydrocarbon degradation bacteria first convert petroleum hydrocarbon pollutants to intermediates through the process of intracellular oxidation. In the next step, the products obtained in the previous step are used for the degradation reaction. Petroleum hydrocarbon pollutants are finally degraded into environmentally friendly water and carbon dioxide [27]. When the number of native petroleum hydrocarbon degradation bacteria in petroleum contaminated sites is low, exogenous microorganisms are inoculated into the contaminated sites to be remediated [28,29]. Mishra et al. [30] used Pseudomonas aeruginosa PSA5 and Rhodococcus sp. NJ2 for the studies and found that these bacteria could degrade n-hexadecane in petroleum sludge via the terminal oxidation pathway. The degradation rates reached 99% and 95% in 10 days. Zhang et al. [31] found that Pseudomonas aeruginosa DQ8 degraded 34.5% of pyrene via the singlet and dioxygen pathways in 12 days. Yu et al. [32] isolated and domesticated two highly efficient oil-degrading strains, Bacillus subtilis SWH-1 and Sphingomonas polyphila SWH-2, from different oil fields, and prepared them into highly efficient microbial formulations to be put into the contaminated site of Shengli oil field. However, most researchers have only investigated the remediation of petroleum hydrocarbon contaminated sites in the presence of one or more petroleum degradation bacteria following specific reaction steps in the degradation pathway. Results on the remediation of petroleum hydrocarbon contaminated sites in the presence of multiple petroleum hydrocarbon degradation bacteria, a complete analysis of the degradation pathway of the petroleum hydrocarbon degradation bacteria, and all the reaction steps have not been reported.
This paper presents the results of the analysis of the degradation pathways of petroleum hydrocarbon pollutants in the presence of petroleum degradation bacteria. A variety of degradation enzymes according to the degradation path was selected to design the inoculation program of the petroleum hydrocarbon degradation bacteria. Firstly, the molecular dynamics method was used to calculate the binding energy of the degradation enzymes and petroleum hydrocarbon pollutants in each step to determine the degradation ability. Following this, a Java program was developed to arrange and combine the degradation enzymes to develop a variety of petroleum hydrocarbon degradation bacteria inoculation programs. Finally, a variety of inoculation programs of petroleum hydrocarbon degradation bacteria were scored by the Copeland method, and the best inoculation program was selected according to the score of each program. Our aim is to design a petroleum hydrocarbon degradation bacterial inoculation program for the remediation of petroleum hydrocarbon contaminated sites. This study will consider the main pathways of petroleum hydrocarbon contaminant degradation and provide a new direction for further research on the microbial remediation of petroleum hydrocarbon contaminated sites.
2. Materials and Methods
2.1. Selection of Degradation Enzymes of Petroleum Hydrocarbon Degradation Bacteria
The degradation pathways of the three petroleum hydrocarbon pollutants (n-tetradecane, norphytane, and cyclopentane) were studied. It was found that the degradation pathways of n-tetradecane, norphytane, and cyclopentane similarly involved the participation of similar enzymes. The degradable proteins of the same petroleum hydrocarbon degradation bacteria were selected to study the degradation ability and the degradation pathways of n-tetradecane, norphytane, and cyclopentane. For the single oxygenation pathway, five types of monooxygenases, membrane-integrated methane monooxygenase of Methylomicrobium alcaliphilum 20Z (PDB ID: 6CXH) [33], flavin-dependent monooxygenase of Cellvibrio sp. BR (PDB ID: 4USQ) [34], flavin-dependent monooxygenase of Nitrincola lacisaponensis (PDB ID: 6HNS) [35], bacterial cytochrome P450 enzyme of Bacillus megaterium (PDB ID: 1SMJ) [36], and cytochrome P450 enzyme of Streptomyces avermitilis (PDB ID: 3E5K) [37] were selected. The hydroxylation pathway was conducted using five kinds of hydroxylases: methane monooxygenase hydroxylase of Methylococcus capsulatus (PDB ID: 1FZI) [38], methane monooxygenase hydroxylase of Methylosinus trichosporium (PDB ID: 1MHZ) [39], long-chain alkane monooxygenase of Geobacillus thermodenitrificans (PDB ID: 3B9N) [40], methane monooxygenase hydroxylase of Methylosinus sporium (PDB ID: 6D7K) [41], and methane monooxygenase hydroxylase of Methylosinus trichosporium OB3b (PDB ID: 6VK6) [42]. The secondary single oxygenation pathway was carried out in the presence of the cytochrome P450 enzyme of Streptomyces avermitilis (PDB ID: 3E5K) [37], alcohol dehydrogenase of Escherichia coli (PDB ID: 7BU3) [43], alcohol dehydrogenase of Saccharomyces cerevisiae (PDB ID: 1Q1N) [44], aldehyde dehydrogenase of Staphylococcus aureus (PDB ID: 6K10) [45], and aldehyde dehydrogenase of Pseudomonas aeruginosa (PDB ID: 4CAZ) [46]. The secondary hydroxylation pathway was conducted in the presence of the methane monooxygenase hydroxylase of Methylococcus capsulatus (PDB ID: 1FZI) [38], methane monooxygenase hydroxylase of Methylosinus sporium (PDB ID: 6D7K) [41], and methane monooxygenase hydroxylase of Methylosinus trichosporium OB3b (PDB ID: 6VK6) [42].
The degradation pathways of benzene and the types of degradation enzymes used were different from those used for the degradation of the other three petroleum hydrocarbon pollutants. The degraded proteins selected in each step of the benzene degradation path were identified: the hydroxylation pathway involved the participation of the methane monooxygenase hydroxylase of Methylococcus capsulatus (PDB ID: 1FZI) [38], methane monooxygenase hydroxylase of Methylosinus trichosporium (PDB ID: 1MHZ) [39], long-chain alkane monooxygenase of Geobacillus thermodenitrificans (PDB ID: 3B9N) [40], methane monooxygenase hydroxylase of Methylosinus sporium (PDB ID: 6D7K) [41], and methane monooxygenase hydroxylase of Methylosinus trichosporium OB3b (PDB ID: 6VK6) [42]. The secondary single oxygenation pathway involved the participation of five kinds of dioxygenases: procatechuic acid 2,3-dioxygenase of Brevibacterium fuscum (PDB ID: 2IGA) [47], catechol 1,2-dioxygenase of Acinetobacter baylyi ADP1 (PDB ID: 1DLT) [48], catechin acid 1,2-dioxygenase of Pseudomonas putida (PDB ID: 2AZQ) [49], 2,3-dioxygenase of Pseudomonas alkylphenolica (PDB ID: 3HPY) [50], and catechol 1,2-dioxygenase of Acinetobacter radioresistens OB3b (PDB ID: 2XSR) [51]. The hydrolysis pathway involved the participation of three kinds of hydrolases: epoxide hydrolase of Aspergillus niger (PDB ID: 3G02) [52], epoxide hydrolase of Rhodococcus erythropolis (PDB ID: 4XBT) [53], and epoxide hydrolase of Bacillus megaterium (PDB ID: 4IO0) [54]. The CoA pathway selected the benzene CoA reductase of Geobacter metallireducens GS-15 (PDB ID: 4Z3W) [55].
The proteins used for the studies were all from the PDB database (http://www.rcsb.org/pdb accessed on 9 June 2021).
2.2. Ability to Characterize Microbial Remediation of Petroleum Hydrocarbon Contaminated Sites Using the Molecular Dynamics Method
Previous studies have shown that the binding energy between receptors and molecules can indicate the degree of binding between them. The greater the absolute value of binding energy, the stronger the binding ability of the petroleum hydrocarbon degradation enzymes. In this paper, the Libdock docking module in Discovery Studio 2020 software version (Biovia, Waltham, MA, USA) was used to dock petroleum hydrocarbon contaminant molecules and study their degradation process intermediates. Protein molecules were pretreated prior to docking to remove ligands and water from the protein samples. According to the degradation path of each step and the degradation intermediates produced, the degradation intermediates were docked with the petroleum hydrocarbon degradation enzyme corresponding to each step of the degradation path. The docked complex of petroleum hydrocarbon degradation enzyme-petroleum hydrocarbon contaminant molecule/petroleum hydrocarbon contaminant degradation intermediate was moved to GROMACS 5.1.4 software version (University of Groningen, city, Groningen, the Netherlands) on the Dell PowerEdge R7425 server to conduct the molecular dynamics simulation calculations. The complex was placed in a periodic dodeca-cube with 10 nm-long sides in molecularly constrained spaces using a GROMOS96 43a1 force field. The Na+ ions were added to neutralize the charge of the system. The petroleum hydrocarbon degradation enzyme-petroleum hydrocarbon contaminant molecule/petroleum hydrocarbon contaminant degradation intermediate product molecule complex system was set up as a group and subjected to energy minimization simulations based on the steepest gradient method. The number of simulation steps was set at 50,000. Both the heat and pressure bath simulation times for the system were 100 ps, and the pressure bath size was set to a constant standard atmospheric pressure of 1 bar. The kinetic simulation operation time of different groups was set to 20 ps, and the binding energy of petroleum hydrocarbon degradation enzyme-petroleum hydrocarbon pollutant molecule/petroleum hydrocarbon pollutant degradation intermediate molecular complex system under different external environment stimulation was calculated.
2.3. Design of Inoculation Program for Petroleum Hydrocarbon Degradation Bacteria: Use of Java
For n-alkanes, branched-chain alkanes, and cycloalkanes, it is necessary to select a degradable protein that exhibits good degradability for the three petroleum hydrocarbon pollutants in each degradation path. Therefore, this requires the selection (via permutation and combination) of a variety of degraded proteins that can participate in each of the selected degradation paths to design the best inoculation plan for the petroleum hydrocarbon degradation bacteria. The single oxygenation pathway, hydroxylation pathway and secondary single oxygenation pathway involved the selection of one enzyme from each of the five enzymes. One enzyme was selected from the three enzymes in secondary hydroxylation pathway. A permutation and combination program was developed based on the Java language to simplify the calculation method (see the Supplementary Material File S1 for the specific program code).
For the design of the inoculation program of aromatic hydrocarbon degradation bacteria, the petroleum hydrocarbon degradation enzyme with the strongest binding ability was directly selected for inoculation and matching.
2.4. Optimization of the Microbial Remediation Inoculation Screening Method in Petroleum Hydrocarbon Contaminated Sites—Copeland Method
Copeland, A.H., University of Michigan, first proposed the Copeland Method in the seminar “On the Application of Mathematics in Social Sciences” [56]. It was initially applied to the field of election. Now it is widely used in logistics and transportation, event statistics, agriculture, and environmental science, among other fields. Caillaux et al. used the Copeland method to select the most suitable shipping route for container cargo between different coupling ports to save transportation cost and time [57]. Li et al. used the Copeland method to evaluate the ecological hazards of 61 environmental priority pollutants from two aspects of environmental behavior and biological toxicity index and predicted the ecological hazards of the new chemicals [58].
The core idea of the Copeland method is that “the minority is subordinate to the majority”. The calculation process is simple and convenient. Hence, this method was selected to design the best inoculation program for the petroleum hydrocarbon degradation bacteria. Firstly, each degradation enzyme in each degradation pathway was taken as the evaluation factor, and the degradation ability for each kind of petroleum hydrocarbon pollutant was taken as the evaluation index to determine the individual score. Following this, the evaluation scores of each degradation enzyme for the three kinds of petroleum hydrocarbon pollutants were added to obtain the overall score of each degradation enzyme. Finally, the overall evaluation score of each program was obtained by synthesizing the overall score of each degradation enzyme in the four-step degradation pathway. The best inoculation program for the petroleum hydrocarbon degradation bacteria was selected based on the score results. The specific evaluation steps of the Copeland method were determined: X evaluation factors were set, and each evaluation factor had N multiple evaluation indexes. The same evaluation index of each evaluation factor was compared in pairs. In the comparison process, the one with the good evaluation index was marked as +1, the one with the bad evaluation index was marked as −1, and the one with the same evaluation index was marked as 0. Following the evaluation of each of the evaluation factors and evaluation indices, the evaluation scores were added to find the final score. Its definition is as follows:
Finally, the score T for each factor was added to determine the final ranking. When the sum of the scores of multiple factors is the same, the factor with the smallest variance is selected as the best program.
2.5. Verification of the Effect of the Best Inoculation Program for Petroleum Hydrocarbon Degradation Bacteria—The Molecular Dynamics Method
The degradation effect of the optimal inoculation program of petroleum hydrocarbon degradation bacteria was verified using the molecular docking method. Firstly, the four enzymes used in each program were docked by the ZDOCK module in Discovery Studio 2020 software version (Biovia, Waltham, MA, USA). The comprehensive petroleum hydrocarbon degradation enzymes were obtained by setting the RMSD Cut-off to cluster radius and Interface Cutoff to 9.0. ZRank was set to False to dock the four degradation enzymes one by one. The Cdocker module was then used to dock n-tetradecane, norphytane, cyclopentane, and their respective three intermediate degradation products to obtain the Cdocker energy values to characterize the overall degradation capacity of the four degradation enzymes.
3. Results and Discussion
3.1. Identification of the Main Degradation Pathways for Microbial Degradation of Petroleum Hydrocarbon Pollutants
It has been previously reported that the microbial degradation of straight-chain alkanes is primarily carried out via four metabolic pathways: single-terminal oxidation, double-terminal oxidation, sub-terminal oxidation, and direct dehydrogenation [59]. Branched-chain alkanes are primarily degraded through the single-terminal oxidation and double-terminal oxidation processes. Some microorganisms degrade branched-chain alkanes through the sub-terminal oxidation pathway [60]. Cycloalkanes are mainly metabolized through the processes of single-terminal oxidation, sub-terminal oxidation, and direct dehydrogenation [61]. When microorganisms degrade aromatic hydrocarbons, they first degrade the aromatic hydrocarbons into catechins, and then break the benzene ring through the processes of orthotopic cracking and eccentricity cracking under the action of dioxygenase [62]. In this paper, the representative contaminant molecules, n-tetradecane, norphytane, cyclopentane, and benzene, were selected as the research objects. The molecular structures of the pollutants were downloaded from the PubChem molecular library (http://pubchem.ncbi.nlm.nih.gov accessed on 7 June 2021). As the degradation pathway of the petroleum hydrocarbon pollutants is complex, we studied the main steps in the degradation pathway. The main degradation pathways for n-tetradecane, norphytane, cyclopentane, and benzene are shown in Figure 1, Figure 2, Figure 3 and Figure 4.
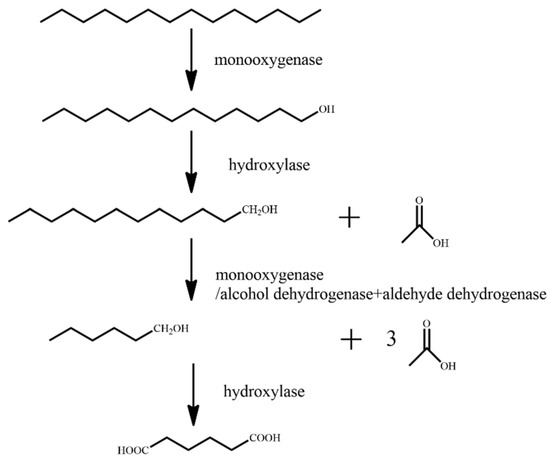
Figure 1.
Degradation pathway of n-tetradecane [63].
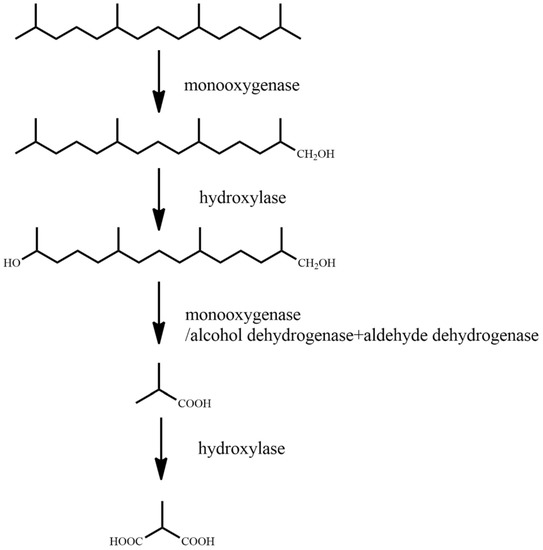
Figure 2.
Degradation pathway of norphytane [64].
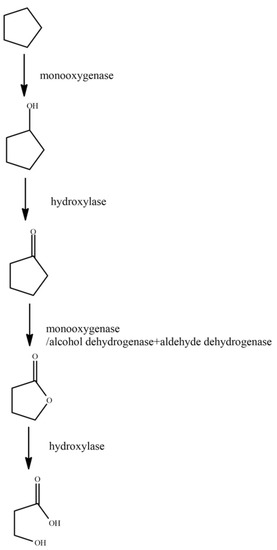
Figure 3.
Degradation pathway of cyclopentane [65].
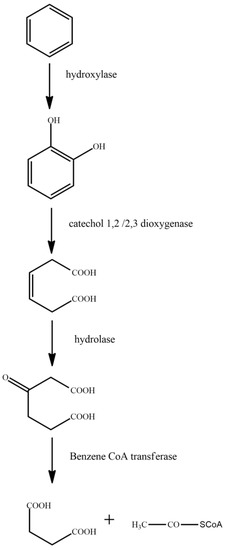
Figure 4.
Degradation pathway of benzene [66].
Firstly, under the action of monooxygenase, n-tetradecane is single-terminally oxidized, and the terminal methyl group is oxidized to hydroxyl to generate tridecane-1-ol. Following this, under the action of hydroxylase, tridecane-1-ol is oxidized to dodecan-1-ol, and acetic acid is generated. Under the action of monooxygenase/alcohol dehydrogenase and aldehyde dehydrogenase, dodecan-1-ol is oxidized to hexane-1-ol, and three molecules of acetic acid is generated. Finally, hexan-1-ol is oxidized to adipic acid under the action of hydroxylase.
For norphytane, single-terminal oxidation is performed on norphytane under the action of monooxygenase, and the terminal methyl group is oxidized to hydroxyl to generate 2,6,10,14-tetramethylpentadecane-1-alcohol. Following this, under the action of hydroxylase, 2,6,10,14-tetramethylpentadecan-1-ol is oxidized to 2,6,10-trimethylpentadecane-1,14-diol, and under the repeated action of monooxygenase/alcohol dehydrogenase and aldehyde dehydrogenase, 2,6,10-trimethylpentadecane-1,14-diol is oxidized to 2-methyl-1-propanol. Finally, under the action of hydroxylase, 2-methyl-1-propanol is oxidized to 2-methyl-1,3-propanediol.
Cyclopentane is firstly oxidized at one end under the action of monooxygenase. The terminal methyl group is oxidized to the hydroxyl group to generate cyclopentanol. Under the action of hydroxylase, cyclopentanol is oxidized to cyclopentanone, and under the action of monooxygenase/alcohol dehydrogenase and aldehyde dehydrogenase, cyclopentanone is oxidized to dihydrofuran-2(3H)-one. Finally, under the action of hydroxylase, dihydrofuran-2(3H)-one is oxidized to 3-hydroxypropionic acid.
Benzene is first degraded to catechol under the action of hydroxylase. Following this, under the action of catechol 1,2/2,3 dioxygenase, the benzene ring in catechol is broken to form (Z)-hexa-3-enedione acid. The produced (Z)-hexa-3-enedione acid is converted to 3-oxoadipate under the action of hydrolase. Finally, 3-oxoadipate is converted to succinic acid by the action of benzene-CoA transferase.
As petroleum hydrocarbon degradation is a very complicated process, it usually takes dozens or even hundreds of steps to degrade petroleum hydrocarbon pollutants into non-toxic and harmless water and carbon dioxide molecules. The bioconcentration and long-distance migration of the final product were predicted using the EPI database. The results are shown in Table 1. The bioconcentration factor logBCF [67] was used to characterize petroleum hydrocarbon pollutants and the bioconcentration of the degradation products. The larger the logBCF value, the substance is more likely to accumulate in the organism. In other words, the substance is more biohazardous. The N-octanol-air partition coefficient (logKOA) [68] was used to characterize the long-distance migration of petroleum hydrocarbon pollutants and their degradation products. The larger the log Koa value, the easier it is for the substance to migrate over long distances. Under these conditions, it is difficult to carry out centralized degradation and treatment. We observed that, compared with the original petroleum hydrocarbon pollutants, the bioconcentration and long-distance migration of the final product were significantly reduced. Therefore, only the single oxygenation pathway, hydroxylation pathway, secondary single oxygenation pathway, and secondary hydroxylation pathway were selected as the main degradation pathways of n-alkanes, branched-chain alkanes, and cycloalkanes. The hydroxylation pathway, 9eoxygenation pathway, hydrolysis pathway, and CoA pathway were selected as the main degradation pathways of aromatic hydrocarbons. The subsequent degradation pathways were not considered for the studies.

Table 1.
Comparison of bioconcentration and long-distance migration of petroleum hydrocarbon pollutants and their degradation products.
3.2. Calculation of Binding Energy of the Degraded Protein of Petroleum Hydrocarbon Degradation Bacteria and Petroleum Hydrocarbon Pollutants
We used the GROMACS 5.1.4 software version (University of Groningen, city, Groningen, the Netherlands) to perform the molecular dynamics simulation calculations on the Dell PowerEdge R7425 server. The binding energy of the degraded protein and petroleum hydrocarbon pollutants under each degradation pathway were calculated to characterize the degradation ability of degradation enzymes. A study of the main degradation pathways identified by us revealed that each degradation pathway corresponded to different degradation enzymes and intermediate degradation products. The calculation results of the binding energy of the degraded protein and petroleum hydrocarbon pollutants and the corresponding intermediate degradation products under each degradation path are shown in Table 2, Table 3, Table 4, Table 5 and Table 6.

Table 2.
Calculation results of the binding energy of each degraded protein and petroleum hydrocarbon pollutants under single oxygenation pathway.

Table 3.
Calculation results of binding energy of each degraded protein and intermediate degradation products of petroleum hydrocarbon pollutants under hydroxylation pathway.

Table 4.
Calculation results of binding energy of each degraded protein and intermediate degradation products of petroleum hydrocarbon pollutants under secondary single oxygenation pathway.

Table 5.
Calculation results of binding energy of each degraded protein and intermediate degradation products of petroleum hydrocarbon pollutants under secondary hydroxylation pathway.

Table 6.
Number of petroleum hydrocarbon degradation enzymes.
The binding energy of each degraded protein with petroleum hydrocarbon pollutants can be used to understand the degradation ability of the degraded protein. As shown in Table 2, under conditions of the single oxygenation pathway, the cytochrome P450 enzyme 3E5K exhibits the strongest degradation ability for n-tetradecane and norphytane. The bacterial cytochrome P450 enzyme 1SMJ exhibits the strongest degradation ability for cyclopentane. As shown in Table 3, under conditions of the hydroxylation pathway, the methane monooxygenase hydroxylase 1FZI exhibits the strongest degradation ability for tridecane-1-ol, the methane monooxygenase hydroxylase 1MHZ exhibits the strongest degradation ability for the degradation products of 2,6,10,14-tetramethylpentadecan-1-ol, and the methane monooxygenase hydroxylase 6D7K exhibits the strongest degradation ability for cyclopentanol. As shown in Table 4, under conditions of the secondary single oxygenation pathway, the aldehyde dehydrogenase 4CAZ exhibits the strongest degradation ability for dodecan-1-ol and cyclopentanone and cytochrome P450 enzyme 3E5K exhibits the strongest degradation ability for 2,6,10-trimethylpentadecane-1,14-diol. As shown in Table 5, under the conditions of the secondary hydroxylation pathway, the methane monooxygenase hydroxylase 6D7K exhibits the strongest degradation ability for hexan-1-ol, the methane monooxygenase hydroxylase 6VK6 exhibits the strongest degradation ability for 2-methyl-1-propanol, and the methane monooxygenase hydroxylase 1FZI exhibits the strongest degradation ability for dihydrofuran-2(3H)-one. As shown in Table 6, the strongest ability to degrade benzene is exhibited by the methane monooxygenase hydroxylase 6VK6 under the conditions of the hydroxylation pathway. The strongest ability to degrade catechol is exhibited by the catechol 1,2-dioxygenase 1DLT under the conditions of the double oxygenation pathway. The strongest ability to degrade (Z)-hexa-3-enedione acid is exhibited by the epoxide hydrolase 4XBT under the conditions of the hydrolysis pathway. Under conditions of the CoA pathway, only the benzene CoA reductase 4Z3W was selected to degrade 3-oxohexanedioic acid.
Figure 5a–d show the heat maps of the degradation ability of various degradation enzymes. Based on the above results for each petroleum hydrocarbon pollutant, it can be concluded that the enzymes with the best degradation efficiency vary from pathway to pathway. Therefore, the comprehensive degradation ability of the degradation enzymes corresponding to each degradation path should be comprehensively studied to obtain an inoculation program that exhibits high degradability for all three petroleum hydrocarbon pollutants.
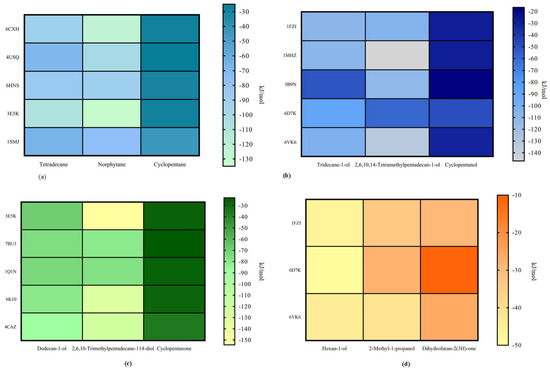
Figure 5.
Degradability of petroleum hydrocarbon pollutants and intermediate degradation products by each degradation enzymes (a) single oxygenation pathway, (b) hydroxylation pathway, (c) single oxygenation pathway, and (d) secondary hydroxylation.
3.3. Design of Inoculation Program for Petroleum Hydrocarbon Degradation Bacteria Based on the Java Language: A Permutation and Combination Program
For the four-step degradation pathway, one of the five monooxygenases in the monooxygenase pathway, one of the five hydroxylases in the hydroxylation pathway, one of the five monooxygenases/aldehyde dehydrogenases/alcohol dehydrogenases in the secondary monooxygenation pathway, and one of the enzymes from the three hydroxylases in the secondary hydroxylation pathway for permutation and combination were selected to formulate the inoculation plan for the petroleum hydrocarbon degradation bacteria that degrade normal alkanes, branched-chain alkanes, and cycloalkanes. The designed program was used for the permutation and combination studies of the inoculation programs of petroleum hydrocarbon degradation bacteria. Finally, 375 inoculation programs of petroleum hydrocarbon degradation bacteria were obtained. The number of each petroleum hydrocarbon degradation enzyme is shown in Table 6. Although the hydroxylation pathway and the secondary hydroxylation pathway involve the presence of the same degradation enzymes, the degradation products of petroleum hydrocarbons degraded following the two degradation pathways are different. The two parts of the same degradation enzymes were numbered in two ways to explore the degradation ability of these enzymes.
3.4. Determination of the Optimal Inoculation Program of Petroleum Hydrocarbon Degradation Bacteria Based on the Copeland Method
In this paper, the Copeland method was used to determine the best petroleum hydrocarbon degradation bacteria inoculation plan for the degradation of n-alkanes, branched-chain alkanes, and cycloalkanes. First, the three petroleum hydrocarbon pollutants were evaluated one by one in the presence of each degradation enzyme for each degradation path. Following this, the evaluation scores of the three petroleum hydrocarbon pollutants were summed to obtain the comprehensive degradability evaluation score for each enzyme. The comprehensive degradation ability evaluation score of each degradation enzyme was substituted into 375 petroleum hydrocarbon degradation bacteria inoculation programs for program evaluation, and the evaluation score of each program was obtained to select the best petroleum hydrocarbon degradation bacteria inoculation program. The evaluation scores of 375 kinds of petroleum hydrocarbon degradation bacteria inoculation programs are shown in Figure 2.
As shown in Figure 6, programs [4, 7, 15, 16] and [4, 10, 15, 16] exhibit the highest evaluation scores, both of which are 22. No. 4, No. 15, and No. 16 degradation enzymes were selected for the other three steps in the two groups. To screen out the best petroleum hydrocarbon degradation bacteria inoculation plan, the binding energies of No. 7 and No. 10 degradation enzymes and petroleum hydrocarbon pollutants in the hydroxylation path in these two groups of plans were calculated. The best inoculation program was determined by comparing the variance of degradation enzymes in the hydroxylating pathway. The sample variance calculation formula is represented as follows:
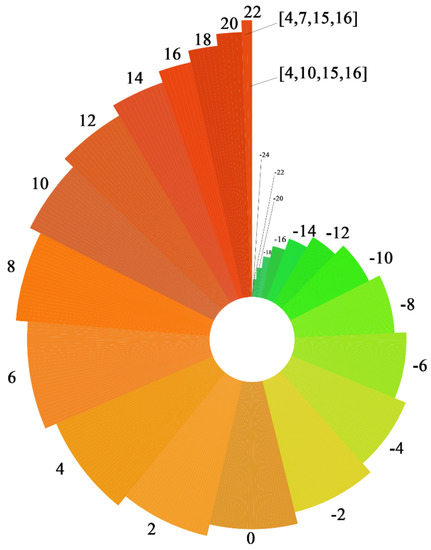
Figure 6.
Score for 375 petroleum hydrocarbon degradation bacteria inoculation programs.
The variances of degradation enzymes No. 7 and No. 10 of the hydroxylation pathway with petroleum hydrocarbon pollutants are shown in Table 7. It can be seen from Table 7 that the variance of the program [4, 10, 15, 16] is the least. Therefore, program [4, 10, 15, 16] is the best program for the inoculation of petroleum hydrocarbon-degradation bacteria.

Table 7.
Variance of degradation enzyme in the second step degradation pathway.
Therefore, the optimal combination of the degradation enzymes for n-alkanes, branched-chain alkanes, and cycloalkanes was as follows: the single oxygen route involved the participation of the cytochrome P450 enzyme (PDB ID: 3E5K) from Streptomyces avermitilis, the hydroxylation path used the methane monooxygenase hydroxylase of Methylosinus trichosporium OB3b (PDB ID: 6VK6), and the second single oxygenation route used the aldehyde dehydrogenase (PDB ID: 4CAZ) of Pseudomonas aeruginosa. The methane monooxygenase hydroxylase (PDB ID: 1FZI) of Methylococcus capsulatus was selected for secondary hydroxylation. The combination of the degradation enzyme to realize the best petroleum hydrocarbon degradation bacteria inoculation plan for the degradation of aromatic hydrocarbons is as follows: Methane monooxygenase hydroxylase (PDB ID: 6VK6) of Methylosinus trichosporium OB3B was selected for the hydroxylation pathway, catechol 1, 2-dioxygenase (PDB ID: 6VK6) of Acinetobacter baylyi ADP1 was selected for the 14eoxygenation pathway (PDB ID: 1DLT), epoxide hydrolase of Rhodococcus erythropolis (PDB ID: 4XBT) was used for hydrolysis, and benzene CoA reductase of Geobacter metallireducens GS-15 (PDB ID: 4Z3W) was used for the CoA route.
In summary, the optimal inoculation plan for petroleum hydrocarbon degradation bacteria was determined. The single oxygenation path primarily involved the participation of Streptomyces avermitilis, the hydroxylation path primarily involved the participation of Methylosinus trichosporium OB3b, and the secondary single oxygenation path primarily involved the participation of Pseudomonas aeruginosa. The secondary hydroxylation path involves the participation of Methylococcus capsulatus, the double oxygenation path involves the activity of Acinetobacter baylyi ADP1, the hydrolysis path primarily involves the participation of Rhodococcus erythropolis, and the CoA path primarily involves the participation of Geobacter metallireducens GS-15.
3.5. Verification of the Degradation Effect of the Optimal Inoculation Program of Petroleum Hydrocarbon Degradation Bacteria Based on the Molecular Docking Method
The degradation effect of the petroleum hydrocarbon degradation bacteria inoculation program was verified using the Cdocker module in Discovery Studio 2020 software version (Biovia, Waltham, MA, USA). The inoculation program of petroleum hydrocarbon degradation bacteria in the first 13 points of the Copeland method was selected to simplify the calculation process and verify the degradation effect. The Cdocker energy values of the 13 inoculation programs of petroleum hydrocarbon degradation bacteria are shown in Figure 3.
As can be seen from Figure 7, the absolute Cdocker energy value of the best inoculation program [4, 10, 15, 16] is the maximum, indicating that the comprehensive degradation effect of this program is the best. The linear fit between the degradation effect of the remaining 12 petroleum hydrocarbon degradation bacteria and their Copeland method score rank is good (Pearson’s r = 0.907 > 0.9). This indicates that the scores of the petroleum hydrocarbon degradation bacteria inoculated by the Copeland method are positively proportional to the degradation effect of the petroleum hydrocarbon pollutants, confirming the best inoculation program of the petroleum hydrocarbon degradation bacteria. This confirms the rationality of the optimal inoculation program determined using the Copeland method.
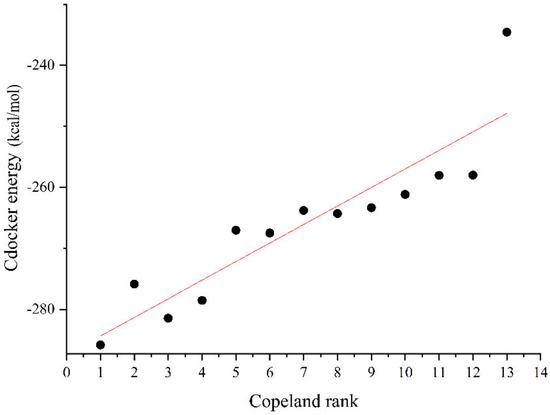
Figure 7.
Plot of the score rankings of the 13 petroleum hydrocarbon degradation bacteria inoculation programs against the Cdocker energy values.
3.6. Prospect of the Optimal Inoculation Program of Petroleum Hydrocarbon Degradation Bacteria
The optimal petroleum hydrocarbon degradation bacteria inoculation program devised in this paper yielded good results at the theoretical simulation level. Streptomyces avermitilis, Methylosinus trichosporium OB3b, Pseudomonas aeruginosa, Methylococcus capsulatus, Acinetobacter baylyi ADP1, Rhodococcus erythropolis, and Geobacter metallireducens GS-15 of the optimal petroleum hydrocarbon degradation bacteria inoculation program have all been used individually in the remediation of actual petroleum contaminated sites and have been effective in the remediation of petroleum hydrocarbon contaminated sites [30]. The best petroleum hydrocarbon degradation bacteria inoculation program designed in this paper will put into practical remediation application, and the combined application of these degradation bacteria to remediate petroleum contaminated sites is the next step of our research. The theoretical scheme designed in this paper will put into practical application to achieve practical results. In practical applications, we will further refine the inoculation conditions to minimize the impact of uncertainties. In the practical application process, the inoculated petroleum hydrocarbon degradation bacteria will rapidly degrade petroleum hydrocarbon pollutants, and the content of petroleum hydrocarbon pollutants in soil will decrease rapidly due to the degradation of petroleum hydrocarbon degradation bacteria [69], and the uniformity and abundance of petroleum hydrocarbon degradation bacteria will tend to be stable as time goes by [70]. The degradation function of petroleum hydrocarbon degradation bacteria can continue to exist for a long time when the external conditions are controlled and the bacteria are in a suitable survival condition. When the uniformity and abundance of petroleum hydrocarbon degradation bacteria decreases, it could be adjusted by adjusting the external conditions or adding the nutrients necessary for petroleum hydrocarbon degradation bacteria to restore the good survival state; if necessary, it could also be selected to re-inoculate a small number of bacteria to adjust the proportion of bacteria in the field.
4. Conclusions
The degradation path of the petroleum hydrocarbon pollutants by petroleum hydrocarbon degradation bacteria was analyzed. A variety of degradation enzymes corresponding to each step were selected for the main degradation path, and the binding energy of each degradation enzyme was calculated suing the molecular dynamics method to characterize the degradation ability. We used the developed Java program to identify the degradation enzymes in each step of the degradation path to obtain multiple groups of petroleum hydrocarbon degradation bacteria inoculation programs. The permutation and combination method was used for the same. The Copeland method was used to determine the best petroleum hydrocarbon degradation bacteria inoculation program. The best petroleum hydrocarbon degradation bacteria inoculation program can effectively repair the petroleum hydrocarbon pollution in the site and provide a reliable theoretical program for the bioremediation of petroleum hydrocarbon contaminated sites.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijerph18168794/s1. File S1: Permutation combination program code based on Java language development.
Author Contributions
Conceptualization, X.L. and W.H.; methodology, M.D.; software, W.H.; validation, X.L., M.D. and J.Z.; formal analysis, X.L. and W.H.; investigation, X.L. and W.H.; resources, X.D.; data curation, M.D.; writing—original draft preparation, X.L. and W.H.; writing—review and editing, W.H.; visualization, J.Z.; supervision, X.D.; and funding acquisition, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Open Project Program of State Key Laboratory of Petroleum Pollution Control, CNPC Research Institute of Safety and Environmental Technology., grant number PPC2019018.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, T.; Liu, Y.; Zhong, S.; Zhang, L. AOPs-based remediation of petroleum hydrocarbons-contaminated soils: Efficiency, influencing factors and environmental impacts. Chemosphere 2019, 246, 125726. [Google Scholar] [CrossRef]
- Falkova, M.; Vakh, C.; Shishov, A.; Zubakina, E.; Moskvin, A.; Moskvin, L.; Bulatov, A. Automated IR determination of petroleum products in water based on sequential injection analysis. Talanta 2016, 148, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Truskewycz, A.; Gundry, T.D.; Khudur, L.S.; Kolobaric, A.; Taha, M.; Aburto-Medina, A.; Ball, A.S.; Shahsavari, E. Petroleum Hydrocarbon Contamination in Terrestrial Ecosystems—Fate and Microbial Responses. Molecules 2019, 24, 3400. [Google Scholar] [CrossRef]
- Song, X.; Yan, X.; Li, G.; Ying, Z.; Huang, T.; Zhong, H. Isolation, characterization of Rhodococcus sp. P14 capable of degradation high-molecular-weight polycyclic aromatic hydrocarbons and aliphatic hydrocarbons. Mar. Pollut. Bull. 2011, 62, 2122–2128. [Google Scholar] [CrossRef] [PubMed]
- Gidudu, B.; Chirwa, E.M. Biosurfactants as demulsification enhancers in bio-electrokinetic remediation of petroleum contaminated soil. Process Saf. Environ. Prot. 2020, 143, 334–339. [Google Scholar] [CrossRef]
- Obire, O.; Nwaubeta, O.; Oofojekwu, P.C.; Ezenwaka, I.S.; Alegbeleye, W.O. Effects of Refined Petroleum Hydrocarbon on Soil Physicochemical and Bacteriological Characteristics. J. Appl. Sci. Environ. Manag. 2002, 6, 39–44. [Google Scholar] [CrossRef][Green Version]
- Wei, Z.; Wang, J.J.; Gaston, L.A.; Li, J.; Dodla, J.K. Remediation of crude oil-contaminated coastal marsh soil: Integrated effect of biochar, rhamnolipid biosurfactant and nitrogen application. J. Hazard. Mater. 2020, 396, 122595. [Google Scholar] [CrossRef] [PubMed]
- Ossai, I.C.; Ahmed, A.; Hassan, A.; Hamid, F.S. Remediation of soil and water contaminated with petroleum hydrocarbon: A review. Environ. Technol. Innov. 2020, 17, 100526. [Google Scholar] [CrossRef]
- Alvarez, P.; Vogel, T.M. Substrate interactions of benzene, toluene, and para-xylene during microbial degradation by pure cultures and mixed culture aquifer slurries. Appl. Environ. Microbiol. 1991, 57, 2981–2985. [Google Scholar] [CrossRef]
- Yen, C.H.; Chen, K.F.; Kao, C.M.; Liang, S.H.; Chen, T.Y. Application of persulfate to remediate petroleum hydrocarbon-contaminated soil: Feasibility and comparison with common oxidants. J. Hazard. Mater. 2011, 186, 2097–2102. [Google Scholar] [CrossRef]
- Hentati, O.; Lachhab, R.; Ayadi, M.; Ksibi, M. Toxicity assessment for petroleum-contaminated soil using terrestrial invertebrates and plant bioassays. Environ. Monit. Assess. 2013, 185, 2989–2998. [Google Scholar] [CrossRef] [PubMed]
- Sammarco, P.W.; Kolian, S.R.; Warby, R.A.F.; Bouldin, J.L.; Subra, W.A.; Porter, S.A. Concentrations in human blood of petroleum hydrocarbons associated with the BP/Deepwater Horizon oil spill, Gulf of Mexico. Arch. Toxicol. 2016, 90, 829–837. [Google Scholar] [CrossRef]
- Azeez, O.M.; Anigbogu, C.N.; Akhigbe, R.E.; Saka, W.A. Cardiotoxicity induced by inhalation of petroleum products. J. Afr. Assoc. Physiol. Sci. 2015, 3, 14–17. [Google Scholar]
- Lawal, A.T. Polycyclic aromatic hydrocarbons. A review. Cogent Environ. Sci. 2017, 3. [Google Scholar] [CrossRef]
- Hunt, L.J.; Duca, D.; Dan, T.; Knopper, L.D. Petroleum hydrocarbon (PHC) uptake in plants: A literature review. Environ. Pollut. 2019, 245, 472–484. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Raju, M.N.; Mallavarapu, M.; Kadiyala, V. Total Petroleum Hydrocarbon—Environmental Fate, Toxicity and Remediation, 1st ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2020; pp. 217–218. [Google Scholar]
- Sam, K.; Coulon, F.; Prpich, G. Management of petroleum hydrocarbon contaminated sites in Nigeria: Current challenges and future direction. Land Use Policy 2017, 64, 133–144. [Google Scholar] [CrossRef]
- Vidonish, J.E.; Zygourakis, K.; Masiello, C.A.; Sabadell, G.; Alvarez, P.J.J. Thermal Treatment of Hydrocarbon-Impacted Soils: A Review of Technology Innovation for Sustainable Remediation. Engineering 2016, 2, 426–437. [Google Scholar] [CrossRef]
- Chang, K.S.; Lo, W.H.; Lin, W.M.; Wen, J.X.; Yang, S.C.; Huang, C.J.; Hsieh, H.Y. Microwave-Assisted Thermal Remediation of Diesel Contaminated Soil. Eng. J. 2016, 20, 93–100. [Google Scholar] [CrossRef][Green Version]
- Akbari, A.; Ghoshal, S. Bioaccessible Porosity in Soil Aggregates and Implications for Biodegradation of High Molecular Weight Petroleum Compounds. Environ. Sci. Technol. 2015, 49, 14368–14375. [Google Scholar] [CrossRef] [PubMed]
- Zubaidy, E.; Abouelnasr, D.M. Fuel recovery from waste oily sludge using solvent extraction. Process Saf. Environ. Prot. 2010, 88, 318–326. [Google Scholar] [CrossRef]
- Abdulbari, H.A.; Abdurahman, N.H.; Rosli, Y.M.; Mahmood, W.K.; Azhari, H.N. Demulsification of petroleum emulsions Using microwave separation method. Exp. Hematol. 2011, 6, 5376–5382. [Google Scholar]
- Pilli, S.; Bhunia, P.; Yan, S.; Leblanc, R.J.; Tyagi, R.D.; Surampalli, R.Y. Ultrasonic pretreatment of sludge: A review. Ultrason. Sonochem. 2011, 18, 1–18. [Google Scholar] [CrossRef]
- April, T.M.; Foght, J.M.; Currah, R.S. Hydrocarbon-degradation filamentous fungi isolated from flare pit soils in northern and western Canada. Can. J. Microbiol. 1999, 46, 38–49. [Google Scholar] [CrossRef]
- Lal, B.; Khanna, S. Degradation of crude oil by Acinetobacter calcoaceticus and Alcaligenes odorans. J. Appl. Microbiol. 2010, 81, 355–362. [Google Scholar] [CrossRef]
- Leahy, J.G.; Colwell, R.R. Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 1990, 54, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Das, N.; Chandran, P. Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview. Biotechnol. Res. Int. 2011, 2011, 941810. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, M.R.; Giorgetti, L.; Becarelli, S.; Siracusa, G.; Gregorio, S.D. Polycyclic aromatic hydrocarbon-contaminated soils: Bioaugmentation of autochthonous bacteria and toxicological assessment of the bioremediation process by means of Vicia faba L. Environ. Sci. Pollut. Res. Int. 2016, 23, 7930–7941. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, M. Bioremediation of crude oil-contaminated soil: Comparison of different biostimulation and bioaugmentation treatments. J. Hazard. Mater. 2010, 183, 395–401. [Google Scholar] [CrossRef]
- Mishra, S.; Singh, S.N. Microbial degradation of n-hexadecane in mineral salt medium as mediated by degradative enzymes. Bioresour. Technol. 2012, 111, 148–154. [Google Scholar] [CrossRef]
- Zhang, Z.Z.; Hou, Z.W.; Yang, C.Y.; Ma, C.Q.; Tao, F.; Xu, P. Degradation of n-alkanes and polycyclic aromatic hydrocarbons in petroleum by a newly isolated Pseudomonas aeruginosa DQ8. Bioresour. Technol. 2011, 102, 4111–4116. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, W.; Chen, G.; Gao, Y.; Wang, J. Preparation of petroleum-degrading bacterial agent and its application in remediation of contaminated soil in Shengli Oil Field, China. Environ. Sci. Pollut. Res. 2014, 21, 7929–7937. [Google Scholar] [CrossRef]
- Ro, S.Y.; Ross, M.O.; Yue, W.D.; Batelu, S.; Rosenzweig, A.C. From micelles to bicelles: Effect of the membrane on particulate methane monooxygenase activity. J. Biol. Chem. 2018, 293, 10457–10465. [Google Scholar] [CrossRef]
- Jensen, C.N.; Ali, S.T.; Allen, M.J.; Grogan, G. Exploring nicotinamide cofactor promiscuity in NAD(P)H-dependent flavin containing monooxygenases (FMOs) using natural variation within the phosphate binding loop. Structure and activity of FMOs from Cellvibrio sp. BR and Pseudomonas stutzeri NF13. J. Mol. Catal. B Enzym. 2014, 109, 191–198. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lončar, N.; Fiorentini, F.; Bailleul, G.; Savino, S.; Romero, E.; Mattevi, A.; Fraaije, M.W. Characterization of a thermostable flavin-containing monooxygenase from Nitrincola lacisaponensis (NiFMO). Appl. Microbiol. Biotechnol. 2019, 103, 1755–1764. [Google Scholar] [CrossRef]
- Joyce, M.G.; Girvan, H.M.; Munro, A.W.; Leys, D. A Single Mutation in Cytochrome P450 BM3 Induces the Conformational Rearrangement Seen upon Substrate Binding in the Wild-type Enzyme. J. Biol. Chem. 2004, 279, 23287–23293. [Google Scholar] [CrossRef]
- Xu, L.H.; Fushinobu, S.; Ikeda, H.; Wakagi, T.; Shoun, H. Crystal Structures of Cytochrome P450 105P1 from Streptomyces avermitilis: Conformational Flexibility and Histidine Ligation State. J. Bacteriol. 2009, 191, 1211–1219. [Google Scholar] [CrossRef]
- Whittington, D.A.; Rosenzweig, A.C.; Frederick, C.A.; Lippard, S.J. Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase. Biochemistry 2001, 40, 3476–3482. [Google Scholar] [CrossRef] [PubMed]
- Elango, N.A.; Radhakrishnan, R.; Froland, W.A.; Wallar, B.J.; Earhart, C.A.; Lipscomb, J.D.; Ohlendorf, D.H. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 1997, 6, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, X.; Yang, W.; Feng, X.; Wang, W.; Feng, L.; Bartlam, M.; Wang, L.; Rao, Z. Crystal Structure of Long-Chain Alkane Monooxygenase (LadA) in Complex with Coenzyme FMN: Unveiling the Long-Chain Alkane Hydroxylase. J. Mol. Biol. 2008, 376, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; An, S.; Park, Y.R.; Jang, H.; Cho, U.S. MMOD-induced structural changes of hydroxylase in soluble methane monooxygenase. Sci. Adv. 2019, 5, eaax0059. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Banerjee, R.; Shi, K.; Aihara, H.; Lipscomb, J.D. Structural Studies of the Methylosinus trichosporium OB3b Soluble Methane Monooxygenase Hydroxylase and Regulatory Component Complex Reveal a Transient Substrate Tunnel. Biochemistry 2020, 59, 2946–2961. [Google Scholar] [CrossRef]
- Nguyen, G.T.; Kim, Y.G.; Ahn, J.W.; Chang, J.H. Structural Basis for Broad Substrate Selectivity of Alcohol Dehydrogenase YjgB from Escherichia coli. Molecules 2020, 25, 2404. [Google Scholar] [CrossRef] [PubMed]
- Valencia, E.; Larroy, C.; Ochoa, W.F.; Parés, X.; Fita, I.; Biosca, J.A. Apo and Holo structures of an NADPH-dependent cinnamyl alcohol dehydrogenase from Saccharomyces cerevisiae. J. Mol. Biol. 2004, 341, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Zhang, Z.; Zhang, X.; Li, H.Y.; Sun, H.Z.; Mao, Z.W.; Xia, W. Structural Insight into the Substrate Gating Mechanism by Staphylococcus aureus Aldehyde Dehydrogenase. CCS Chem. 2020, 2, 946–954. [Google Scholar] [CrossRef]
- Díaz-Sánchez, Á.G.; González-Segura, L.; Mújica-Jiménez, C.; Rudiño-Piñera, E.; Montiel, C.; Martínez-Castilla, L.P.; Muñoz-Clares, R.A. Amino Acid Residues Critical for the Specificity for Betaine Aldehyde of the Plant ALDH10 Isoenzyme Involved in the Synthesis of Glycine Betaine. Plant Physiol. 2012, 158, 1570–1582. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, E.G.; Lipscomb, J.D. Crystal Structures of Fe2+ Dioxygenase Superoxo, Alkylperoxo and Bound Product Intermediates. Science 2007, 316, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Vetting, M.W.; Ohlendorf, D.H. The 1.8 crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker. Structure 2000, 8, 429–440. [Google Scholar] [CrossRef]
- Earhart, C.A.; Vetting, M.W.; Gosu, R.; Michaud-Soret, I.; Que, L.; Ohlendorf, D.H. Structure of catechol 1,2-dioxygenase from Pseudomonas arvilla. Biochem. Biophys. Res. Commun. 2005, 338, 198–205. [Google Scholar] [CrossRef]
- Cho, J.H.; Jung, D.K.; Lee, K.; Rhee, S. Crystal Structure and Functional Analysis of the Extradiol Dioxygenase LapB from a Long-chain Alkylphenol Degradation Pathway in Pseudomonas. J. Biol. Chem. 2009, 284, 34321–34330. [Google Scholar] [CrossRef]
- Micalella, C.; Martignon, S.; Bruno, S.; Pioselli, B.; Caglio, R.; Valetti, F.; Pessione, E.; Giunta, C.; Rizzi, R.M. X-ray crystallography, mass spectrometry and single crystal microspectrophotometry: A multidisciplinary characterization of catechol 1,2 dioxygenase. BBA Proteins Proteom. 2011, 1814, 817–823. [Google Scholar] [CrossRef]
- Reetz, M.T.; Bocola, M.; Wang, L.W.; Sanchis, J.; Cronin, A.; Arand, M.; Zou, J.; Archelas, A.; Bottalla, A.-L.; Naworyta, A.; et al. Directed Evolution of an Enantioselective Epoxide Hydrolase: Uncovering the Source of Enantioselectivity at Each Evolutionary Stage. J. Am. Chem. Soc. 2009, 131, 7334–7343. [Google Scholar] [CrossRef]
- Sun, Z.; Lonsdale, R.; Kong, X.D.; Xu, J.H.; Zhou, J.; Reetz, M.T. Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution. Angew. Chem. 2015, 54, 12410–12415. [Google Scholar] [CrossRef]
- Kong, X.D.; Yuan, S.; Li, L.; Chen, S.; Xu, J.H.; Zhou, J.H. Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates. Proc. Natl. Acad. Sci. USA 2014, 111, 15717–15722. [Google Scholar] [CrossRef]
- Weinert, T.; Huwiler, S.G.; Kung, J.W.; Weidenweber, S.; Hellwig, P.; Stark, H.J.; Biskup, T.; Weber, S.; Cotelesage, J.J.; George, G.N.; et al. Structural basis of enzymatic benzene ring reduction. Nat. Chem. Biol. 2015, 11, 586–591. [Google Scholar] [CrossRef]
- Saari, D.G.; Merlin, V.R. The Copeland method. Econ. Theory 1996, 8, 51–76. [Google Scholar] [CrossRef]
- Caillaux, M.A.; Sant’Anna, A.P.; Meza, L.A.; Mello, J.C. Container logistics in Mercosur: Choice of a transshipment port using the ordinal Copeland method, data envelopment analysis and probabilistic composition. Marit. Econ. Logist. 2011, 13, 355–370. [Google Scholar] [CrossRef]
- Li, C.D.; Jin, Q.; Huang, Y.; Liu, J.N.; Chen, G.S. Application of copeland scoring and ranking method in the ecological hazard evaluation of chemical substances. Res. Environ. Sci. 2011, 24, 1161–1165. [Google Scholar]
- Ashraf, W.; Mihdhir, A.; Colin Murrell, J. Bacterial oxidation of propane. FEMS Microbiol. Lett. 1994, 122, 1–6. [Google Scholar] [CrossRef]
- Mckenna, E.J.; Coon, M.J. Enzymatic omega-oxidation. IV. Purification and properties of the omega-hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 1970, 245, 3882–3889. [Google Scholar] [CrossRef]
- Yi, T.; Lee, E.H.; Yun, G.A.; Hwang, G.S.; Cho, K.S. Novel biodegradation pathways of cyclohexane by Rhodococcus sp. EC1. J. Hazard. Mater. 2011, 191, 393–396. [Google Scholar] [CrossRef]
- Seo, J.S.; Keum, Y.S.; Hu, Y.; Lee, S.E.; Li, Q.X. Phenanthrene degradation in Arthrobacter sp. P1-1: Initial 1,2-, 3,4- and 9,10-dioxygenation, and meta- and ortho-cleavages of naphthalene-1,2-diol after its formation from naphthalene-1,2-dicarboxylic acid and hydroxyl naphthoic acids. Chemosphere 2006, 65, 2388–2394. [Google Scholar] [CrossRef] [PubMed]
- Beilen, J.B.V.; Li, Z.; Duetz, W.A.; Smits, T.H.M.; Witholt, B. Diversity of Alkane Hydroxylase Systems in the Environment. Oil Gas Sci. Technol. 2003, 58, 427–440. [Google Scholar] [CrossRef]
- Le, N.C.; Mikolasch, A.; Klenk, H.P.; Schauer, F. Degradation of the multiple branched alkane 2,6,10,14-tetramethyl-pentadecane (pristane) in Rhodococcus ruber and Mycobacterium neoaurum. Int. Biodeterior. Biodegrad. 2009, 63, 201–207. [Google Scholar]
- Zhao, Q. Construction of an Enhanced Degradation System for Hydrophobic Petroleum Hydrocarbon Degradation Bacteria and Its Degradation Capacity. Master’s Thesis, Wuhan University, Wuhan, China, 2005. [Google Scholar]
- Táncsics, A.; Szabó, I.; Baka, E.; Szoboszlay, S.; Kukolya, J.; Kriszt, B.; Márialigeti, K. Investigation of catechol 2,3-dioxygenase and 16S rRNA gene diversity in hypoxic, petroleum hydrocarbon contaminated groundwater. Syst. Appl. Microbiol. 2010, 33, 398–406. [Google Scholar] [CrossRef]
- Li, M.H.; Wang, X.L.; Chu, Z.H.; Li, Y. Multiple-Site Molecular Modification of Dioxin-Like PCBs to Eliminate Bioconcentration. Pol. J. Environ. Stud. 2021, 30, 1655–1675. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, X.; Jiang, L.; Li, Y. Prediction of octanol-air partition coefficients for polychlorinated biphenyls (PCBs) using 3D-QSAR models. Ecotoxicol. Environ. Saf. 2016, 124, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.R.; Wang, J.A.; Han, Z.J. Bioremediation Mode of Saline Soil Contaminated by Petroleum. J. Anhui Agric. Sci. 2015, 43, 278–283. [Google Scholar]
- Wu, M.; Ye, X.; Chen, K.; Li, W.; Yuan, J.; Jiang, X. Bacterial community shift and hydrocarbon transformation during bioremediation of short-term petroleum-contaminated soil. Environ. Pollut. 2017, 223, 657–664. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).