LSTM Networks to Improve the Prediction of Harmful Algal Blooms in the West Coast of Sabah
Abstract
1. Introduction
2. Materials and Methods
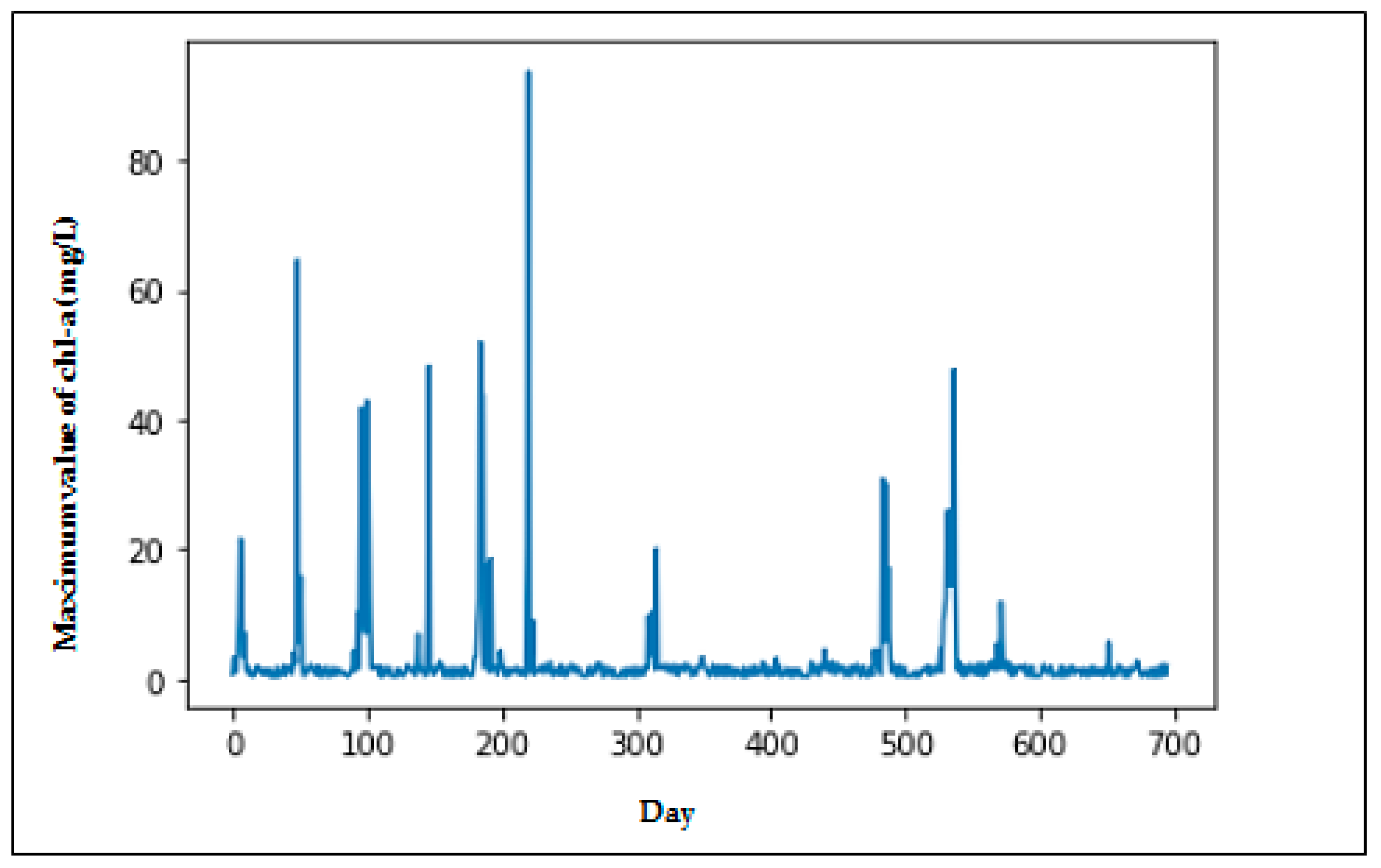
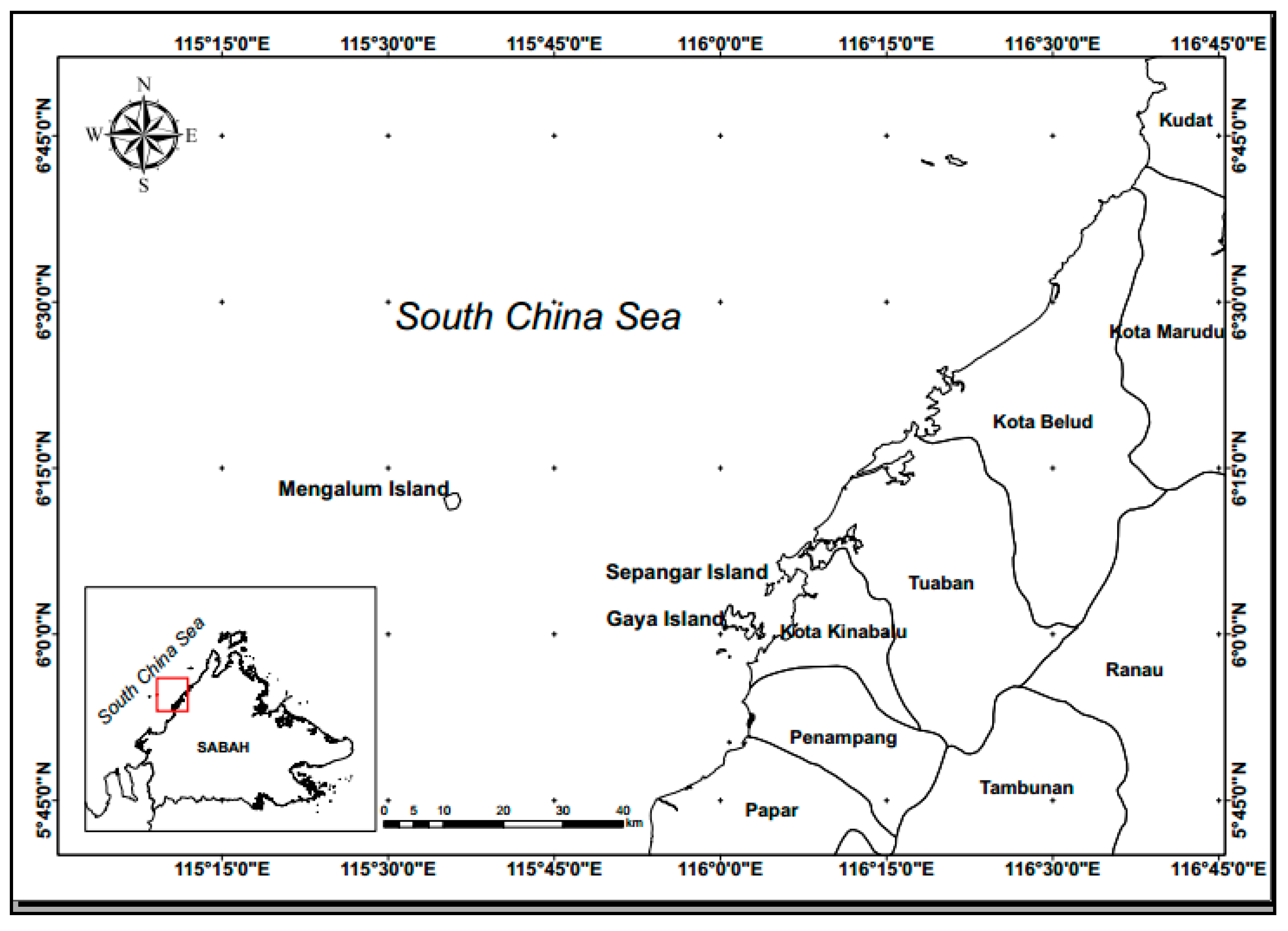
2.1. Data
2.2. Long Short-Term Memory (LSTM)
- Internal memory of cell state
- Element wise input
- Output of the hidden state
- Previous hidden state
- Element of the output gate
- Element of input gate
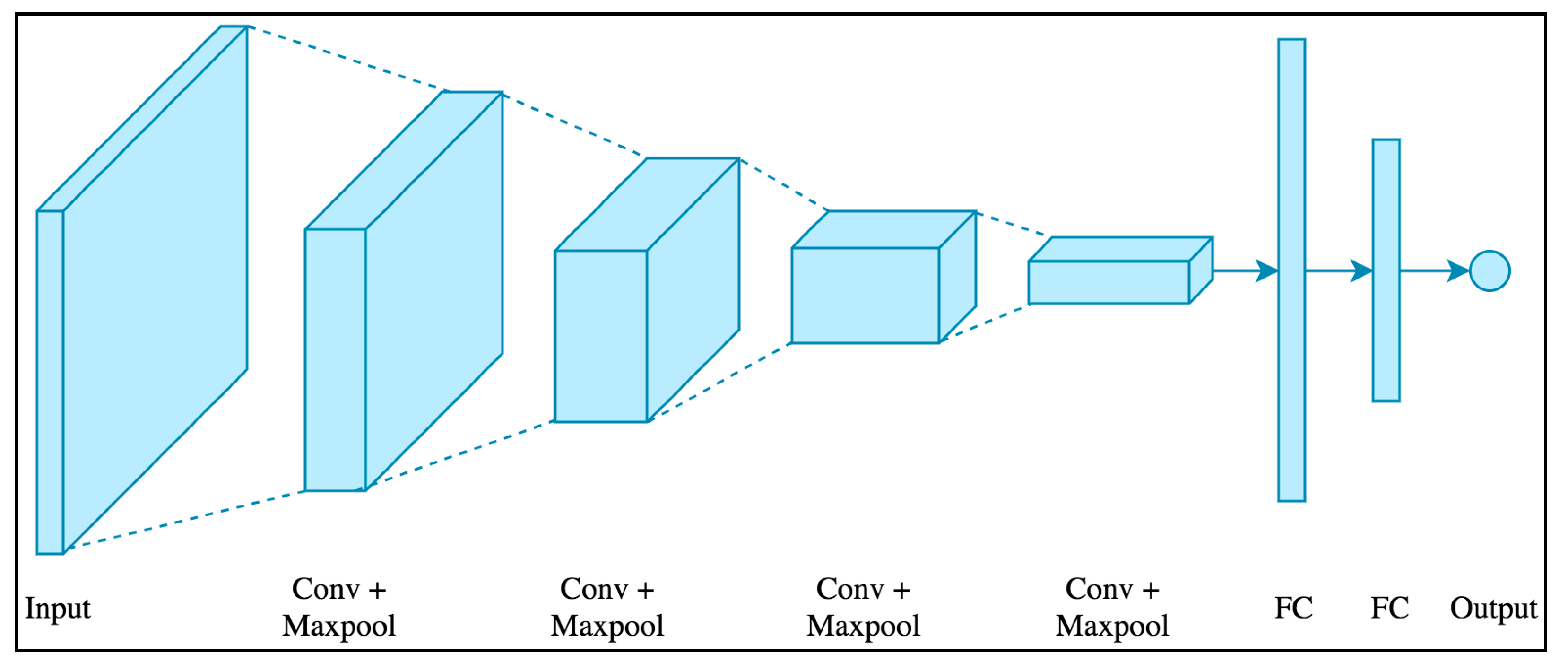
2.3. Convolutional Neural Network (CNN)
2.4. Evaluation Criteria
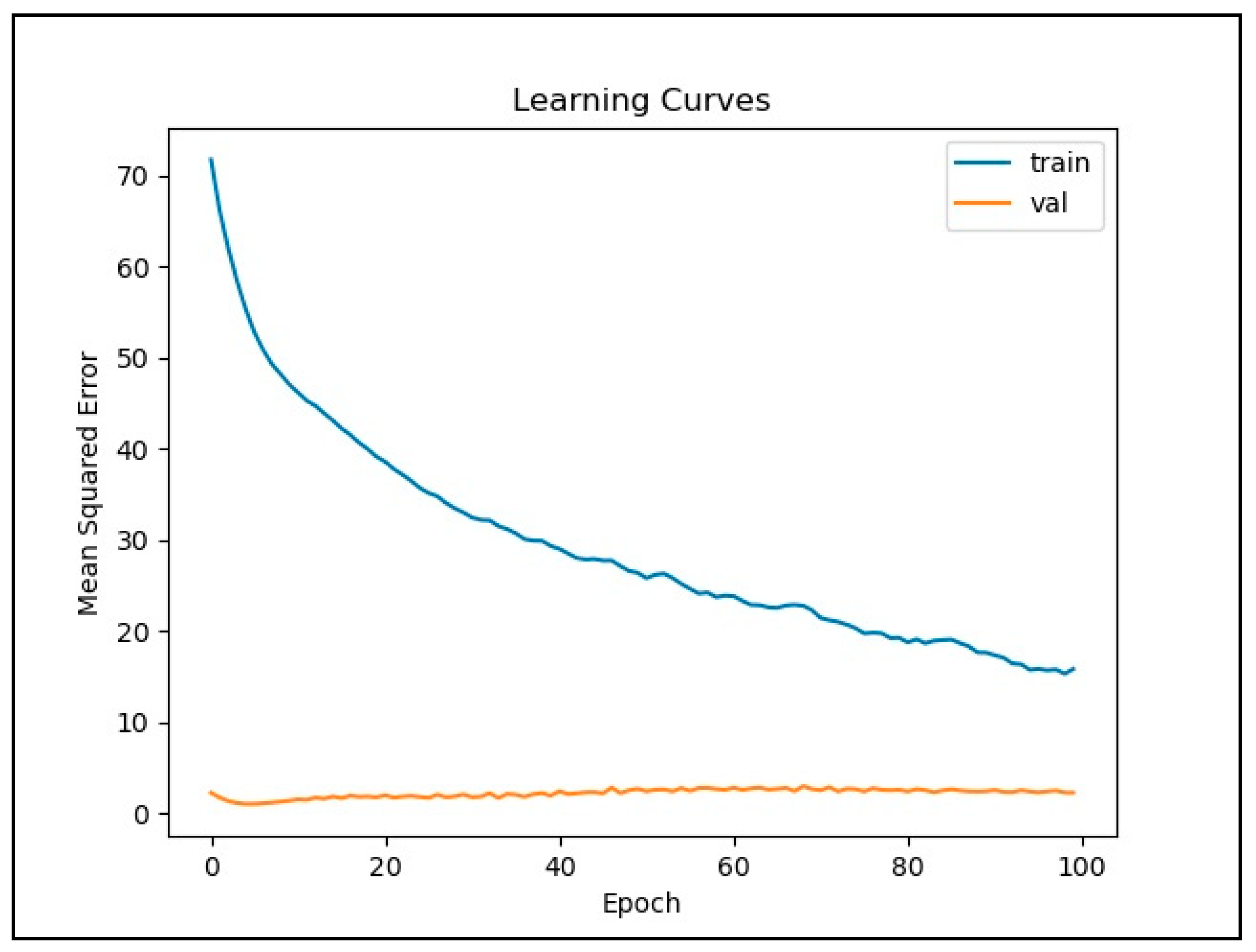
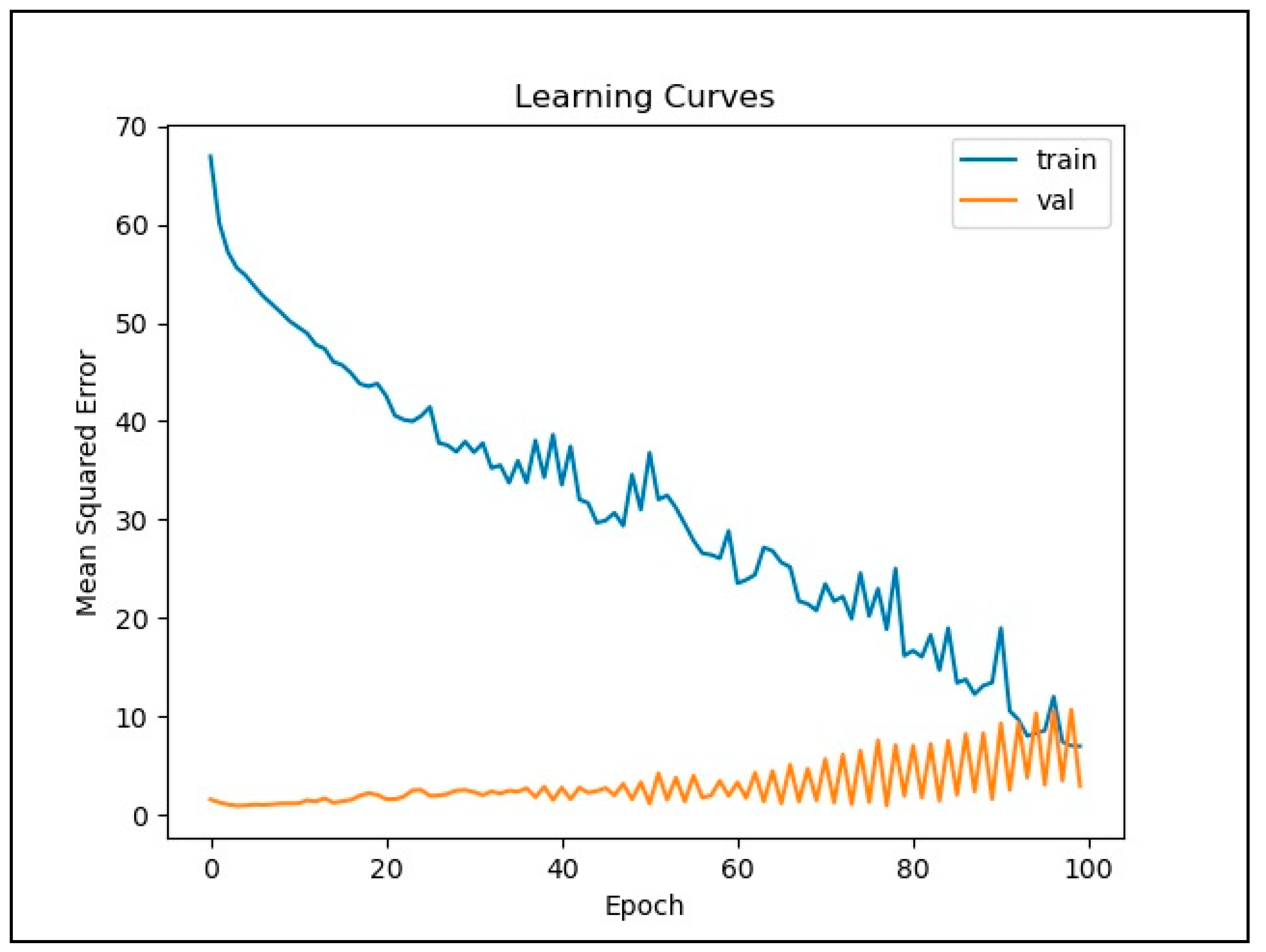
2.5. Training LSTM Network
2.6. Prediction Based on Trained LSTM
2.7. Prediction Based on the CNN
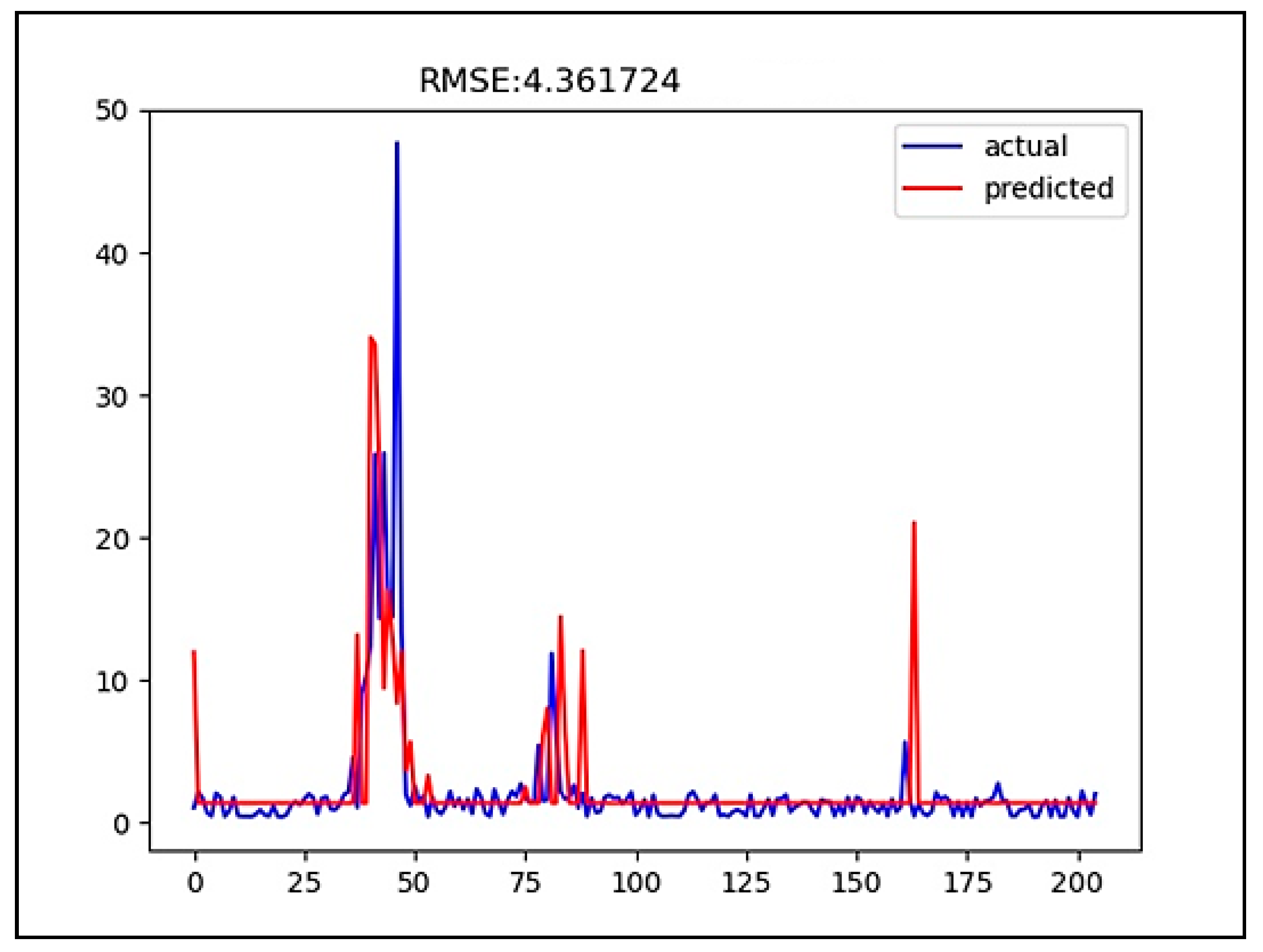
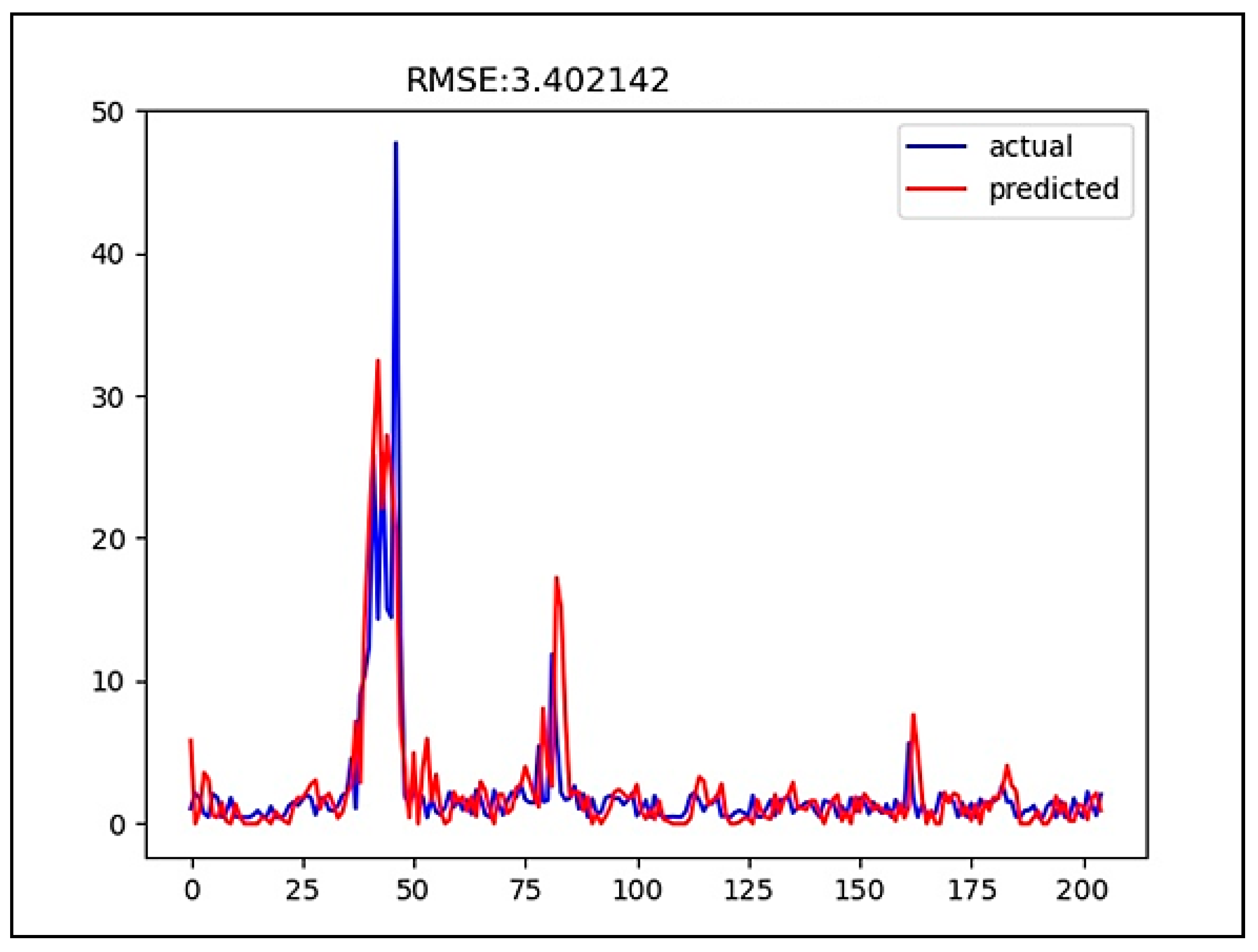
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lim, P.T.; Gires, U.; Leaw, C.P. Harmful Algal Blooms in Malaysia. Sains Malays. 2012, 41, 1509–1515. [Google Scholar]
- Blondeau-Patissier, D.; Gower, J.; Dekker, A.; Phinn, S.R.; Brando, V.E. A review of ocean color remote sensing methods and statistical techniques for the detection, mapping and analysis of phytoplankton blooms in coastal and open oceans. Prog. Oceanogr. 2014, 123, 123–144. [Google Scholar] [CrossRef]
- Stumpf, R.P.; Culver, M.E.; Tester, P.A.; Tomlinson, M.; Kirkpatrick, G.J.; Pederson, B.A.; Truby, E.; Ransibrahmanakul, V.; Soracco, M. Monitoring karenia brevis blooms in the gulf of mexico using satellite ocean color imagery and other data. Harmful Algae 2003, 2, 147–160. [Google Scholar] [CrossRef]
- Tomlinson, M.C.; Stumpf, R.P.; Ransibrahmanakul, V.; Truby, E.W.; Kirkpatrick, G.J.; Pederson, B.A.; Vargo, G.A.; Heil, C.A. Evaluation of the use of seawifs imagery for detecting karenia brevis harmful algal blooms in the eastern gulf of mexico. Remote Sens. Environ. 2004, 91, 293–303. [Google Scholar] [CrossRef]
- Gokaraju, B.; Durbha, S.S.; King, R.L.; Younan, N.H. A machine learning based spatio-temporal data mining approach for detection of harmful algal blooms in the gulf of mexico. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2011, 4, 710–720. [Google Scholar] [CrossRef]
- Gokaraju, B.; Durbha, S.S.; King, R.L.; Younan, N.H. Ensemble methodology using multistage learning for improved detection of harmful algal blooms. IEEE Geosci. Remote Sens. Lett. 2012, 9, 827–831. [Google Scholar] [CrossRef]
- Song, W.; Dola, J.M.; Cline, D.; Xiong, G. Learning-based algal bloom event recognition for oceanographic decision support system using remote sensing data. Remote Sens. 2015, 7, 13564–13585. [Google Scholar] [CrossRef]
- Lee, G.; Bae, J.; Lee, S.; Jang, M.; Park, H. Monthly chlorophyll—A prediction using neuro-genetic algorithm for water quality management in Lakes. Desalination Water Treat. 2016, 57, 26783–26791. [Google Scholar] [CrossRef]
- Santoleri, R.; Banzon, V.; Marullo, S.; Napolitano, E.; D’Ortenzio, F.; Evans, R. Year-to-year variability of the phytoplankton bloom in the southern adriatic sea (1998–2000): Sea-viewing wide field-of-view sensor observations and modeling study. J. Geophys. Res. 2003, 108, 8122. [Google Scholar] [CrossRef]
- Gohin, F.; Lampert, L.; Guillaud, J.-F.; Herbland, A.; Nézan, E. Satellite and in situ observations of a late winter phytoplankton bloom, in the Northern Bay of Biscay. Cont. Shelf Res. 2003, 23, 1117–1141. [Google Scholar] [CrossRef]
- Thomas, A.C.; Townsend, D.W.; Weatherbee, R. Satellite-measured phytoplankton variability in the Gulf of Maine. Cont. Shelf Res. 2003, 23, 971–989. [Google Scholar] [CrossRef]
- Craig, S.E.; Lohrenz, S.E.; Lee, Z.P.; Mahoney, K.L.; Kirkpatrick, G.J.; Schofield, O.M.; Steward, R.G. Use of hyperspectral remote sensing reflectance for detection and assessment of the harmful alga, karenia brevis. Appl. Opt. 2006, 45, 5414–5425. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, Y.; Cao, M.; Sun, X.; Du, Z.; Liu, R.; Ye, X. Deep-learning-based approach for prediction of algal blooms. Sustainability 2016, 8, 1060. [Google Scholar] [CrossRef]
- Park, Y.; Cho, K.H.; Park, J.; Cha, S.M.; Kim, J.H. Development of early-warning protocol for predicting chlorophyll-a concentration using machine learning models in freshwater and estuarine reservoirs, Korea. Sci. Total. Environ. 2015, 502, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sha, J.; Wang, Z.-L. Application of feature selection and regression models for chlorophyll-a prediction in a shallow lake. Environ. Sci. Pollut. Res. 2018, 25, 19488–19498. [Google Scholar] [CrossRef]
- Yajima, H.; Derot, J. Application of the Random Forest model for chlorophyll-a forecasts in fresh and brackish water bodies in Japan, using multivariate long-term databases. J. Hydroinform. 2018, 20, 206–220. [Google Scholar] [CrossRef]
- McClain, C.R. A decade of satellite ocean color observations. Annu. Rev. Mar. Sci. 2009, 1, 19–42. [Google Scholar] [CrossRef]
- Aghel, B.; Rezaei, A.; Mohadesi, M. Modeling and prediction of water quality parameters using a hybrid particle swarm optimization–neural fuzzy approach. Int. J. Environ. Sci. Technol. 2019, 16, 4823–4832. [Google Scholar] [CrossRef]
- Zhu, S.; Hadzima-Nyarko, M.; Gao, A.; Wang, F.; Wu, J.; Wu, S. Two hybrid data-driven models for modeling water-air temperature relationship in rivers. Environ. Sci. Pollut. Res. 2019, 26, 12622–12630. [Google Scholar] [CrossRef]
- Fijani, E.; Barzegar, R.; Deo, R.; Tziritis, E.; Konstantinos, S. Design and implementation of a hybrid model based on two-layer decomposition method coupled with extreme learning machines to support real-time environmental monitoring. Sci. Total. Environ. 2019, 648, 839–853. [Google Scholar] [CrossRef]
- Najafzadeh, M.; Ghaemi, A. Prediction of the five-day biochemical oxygen demand and chemical oxygen demand in natural streams using machine learning methods. Environ. Monit. Assess. 2019, 191, 380. [Google Scholar] [CrossRef]
- Lee, S.; Lee, D. Improved prediction of harmful algal blooms in four Major South Korea’s Rivers using deep learning models. Int. J. Environ. Res. Public health 2018, 15, 1322. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.L.; Huang, Q.; Wang, C.; Chen, J.; He, D. Long short-term memory neural networks for online disturbance detection in satellite image time series. Remote Sens. 2018, 10, 452. [Google Scholar] [CrossRef]
- Liu, P.; Wang, J.; Sangaiah, A.K.; Xie, Y.; Yin, X. Analysis and prediction of water quality using LSTM deep neural networks in IoT environment. Sustainability 2018, 11, 2058. [Google Scholar] [CrossRef]
- Xayasouk, T.; Lee, H.; Lee, G. Air pollution prediction using long short-term memory (LSTM) and deep autoencoder (DAE) models. Sustainability 2020, 12, 2570. [Google Scholar] [CrossRef]
- Kumar, A.; Islam, T.; Sekimoto, Y.; Mattmann, C.; Wilson, B. Convcast: An embedded convolutional LSTM based architecture for precipitation nowcasting using satellite data. PLoS ONE 2020, 15, e0230114. [Google Scholar] [CrossRef]
- Kim, T.Y.; Cho, S.B. Predicting residential energy consumption using CNN-LSTM neural networks. Energy 2019, 182, 72–81. [Google Scholar] [CrossRef]
- Borovykh, A.; Bohte, S.; Oosterlee, C.W. Conditional time series forecasting with convolutional neural networks. arXiv 2017, arXiv:1703.04691. [Google Scholar]
- Choi, J.; Kim, J.; Won, J.; Min, O. Modelling chlorophyll-a concentration using deep neural networks considering extreme data imbalance and skewness. In Proceedings of the 21st International Conference on Advanced Communication Technology (ICACT), Pyeong-Chang, Kwangwoon Do, Korea, 17–20 February 2019; pp. 631–634. [Google Scholar] [CrossRef]
- Cho, H.; Choi, U.J.; Park, H. Deep learning application to time series prediction of daily chlorophyll-a concentration. WIT Trans. Ecol. Environ. 2018, 215, 157–163. [Google Scholar]
- Li, X.; Peng, L.; Yao, X.; Cui, S.; Hu, Y.; You, C.; Chi, T. Long short term memory neural network for air pollutant concentration predictions: Method development and evaluation. Environ. Pollut. 2017, 231, 997–1004. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Zuo, R.; Xiong, Y.; Wang, J.; Carranza, E.J.M. Deep learning and its application in geochemical mapping. Earth Sci Rev. 2019, 192, 1–14. [Google Scholar] [CrossRef]
- Hoseinzade, E.; Haratizadeh, S. CNN pred: CNN-based stock market prediction using a diverse set of variables. Expert Syst. Appl. 2019, 129, 273–285. [Google Scholar] [CrossRef]
- Sameen, M.I.; Pradhan, B.; Shafri, H.Z.M.; Hamid, H.B. Applications of deep learning in severity prediction of traffic accidents. In Proceedings of the Global Civil Engineering Conference, Kuala Lumpur, Malaysia, 25–28 July 2017; Springer: Singapore, 2017; pp. 793–808. [Google Scholar]
- Xiao, X.; He, J.; Huang, H.; Miller, T.R.; Christakos, G.; Reichwaldt, E.S.; Ghadouani, A.; Lin, S.; Xu, X.; Shi, J. A novel single-parameter approach for forecasting algal blooms. Water Res. 2017, 108, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Xu, Q.; Lv, J.; Liu, W.; Li, Q.; Wang, J. MS-LSTM: A multi-scale LSTM model for BGP anomaly detection. In Proceedings of the 2016 IEEE 24th International Conference on the Network Protocols (ICNP), Singapore, 8–11 November 2016; pp. 1–6, 8–11. [Google Scholar]
- Gu, Y.; Lu, W.; Qin, L.; Li, M.; Shao, Z. Short-term prediction of lane-level traffic speeds: A fusion deep learning model. Transp. Res. Part C Emerg. Technol. 2019, 106, 1–16. [Google Scholar] [CrossRef]
- Barzegar, R.; Aalami, M.T.; Adamowski, J. Short-term water quality variable prediction using a hybrid CNN–LSTM deep learning model. Stoch. Environ. Res. Risk Assess. 2020, 34, 1–19. [Google Scholar] [CrossRef]
- Yussof, F.N.B.M.; Maan, N.B.; Reba, M.N.B.M. Reconstruction of Chlorophyll-a Data by Using DINEOF Approach in Sepanggar Bay, Malaysia. Comput. Sci. 2021, 16, 345–356. [Google Scholar]



| Step | Description |
|---|---|
| 1 | Preprocessing of all fine particulate matter and meteorological data LSTM pre-training |
| 2 |
|
| 3 | Fine-tuning
|
| 4 | Obtain prediction results |
| Model | RMSE | r |
|---|---|---|
| LSTM | 3.402142 | 0.338385 |
| CNN | 4.361724 | 0.111790 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yussof, F.N.; Maan, N.; Md Reba, M.N. LSTM Networks to Improve the Prediction of Harmful Algal Blooms in the West Coast of Sabah. Int. J. Environ. Res. Public Health 2021, 18, 7650. https://doi.org/10.3390/ijerph18147650
Yussof FN, Maan N, Md Reba MN. LSTM Networks to Improve the Prediction of Harmful Algal Blooms in the West Coast of Sabah. International Journal of Environmental Research and Public Health. 2021; 18(14):7650. https://doi.org/10.3390/ijerph18147650
Chicago/Turabian StyleYussof, Fatin Nadiah, Normah Maan, and Mohd Nadzri Md Reba. 2021. "LSTM Networks to Improve the Prediction of Harmful Algal Blooms in the West Coast of Sabah" International Journal of Environmental Research and Public Health 18, no. 14: 7650. https://doi.org/10.3390/ijerph18147650
APA StyleYussof, F. N., Maan, N., & Md Reba, M. N. (2021). LSTM Networks to Improve the Prediction of Harmful Algal Blooms in the West Coast of Sabah. International Journal of Environmental Research and Public Health, 18(14), 7650. https://doi.org/10.3390/ijerph18147650






