The Foraging Gene, a New Environmental Adaptation Player Involved in Xenobiotic Detoxification
Abstract
1. Introduction
2. Materials and Methods
2.1. D. melanogaster Strains
2.2. Toxicological Tests
2.3. Enzymatic Activities Measurements
2.4. Northern Blotting
2.5. Accord Insertion Detection
3. Results
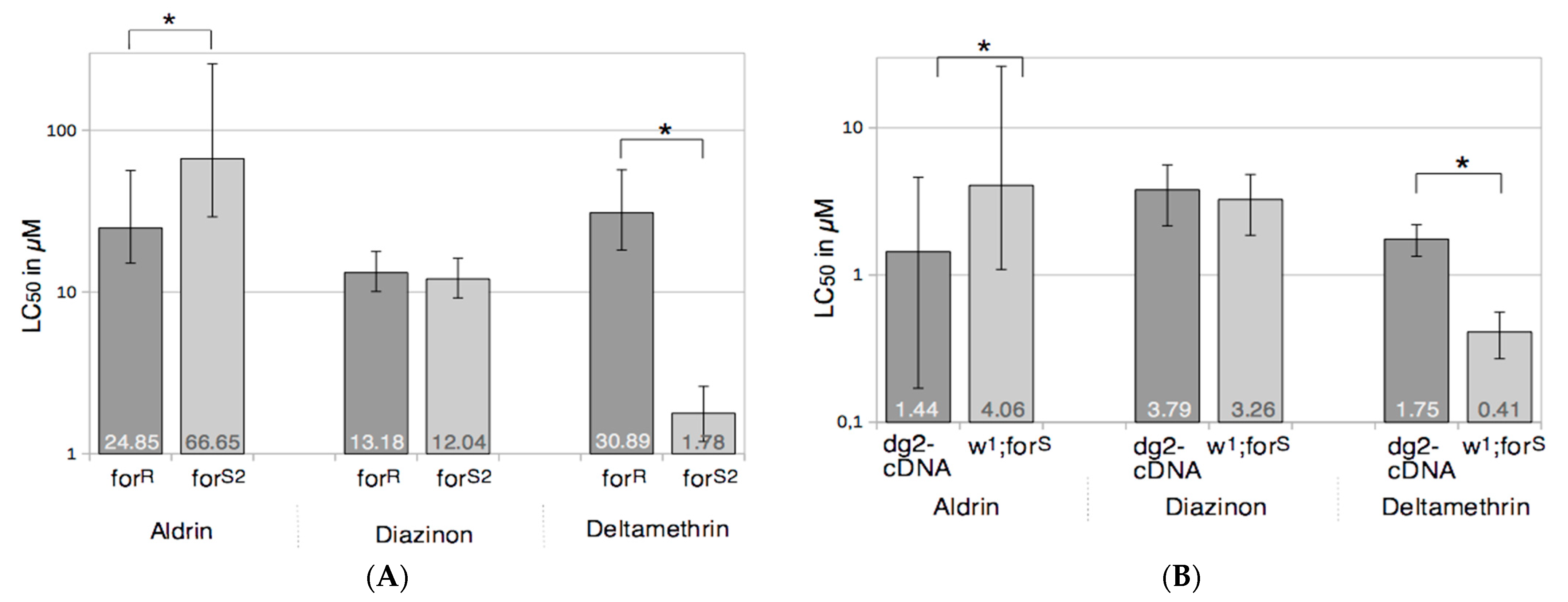
3.1. Foraging and the Response to Chemical Stresses in D. melanogaster
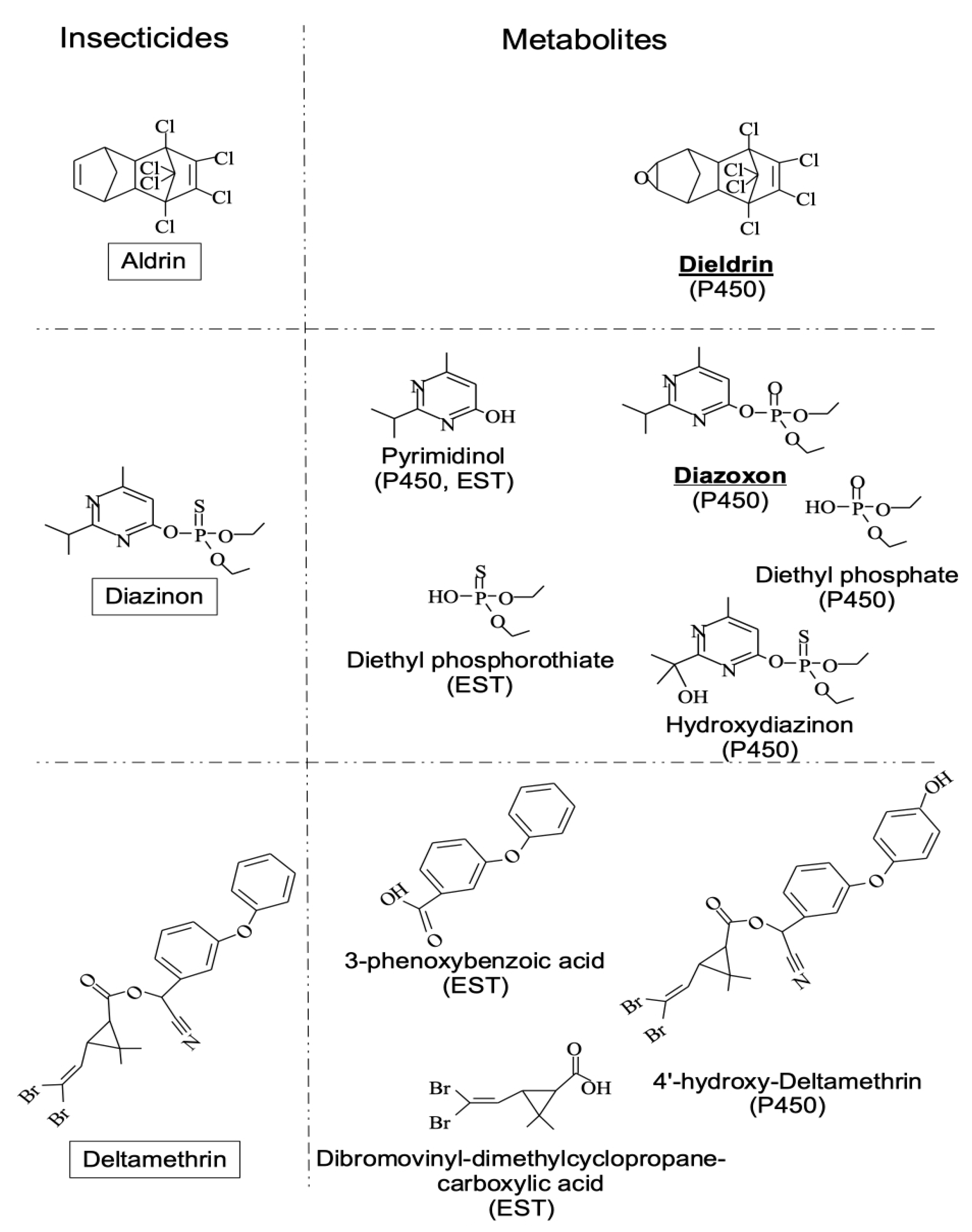
3.2. Xenobiotics Degradative Enzymes
3.3. Cytochrome P450s Activities, Foraging, and Toxicology
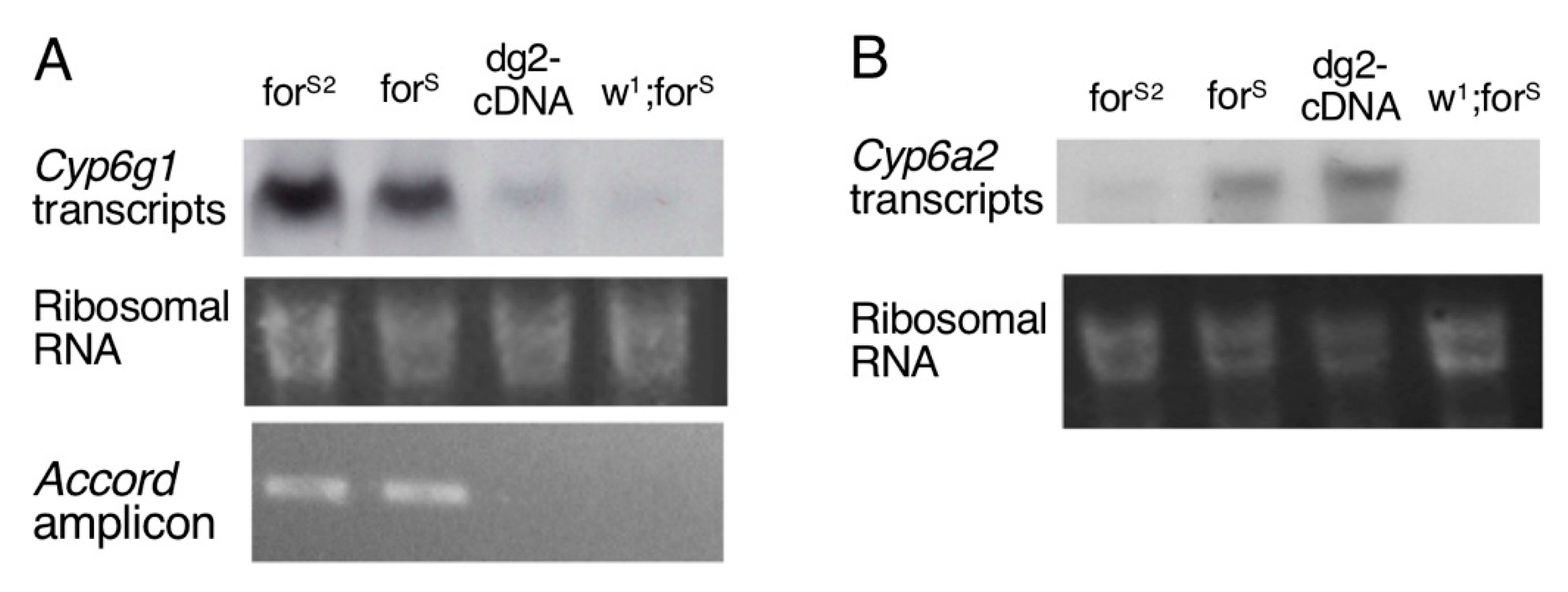
3.4. Cytochrome P450 Genes Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Charnov, E.L. Optimal Foraging, the Marginal Value Theorem. Theor. Popul. Biol. 1976, 9, 129–136. [Google Scholar] [CrossRef]
- Emlen, J.M. The Role of Time and Energy in Food Preference. Am. Nat. 1966, 100, 611–617. Available online: www.jstor.org/stable/2459299 (accessed on 16 August 2020). [CrossRef]
- Fretwell, S.D.; Lucas, H.L. On Territorial Behavior and Other Factors Influencing Habitat Distribution in Birds. I. Theoretical Development. Acta Biotheor. 1970, 19, 16–36. [Google Scholar] [CrossRef]
- MacArthur, R.H.; Pianka, E.R. On Optimal Use of a Patchy Environment. Am. Nat. 1966, 100, 603–609. Available online: www.jstor.org/stable/2459298 (accessed on 1 August 2020). [CrossRef]
- Schoener, T.W. Theory of Feeding Strategies. Ann. Rev. Ecol. Syst. 1971, 2, 369–404. [Google Scholar] [CrossRef]
- Rymer, T.L.; Cruise, M.; Pillay, N. Decision-Making by Bushveld Gerbils (Gerbilliscus Leucogaster). J. Comp. Psychol. 2021, 135, 244–257. [Google Scholar] [CrossRef]
- Stockwell, J.D.; O’Malley, B.P.; Hansson, S.; Chapina, R.J.; Rudstam, L.G.; Weidel, B.C. Benthic Habitat Is an Integral Part of Freshwater Mysis Ecology. Freshw. Biol. 2020, 65, 1997–2009. [Google Scholar] [CrossRef] [PubMed]
- Pereira, H.S.; Sokolowski, M.B. Mutations in the Larval Foraging Gene Affect Adult Locomotory Behavior after Feeding in Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 1993, 90, 5044–5046. [Google Scholar] [CrossRef] [PubMed]
- Kaun, K.R.; Sokolowski, M.B. CGMP-Dependent Protein Kinase: Linking Foraging to Energy Homeostasis. Genome 2009, 52, 1–7. [Google Scholar] [CrossRef][Green Version]
- Osborne, K.A.; Robichon, A.; Burgess, E.; Butland, S.; Shaw, R.A.; Coulthard, A.; Pereira, H.S.; Greenspan, R.J.; Sokolowski, M.B. Natural Behavior Polymorphism Due to a CGMP-Dependent Protein Kinase of Drosophila. Science 1997, 277, 834–836. [Google Scholar] [CrossRef]
- Sokolowski, M.B. Foraging Strategies of Drosophila Melanogaster: A Chromosomal Analysis. Behav. Genet. 1980, 10, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Sokolowski, M.B.; Pereira, H.S.; Hughes, K. Evolution of Foraging Behavior in Drosophila by Density-Dependent Selection. Proc. Natl. Acad. Sci. USA 1997, 94, 7373–7377. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, M.J.; Feder, E.; Rowe, L.; Sokolowski, M.B. Maintaining a Behaviour Polymorphism by Frequency-Dependent Selection on a Single Gene. Nature 2007, 447, 210–212. [Google Scholar] [CrossRef] [PubMed]
- Edelsparre, A.H.; Vesterberg, A.; Lim, J.H.; Anwari, M.; Fitzpatrick, M.J. Alleles Underlying Larval Foraging Behaviour Influence Adult Dispersal in Nature. Ecol. Lett. 2014, 17, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Dason, J.S.; Cheung, A.; Anreiter, I.; Montemurri, V.A.; Allen, A.M.; Sokolowski, M.B. Drosophila melanogaster Foraging regulates a Nociceptive-like Escape Behavior through a Developmentally Plastic Sensory Circuit. Proc. Natl. Acad. Sci. USA 2019, 27, 201820840–201820846. [Google Scholar] [CrossRef]
- Hofmann, F.; Feil, R.; Kleppisch, T.; Schlossmann, J. Function of CGMP-Dependent Protein Kinases as Revealed by Gene Deletion. Physiol. Rev. 2006, 86, 1–23. [Google Scholar] [CrossRef]
- Peng, Q.; Wang, Y.; Li, M.; Yuan, D.; Xu, M.; Li, C.; Gong, Z.; Jiao, R.; Liu, L. cGMP-Dependent Protein Kinase Encoded by Foraging Regulates Motor Axon Guidance in Drosophila by Suppressing Lola Function. J. Neurosci. 2016, 36, 4635–4646. [Google Scholar] [CrossRef]
- Reaume, C.J.; Sokolowski, M.B. CGMP-Dependent Protein Kinase as a Modifier of Behaviour. In cGMP: Generators, Effectors and Therapeutic Implications; Handbook of Experimental Pharmacology; Schmidt, H.H.H.W., Hofmann, F., Stasch, J.-P., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 191, pp. 423–443. [Google Scholar] [CrossRef]
- Liu, N. Insecticide Resistance in Mosquitoes: Impact, Mechanisms, and Research Directions. Annu. Rev. Entomol. 2015, 60, 537–559. [Google Scholar] [CrossRef]
- Bhatt, P.; Zhou, X.; Huang, Y.; Zhang, W.; Chen, S. Characterization of the Role of Esterases in the Biodegradation of Organophosphate, Carbamate, and Pyrethroid Pesticides. J. Hazard. Mater. 2021, 411, 125026. [Google Scholar] [CrossRef]
- Palli, S.R. CncC/Maf-mediated xenobiotic response pathway in insects. Arch. Insect Biochem. Physiol. 2020, 104, e21674. [Google Scholar] [CrossRef]
- Rane, R.V.; Ghodke, A.B.; Hoffmann, A.A.; Edwards, O.R.; Walsh, T.K.; Oakeshott, J.G. Detoxifying Enzyme Complements and Host Use Phenotypes in 160 Insect Species. Curr. Opin. Insect Sci. 2019, 31, 131–138. [Google Scholar] [CrossRef]
- Piechowicz, B.; Sudoł, M.; Grodzicki, P.; Podbielska, M.; Szpyrka, E.; Zwolak, A.; Potocki, L. The dynamics of pyrethroid residues and Cyp P450 gene expression in insects depends on the circadian clock. Environ. Res. 2021, 194. [Google Scholar] [CrossRef]
- Ben-Shahar, Y.; Robichon, A.; Sokolowski, M.B.; Robinson, G.E. Influence of gene action across different time scales on behavior. Science 2002, 296, 741–744. [Google Scholar] [CrossRef]
- Heylen, K.; Gobin, B.; Billen, J.; Hu, T.T.; Arckens, L.; Huybrechts, R. Amfor expression in the honeybee brain: A trigger mechanism for nurse-forager transition. J. Insect Physiol. 2008, 54, 1400–1403. [Google Scholar] [CrossRef]
- Ben-Shahar, Y.; Leung, H.T.; Pak, W.L.; Sokolowski, M.B.; Robinson, G.E. cGMP- dependent changes in phototaxis: A possible role for the foraging gene in honey bee division of labor. J. Exp. Biol. 2003, 206 Pt 14, 2507–2515. [Google Scholar] [CrossRef]
- Ben-Shahar, Y. The foraging gene, behavioral plasticity, and honeybee division of labor. J. Comp. Physiol. 2005, 191, 987–994. [Google Scholar] [CrossRef]
- De-Belle, J.S.; Hilliker, A.J.; Sokolowski, M.B. Genetic Localization of Foraging (for): A Major Gene for Larval Behavior in Drosophila Melanogaster. Genetics 1989, 123, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Johnstone, A.F.M.; Aydin, C. Thermal Stress and Toxicity. Compr. Physiol. 2014, 4, 995–1016. [Google Scholar] [CrossRef] [PubMed]
- Casida, J.E. Pest Toxicology: The Primary Mechanisms of Pesticide Action. Chem. Res. Toxicol. 2009, 22, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, R.L. Encyclopedia of Environmental Science and Engineering, 6th ed.; Pfafflin, J.R., Ziegler, E.N., Eds.; Taylor & Francis: New York, NY, USA, 2012; ISBN 9781439804421. [Google Scholar]
- Pisani-Borg, E.; Cuany, A.; Brun, A.; Amichot, M.; Fournier, D.; Bergé, J.B. Oxidative Degradation of Diazinon by Drosophila: Metabolic Changes Associated with Insecticide Resistance and Induction. Pest. Biochem. Physiol. 1996, 54, 56–64. [Google Scholar] [CrossRef]
- Soto, A.R.; Deichmann, W.B. Major Metabolism and Acute Toxicity of Aldrin, Dieldrin, and Endrin. Environ. Res. 1967, 1, 307–322. [Google Scholar] [CrossRef]
- Stevenson, B.J.; Bibby, J.; Pignatelli, P.; Muangnoicharoen, S.; O’Neill, P.M.; Lian, L.-Y.; Müller, P.; Nikou, D.; Steven, A.; Hemingway, J.; et al. Cytochrome P450 6M2 from the Malaria Vector Anopheles Gambiae Metabolizes Pyrethroids: Sequential Metabolism of Deltamethrin Revealed. Insect Biochem. Mol. Biol. 2011, 41, 492–502. [Google Scholar] [CrossRef]
- Hodgson, E.; Levi, P. Interactions of Piperonyl Butoxide with Cytochrome P450. In Piperonyl Butoxide; Jones, D., Ed.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 41–53. [Google Scholar] [CrossRef]
- de Sousa, G.; Cuany, A.; Brun, A.; Amichot, M.; Rahmani, R.; Bergé, J.-B. A Microfluorometric Method for Measuring Ethoxycoumarin-O-Deethylase Activity on Individual Drosophila Melanogaster Abdomens: Interest for Screening Resistance in Insect Populations. Anal. Biochem. 1995, 229, 86–91. [Google Scholar] [CrossRef]
- Nauen, R.; Stumpf, N. Fluorometric Microplate Assay to Measure Glutathione S-Transferase Activity in Insects and Mites Using Monochlorobimane. Anal. Biochem. 2002, 303, 194–198. [Google Scholar] [CrossRef]
- Pasteur, N.; Raymond, M.; Rousset, F.; Bergé, J.B.; Amichot, M.; Pauron, D.; Cuany, A.; Fournier, D. Cloning and Detection of Insecticide Resistance Genes. In The Molecular Biology of Insect Disease Vectors; Crampton, J.M., Beard, C.B., Louis, C., Eds.; Chapman & Hall: Dordrecht, The Netherlands, 1997; pp. 399–419. [Google Scholar] [CrossRef]
- Zhou, X.; Scharf, M.E.; Meinke, L.J.; Chandler, L.D.; Siegfried, B.D. Characterization of General Esterases from Methyl Parathion-Resistant and -Susceptible Populations of Western Corn Rootworm (Coleoptera: Chrysomelidae). J. Econ. Entomol. 2003, 96, 1855–1863. [Google Scholar] [CrossRef][Green Version]
- Daborn, P.J.; Yen, J.L.; Bogwitz, M.R.; Le Goff, G.; Feil, E.; Jeffers, S.; Tijet, N.; Perry, T.; Heckel, D.; Batterham, P. A Single P450 Allele Associated with Insecticide Resistance in Drosophila. Science 2002, 297, 2253–2256. [Google Scholar] [CrossRef] [PubMed]
- Hemingway, J.; Ranson, H. Insecticide Resistance in Insect Vectors of Human Disease. Annu. Rev. Entomol. 2000, 45, 371–391. [Google Scholar] [CrossRef] [PubMed]
- Dunkov, B.C.; Guzov, V.M.; Mocelin, G.; Shotkoski, F.; Brun, A.; Amichot, M.; Ffrench-Constant, R.H.; Feyereisen, R. The Drosophila Cytochrome P450 Gene Cyp6a2: Structure, Localization, Heterologous Expression, and Induction by Phenobarbital. DNA Cell Biol. 1997, 16, 1345–1356. [Google Scholar] [CrossRef]
- Joußen, N.; Schuphan, I.; Schmidt, B. Metabolism of Methoxychlor by the P450-Monooxygenase CYP6G1 Involved in Insecticide Resistance of Drosophila Melanogaster after Expression in Cell Cultures of Nicotiana Tabacum. Chem. Biodivers. 2010, 7, 722–735. [Google Scholar] [CrossRef]
- Chung, H.; Bogwitz, M.R.; McCart, C.; Andrianopoulos, A.; Ffrench-Constant, R.H.; Batterham, P.; Daborn, P.J. Cis-Regulatory Elements in the Accord Retrotransposon Result in Tissue-Specific Expression of the Drosophila Melanogaster Insecticide Resistance Gene Cyp6g1. Genetics 2007, 175, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Kent, C.F.; Daskalchuk, T.; Cook, L.; Sokolowski, M.B.; Greenspan, R.J. The Drosophila Foraging Gene Mediates Adult Plasticity and Gene-Environment Interactions in Behaviour, Metabolites, and Gene Expression in Response to Food Deprivation. PLoS Genet. 2009, 5, e1000609. [Google Scholar] [CrossRef]
- Castel, H.; Jégou, S.; Tonon, M.C.; Vaudry, H. Regulation of the GABAA Receptor by Nitric Oxide in Frog Pituitary Melanotrophs. Endocrinology 2000, 141, 3451–3460. [Google Scholar] [CrossRef]
- Galisteo, M.; Marc, N.; Fautrel, A.; Guillouzo, A.; Corcos, L.; Lagadic-Gossmann, D. Involvement of Cyclic Nucleotide-and Calcium-Regulated Pathways in Phenobarbital-Induced Cytochrome P-450 3A Expression in Mouse Primary Hepatocytes. J. Pharmacol. Exp. Ther. 1999, 290, 1270–1277. [Google Scholar]
- Marc, N.; Galisteo, M.; Lagadic-Gossmann, D.; Fautrel, A.; Joannard, F.; Guillouzo, A.; Corcos, L. Regulation of Phenobarbital Induction of the Cytochrome P450 2b9/10 Genes in Primary Mouse Hepatocyte Culture. Involvement of Calcium- and CAMP-Dependent Pathways. Eur. J. Biochem. 2000, 267, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Jiang, X.; Gu, S.; Lai, Y.; Liu, Y.; Zhang, Z. Protection of Nrf2 against Arsenite-Induced Oxidative Damage Is Regulated by the Cyclic Guanosine Monophosphate-Protein Kinase G Signaling Pathway. Environ. Toxicol. 2016, 32, 2004–2020. [Google Scholar] [CrossRef] [PubMed]
- Ashino, T.; Yamamoto, M.; Numazawa, S. Nrf2 Antioxidative System is Involved in Cytochrome P450 Gene Expression and Activity: A Delay in Pentobarbital Metabolism in Nrf2-Deficient Mice. Drug Metab. Dispos. 2020, 48, 673–680. [Google Scholar] [CrossRef]
- Misra, J.R.; Lam, G.; Thummel, C.S. Constitutive Activation of the Nrf2/Keap1 Pathway in Insecticide-Resistant Strains of Drosophila. Insect Biochem. Mol. Biol. 2013, 43, 1116–1124. [Google Scholar] [CrossRef]
- Brun, A.; Cuany, A.; Le Mouel, T.; Berge, J.; Amichot, M. Inducibility of the Drosophila Melanogaster Cytochrome P450 Gene, CYP6A2, by Phenobarbital in Insecticide Susceptible or Resistant Strains. Insect Biochem. Mol. Biol. 1996, 26, 697–703. [Google Scholar] [CrossRef]
- Sun, W.; Margam, V.M.; Sun, L.; Buczkowski, G.; Bennett, G.W.; Schemerhorn, B.; Muir, W.M.; Pittendrigh, B.R. Genome-Wide Analysis of Phenobarbital-Inducible Genes in Drosophila Melanogaster. Insect Mol. Biol. 2006, 15, 455–464. [Google Scholar] [CrossRef]
- Chen, A.; Kramer, E.F.; Purpura, L.; Krill, J.L.; Zars, T.; Dawson-Scully, K. The Influence of Natural Variation at the Foraging Gene on Thermotolerance in Adult Drosophila in a Narrow Temperature Range. J. Comp. Physiol. A 2011, 197, 1113–1118. [Google Scholar] [CrossRef]
- Dawson-Scully, K.; Armstrong, G.A.B.; Kent, C.; Robertson, R.M.; Sokolowski, M.B. Natural Variation in the Thermotolerance of Neural Function and Behavior Due to a CGMP-Dependent Protein Kinase. PLoS ONE 2007, 2, e773. [Google Scholar] [CrossRef] [PubMed]
- Dawson-Scully, K.; Bukvic, D.; Chakaborty-Chatterjee, M.; Ferreira, R.; Milton, S.L.; Sokolowski, M.B. Controlling Anoxic Tolerance in Adult Drosophila via the CGMP–PKG Pathway. J. Exp. Biol. 2010, 213, 2410–2416. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.L.; Levine, M.T.; Eckert, M.L.; Begun, D.J. Genomic Analysis of Adaptive Differentiation in Drosophila Melanogaster. Genetics 2008, 179, 455–473. [Google Scholar] [CrossRef]
- Struk, A.A.; Mugon, J.; Huston, A.; Scholer, A.A.; Stadler, G.; Higgins, E.T.; Sokolowski, M.B.; Danckert, J. Self-Regulation and the Foraging Gene (PRKG1) in Humans. Proc. Natl. Acad. Sci. USA 2019, 116, 4434–4439. [Google Scholar] [CrossRef] [PubMed]


| Gene | Forward | Reverse |
|---|---|---|
| Cyp6a2 | CTGGTCAACGACACGATTGC | GTAGGTCATGGCCTTGATGG |
| Cyp6g1 | CGATCATTGCAACACCAAGG | TCGCGTATTATCAAGCCGGG |
| Accord | GGGTGCAACAGAGTTTCAGGTA | CTTTTTGTGTGCTATGGTTTAGTTAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amichot, M.; Tarès, S. The Foraging Gene, a New Environmental Adaptation Player Involved in Xenobiotic Detoxification. Int. J. Environ. Res. Public Health 2021, 18, 7508. https://doi.org/10.3390/ijerph18147508
Amichot M, Tarès S. The Foraging Gene, a New Environmental Adaptation Player Involved in Xenobiotic Detoxification. International Journal of Environmental Research and Public Health. 2021; 18(14):7508. https://doi.org/10.3390/ijerph18147508
Chicago/Turabian StyleAmichot, Marcel, and Sophie Tarès. 2021. "The Foraging Gene, a New Environmental Adaptation Player Involved in Xenobiotic Detoxification" International Journal of Environmental Research and Public Health 18, no. 14: 7508. https://doi.org/10.3390/ijerph18147508
APA StyleAmichot, M., & Tarès, S. (2021). The Foraging Gene, a New Environmental Adaptation Player Involved in Xenobiotic Detoxification. International Journal of Environmental Research and Public Health, 18(14), 7508. https://doi.org/10.3390/ijerph18147508






