Characterization and Transcriptome Analysis of a Long-Chain n-Alkane-Degrading Strain Acinetobacter pittii SW-1
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain, Media, and Chemicals
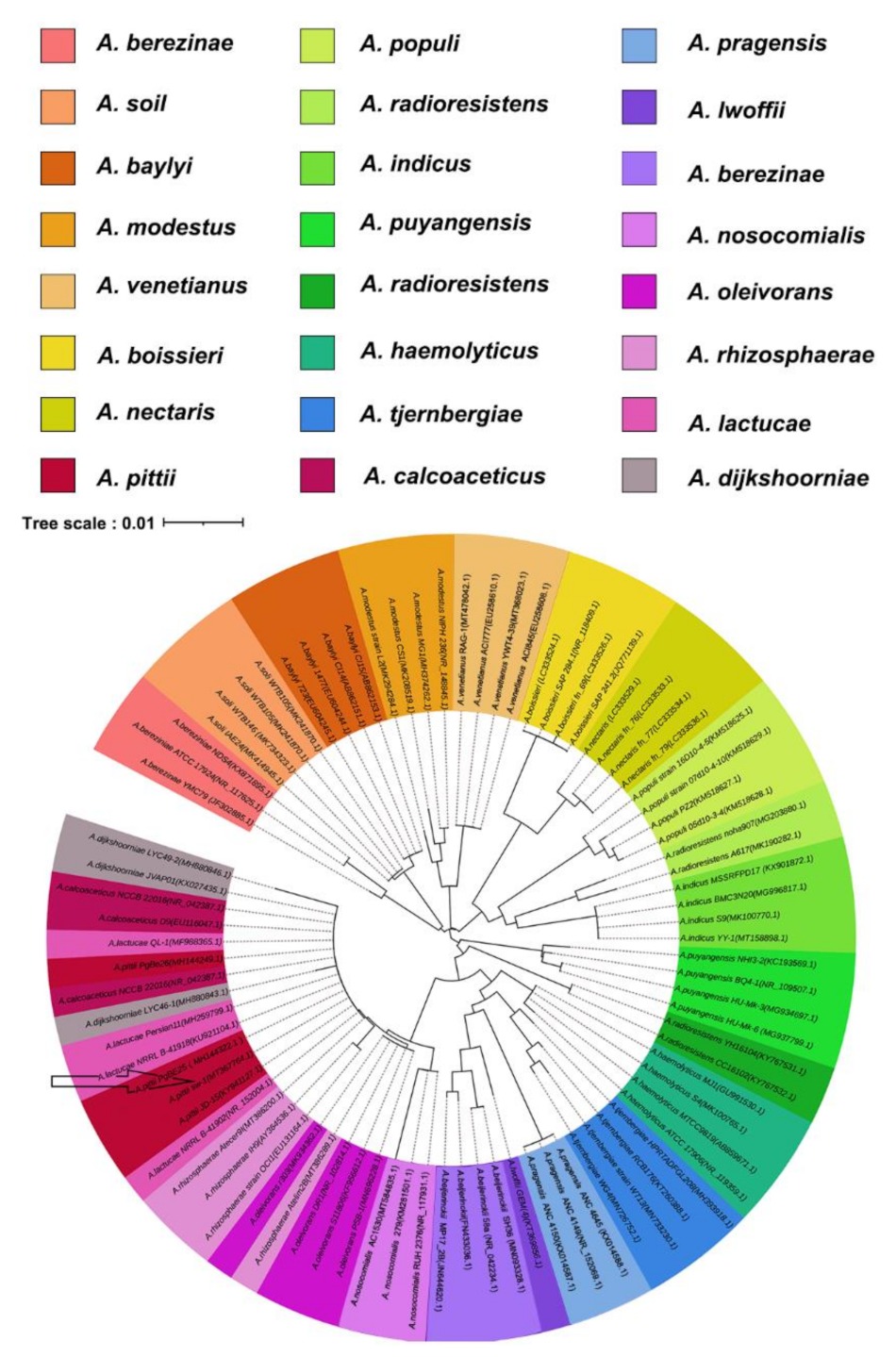
2.2. 16S rRNA Gene Analysis and Phylogenic Tree Construction
2.3. Genome Sequencing and Comparative Genomics Analysis
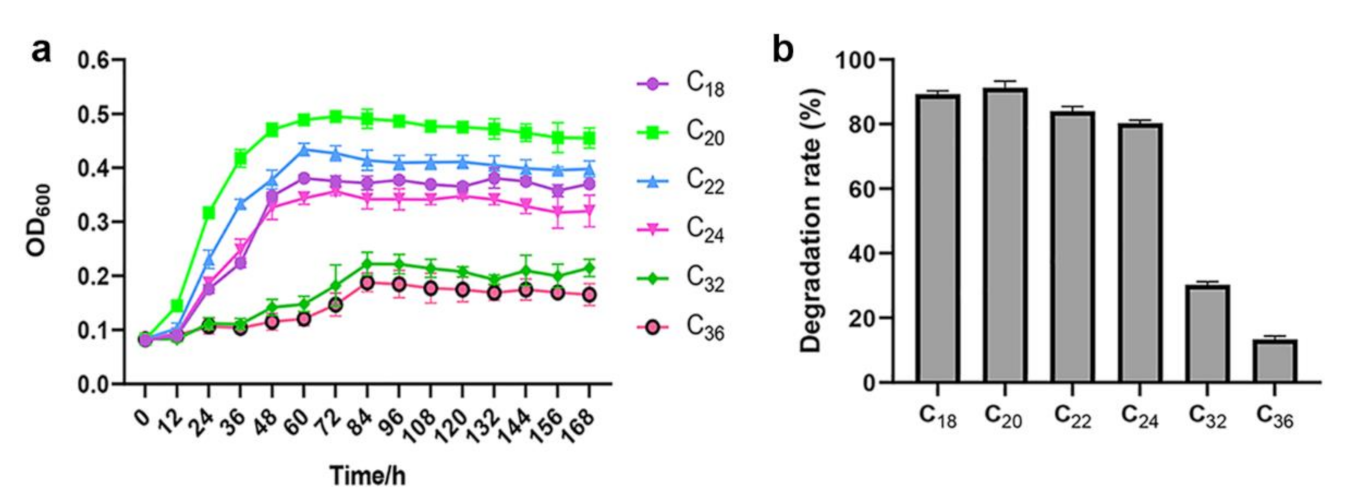
2.4. Growth and Biodegradation Rate of SW-1 in the Presence of Various n-Alkanes
2.5. RNA-Seq and Data Analysis
2.6. RT-qPCR
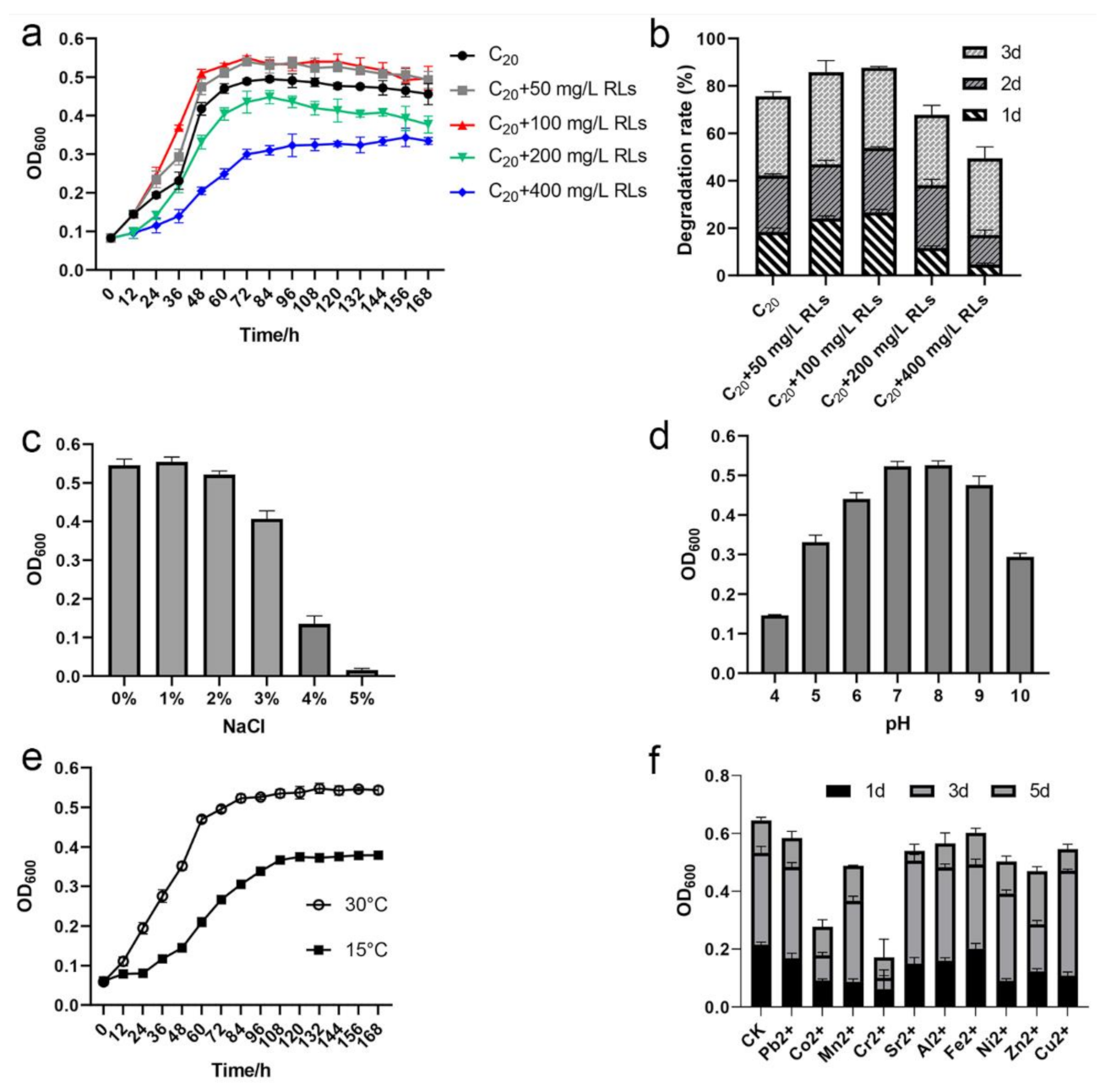
2.7. Effect of Rhamnolipid, NaCl, pH, and Temperature on C20 Utilization by Strain SW-1
2.8. Heavy Metal Tolerance Assays
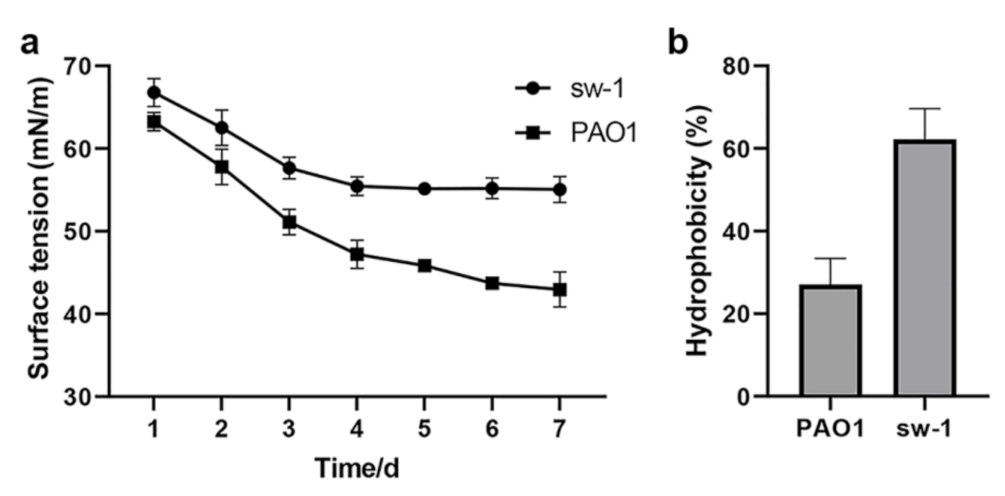
2.9. Determination of the Surface Tension and Cell Surface Hydrophobicity
3. Results
3.1. Isolation and Identification of A. pittii Strain SW-1
3.2. A. pittii SW-1 Degrades LC n-Alkanes
3.3. Analysis of the Genes Involved in Alkane Degradation
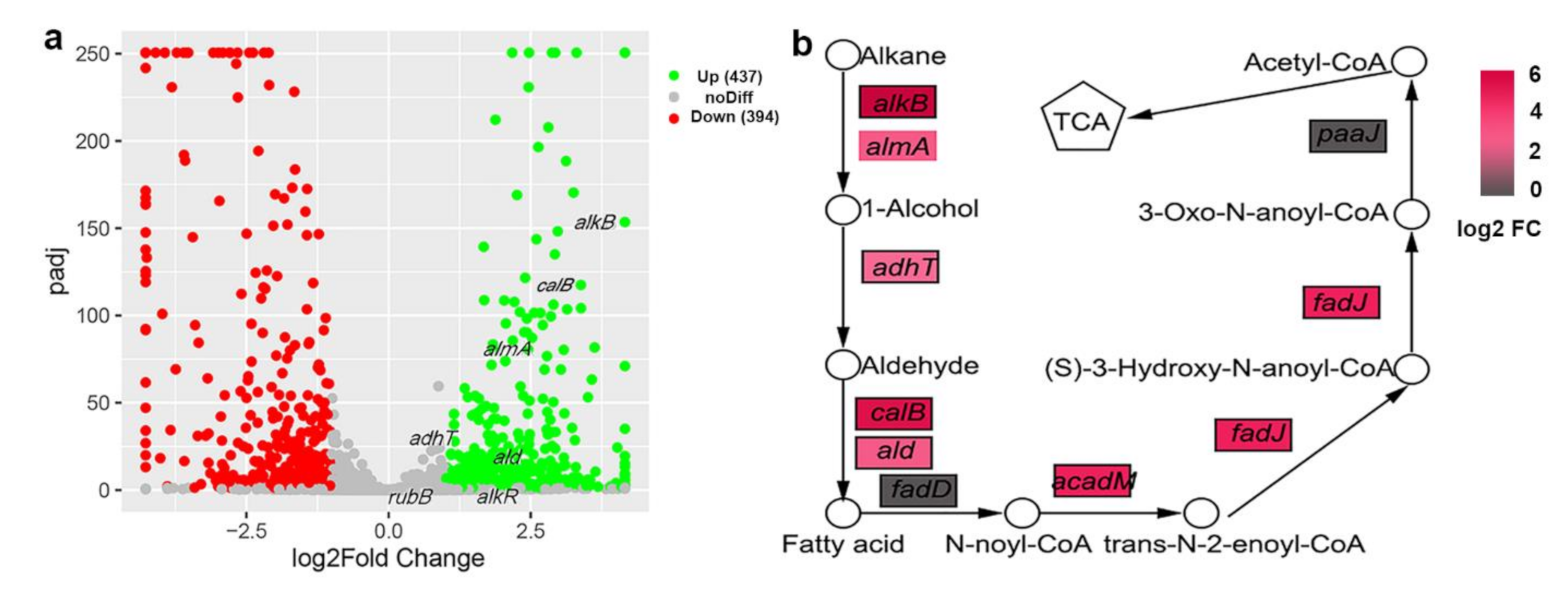
3.4. Alkane Degradation-Related Genes Were Induced by C20
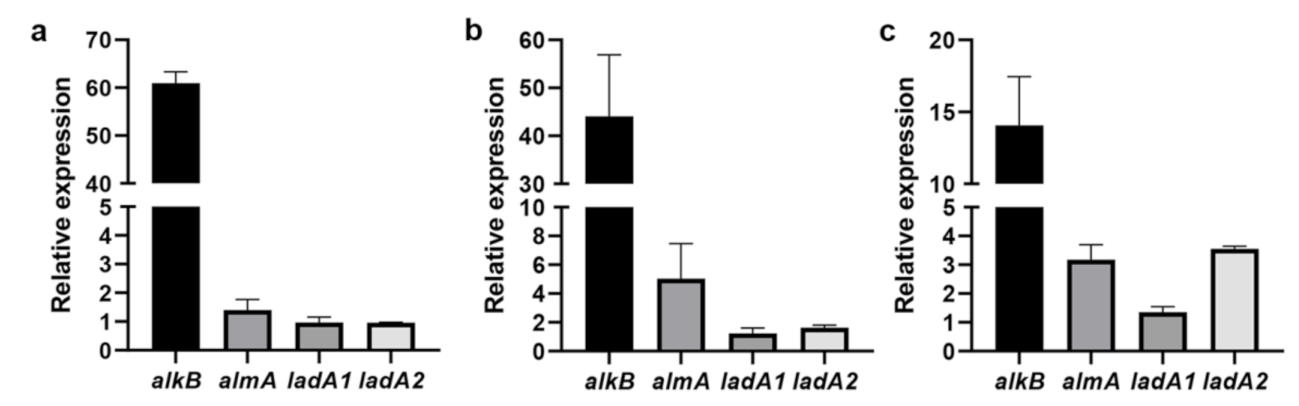
3.5. Detection of the Expression of Alkane Degradation-Related Genes by RT-qPCR
3.6. Effects of Rhamnolipids, NaCl, pH, Low Temperature, and Heavy Metals on A. pittii SW-1 Growth
3.7. Cell Surface Hydrophobicity Might Contribute to the Alkane Uptake in A. pittii SW-1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarkar, P.; Roy, A.; Pal, S.; Mohapatra, B.; Kazy, S.K.; Maiti, M.K.; Sar, P. Enrichment and characterization of hydrocarbon-degrading bacteria from petroleum refinery waste as potent bioaugmentation agent for in situ bioremediation. Bioresour. Technol. 2017, 242, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Throne-Holst, M.; Markussen, S.; Winnberg, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B. Utilization of n-alkanes by a newly isolated strain of Acinetobacter venetianus: The role of two AlkB-type alkane hydroxylases. Appl. Microbiol. Biotechnol. 2006, 72, 353–360. [Google Scholar] [CrossRef]
- Zampolli, J.; Collina, E.; Lasagni, M.; Di Gennaro, P. Biodegradation of variable-chain-length n-alkanes in Rhodococcus opacus R7 and the involvement of an alkane hydroxylase system in the metabolism. AMB Express 2014, 4, 73. [Google Scholar] [CrossRef]
- Al-Hawash, A.B.; Zhang, J.; Li, S.; Liu, J.; Ghalib, H.B.; Zhang, X.; Ma, F. Biodegradation of n-hexadecane by Aspergillus sp. RFC-1 and its mechanism. Ecotoxicol. Environ. Saf. 2018, 164, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Simister, R.; Poutasse, C.; Thurston, A.; Reeve, J.; Baker, M.; White, H. Degradation of oil by fungi isolated from Gulf of Mexico beaches. Mar. Pollut. Bull. 2015, 100, 327–333. [Google Scholar] [CrossRef]
- El-Sheekh, M.M.; Ghareib, M.; El-Souod, G.A. Biodegradation of Phenolic and Polycyclic Aromatic Compounds by Some Algae and Cyanobacteria. J. Bioremediat. Biodegrad. 2012, 3, 13. [Google Scholar] [CrossRef]
- Safonova, E.; Kvitko, K.V.; Kuschk, P.; Moder, M.; Reisser, W. Biodegradation of Phenanthrene by the Green AlgaScenedesmus obliquus ES-55. Eng. Life Sci. 2005, 5, 234–239. [Google Scholar] [CrossRef]
- Margesin, R.; Schinner, F. Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl. Microbiol. Biotechnol. 2001, 56, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Kostka, J.E.; Prakash, O.; Overholt, W.A.; Green, S.J.; Freyer, G.; Canion, A.; Delgardio, J.; Norton, N.; Hazen, T.C.; Huettel, M. Hydro-carbon-degrading bacteria and the bacterial community response in gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 2011, 77, 7962–7974. [Google Scholar] [CrossRef] [PubMed]
- Mahjoubi, M.; Jaouani, A.; Guesmi, A.; Ben Amor, S.; Jouini, A.; Cherif, H.; Najjari, A.; Boudabous, A.; Koubaa, N.; Cherif, A. Hydro-carbonoclastic bacteria isolated from petroleum contaminated sites in Tunisia: Isolation, identification and characterization of the biotechnological potential. New Biotechnol. 2013, 30, 723–733. [Google Scholar] [CrossRef]
- Ratajczak, A.; Geißdörfer, W.; Hillen, W. Expression of Alkane Hydroxylase from Acinetobacter sp. Strain ADP1 Is Induced by a Broad Range of n-Alkanes and Requires the Transcriptional Activator AlkR. J. Bacteriol. 1998, 180, 5822–5827. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Martin, F.L.L.; Zhang, D. Quantification of Chemotaxis-Related Alkane Accumulation inAcinetobacter baylyiUsing Raman Microspectroscopy. Anal. Chem. 2017, 89, 3909–3918. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhao, B.; Lan, Y.; Ma, T. Enhanced degradation of different crude oils by defined engineered consortia of Acinetobacter venetianus RAG-1 mutants based on their alkane metabolism. Bioresour. Technol. 2021, 327, 124787. [Google Scholar] [CrossRef]
- Farinas Edgardo, T.; Schwaneberg, U.; Glieder, A.; Arnold Frances, H. Directed Evolution of a Cytochrome P450 Monooxygenase for Alkane Oxidation. Adv. Synth. Catal. 2001, 343, 601–606. [Google Scholar] [CrossRef]
- Maier, T.; Förster, H.-H.; Asperger, O.; Hahn, U. Molecular Characterization of the 56-kDa CYP153 from Acinetobacter sp. EB104. Biochem. Biophys. Res. Commun. 2001, 286, 652–658. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Eggink, G.; Enequist, H.; Bos, R.; Witholt, B. DNA sequence determination and functional characterization of the OCT-plasmid-encoded alkJKL genes of Pseudomonas oleovorans. Mol. Microbiol. 1992, 6, 3121–3136. [Google Scholar] [CrossRef]
- Park, C.; Shin, B.; Jung, J.; Lee, Y.; Park, W. Metabolic and stress responses ofAcinetobacter oleivoransDR1 during long-chain alkane degradation. Microb. Biotechnol. 2017, 10, 1809–1823. [Google Scholar] [CrossRef]
- Throne-Holst, M.; Wentzel, A.; Ellingsen, T.E.; Kotlar, H.-K.; Zotchev, S.B. Identification of Novel Genes Involved in Long-Chain n -Alkane Degradation by Acinetobacter sp. Strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332. [Google Scholar] [CrossRef]
- Feng, L.; Wang, W.; Cheng, J.; Ren, Y.; Zhao, G.; Gao, C.; Tang, Y.; Liu, X.; Han, W.; Peng, X.; et al. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Natl. Acad. Sci. USA 2007, 104, 5602–5607. [Google Scholar] [CrossRef]
- Liu, H.; Xu, J.; Liang, R.; Liu, J. Characterization of the Medium- and Long-Chain n-Alkanes Degrading Pseudomonas aeruginosa Strain SJTD-1 and Its Alkane Hydroxylase Genes. PLoS ONE 2014, 9, e105506. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Pan, J.C.; Ma, Y.L. Elucidation of multiple alkane hydroxylase systems in biodegradation of crude oil n-alkane pollution by Pseudomonas aeruginosa DN1. J. Appl. Microbiol. 2019, 128, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gai, L.; Hou, Z.; Yang, C.; Ma, C.; Wang, Z.; Sun, B.; He, X.; Tang, H.; Xu, P. Characterization and biotechnological potential of petroleum-degrading bacteria isolated from oil-contaminated soils. Bioresour. Technol. 2010, 101, 8452–8456. [Google Scholar] [CrossRef]
- Zhang, Z.; Hou, Z.; Yang, C.; Ma, C.; Tao, F.; Xu, P. Degradation of n-alkanes and polycyclic aromatic hydrocarbons in petroleum by a newly isolated Pseudomonas aeruginosa DQ8. Bioresour. Technol. 2011, 102, 4111–4116. [Google Scholar] [CrossRef]
- Cecchini, T.; Yoon, E.J.; Charretier, Y.; Bardet, C.; Beaulieu, C.; Lacoux, X.; Docquier, J.D.; Lemoine, J.; Courvalin, P.; Grillot-Courvalin, C.; et al. Deciphering Multifactorial Resistance Phenotypes in Acinetobacter baumannii by Genomics and Targeted La-bel-free Proteomics. Mol. Cell. Proteom. MCP 2018, 17, 442–456. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chettri, B.; Singha, N.A.; Mukherjee, A.; Rai, A.N.; Chattopadhyay, D.; Singh, A.K. Hydrocarbon degradation potential and compet-itive persistence of hydrocarbonoclastic bacterium Acinetobacter pittii strain ABC. Arch. Microbiol. 2019, 201, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, Y.; Omori, T.; Kusudo, H.; Bistafa, C.; Yamaguchi, Y. Wilhelmy equation revisited: A lightweight method to measure liquid–vapor, solid–liquid, and solid–vapor interfacial tensions from a single molecular dynamics simulation. J. Chem. Phys. 2020, 153, 034701. [Google Scholar] [CrossRef]
- Rosenberg, M. Microbial adhesion to hydrocarbons: Twenty-five years of doing MATH. FEMS Microbiol. Lett. 2006, 262, 129–134. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 2, 346–351. [Google Scholar] [CrossRef]
- Geissdörfer, W.; Kok, R.G.; Ratajczak, A.; Hellingwerf, K.J.; Hillen, W. The genes rubA and rubB for alkane degradation in Acineto-bacter sp. strain ADP1 are in an operon with estB, encoding an esterase, and oxyR. J. Bacteriol. 1999, 181, 4292–4298. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tani, A.; Ishige, T.; Sakai, Y.; Kato, N. Gene Structures and Regulation of the Alkane Hydroxylase Complex in Acinetobacter sp. Strain M-1. J. Bacteriol. 2001, 183, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Kothari, A.; Charrier, M.; Wu, Y.-W.; Malfatti, S.; Zhou, C.E.; Singer, S.W.; Dugan, L.; Mukhopadhyay, A. Transcriptomic analysis of the highly efficient oil-degrading bacteriumAcinetobacter venetianusRAG-1 reveals genes important in dodecane uptake and utilization. FEMS Microbiol. Lett. 2016, 363, 224. [Google Scholar] [CrossRef][Green Version]
- Wang, W.; Shao, Z. Genes involved in alkane degradation in the Alcanivorax hongdengensis strain A-11-3. Appl. Microbiol. Biotechnol. 2011, 94, 437–448. [Google Scholar] [CrossRef]
- Doong, R.-A.; Lei, W.-G. Solubilization and mineralization of polycyclic aromatic hydrocarbons by Pseudomonas putida in the presence of surfactant. J. Hazard. Mater. 2003, 96, 15–27. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Piotrowska-Seget, Z.; Cameotra, S.S. Environmental Applications of Biosurfactants: Recent Advances. Int. J. Mol. Sci. 2011, 12, 633–654. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, Z.; Li, Y.; Peng, X.; Li, W.; Yan, Y. Biodegradation of p-nitrophenol by Rhodococcus sp. CN6 with high cell surface hydrophobicity. J. Hazard. Mater. 2009, 163, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Lal, B.; Khanna, S. Degradation of crude oil by Acinetobacter calcoaceticus and Alcaligenes odorans. J. Appl. Bacteriol. 1996, 81, 355–362. [Google Scholar]
- Liu, C.; Wang, W.; Wu, Y.; Zhou, Z.; Lai, Q.; Shao, Z. Multiple alkane hydroxylase systems in a marine alkane degrader, Alcanivorax dieselolei B-5. Environ. Microbiol. 2011, 13, 1168–1178. [Google Scholar] [CrossRef]
- Boonmak, C.; Takahashi, Y.; Morikawa, M. Cloning and expression of three ladA-type alkane monooxygenase genes from an extremely thermophilic alkane-degrading bacterium Geobacillus thermoleovorans B23. Extremophiles 2014, 18, 515–523. [Google Scholar] [CrossRef]
- Bowman, J.S.; Deming, J.W. Alkane hydroxylase genes in psychrophile genomes and the potential for cold active catalysis. BMC Genom. 2014, 15, 1120. [Google Scholar] [CrossRef]
- Banat, I.M.; Makkar, R.S.; Cameotra, S.S. Potential commercial applications of microbial surfactants. Appl. Microbiol. Biotechnol. 2000, 53, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Ron, E.Z.; Rosenberg, E. Biosurfactants and oil bioremediation. Curr. Opin. Biotechnol. 2002, 13, 249–252. [Google Scholar] [CrossRef]
- Shreve, G.S.; Inguva, S.; Gunnam, S. Rhamnolipid biosurfactant enhancement of hexadecane biodegradation by Pseudomonas aeruginosa. Mol. Mar. Boil. Biotechnol. 1995, 4, 331–337. [Google Scholar]
- Song, D.; Liang, S.; Yan, L.; Shang, Y.; Wang, X. Solubilization of Polycyclic Aromatic Hydrocarbons by Single and Binary Mixed Rhamnolipid-Sophorolipid Biosurfactants. J. Environ. Qual. 2016, 45, 1405–1412. [Google Scholar] [CrossRef]
- Yu, H.; Huang, G.H.; Xiao, H.; Wang, L.; Chen, W. Combined effects of DOM and biosurfactant enhanced biodegradation of pol-ycylic armotic hydrocarbons (PAHs) in soil-water systems. Environ. Sci. Pollut. Res. Int. 2014, 21, 10536–10549. [Google Scholar] [CrossRef]
- Jung, J.; Jang, I.-A.; Ahn, S.; Shin, B.; Kim, J.; Park, C.; Jee, S.-C.; Sung, J.-S.; Park, W. Molecular Mechanisms of Enhanced Bacterial Growth on Hexadecane with Red Clay. Microb. Ecol. 2015, 70, 912–921. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Wubbolts, M.G.; Witholt, B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 1994, 5, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Gai, Y.; Guo, X.; Hou, Y.; Zeng, R. Properties and Applications of Extremozymes from Deep-Sea Extremophilic Microor-ganisms: A Mini Review. Mar. Drugs. 2019, 17, 656. [Google Scholar] [CrossRef] [PubMed]
- Perez Calderon, L.J.; Gontikaki, E.; Potts, L.D.; Shaw, S.; Gallego, A.; Anderson, J.A.; Witte, U. Pressure and temperature effects on deep-sea hydrocarbon-degrading microbial communities in subarctic sediments. Microbiologyopen 2019, 8, e00768. [Google Scholar] [CrossRef]
- Abe, F.; Horikoshi, K. The biotechnological potential of piezophiles. Trends Biotechnol. 2001, 19, 102–108. [Google Scholar] [CrossRef]
- Li, W.-L.; Huang, J.-M.; Zhang, P.-W.; Cui, G.-J.; Wei, Z.-F.; Wu, Y.-Z.; Gao, Z.-M.; Han, Z.; Wang, Y. Periodic and Spatial Spreading of Alkanes and Alcanivorax Bacteria in Deep Waters of the Mariana Trench. Appl. Environ. Microbiol. 2018, 85, e02089-18. [Google Scholar] [CrossRef]
- Pavlo, T.; Kamrul Huda, M.D.; Hoong Gin, K.Y. A multiphaseoil spill model. J. Hydraul. Res. 2003, 41, 115–125. [Google Scholar]
- Patton, J.S.; Rigler, M.W.; Boehm, P.D.; Fiest, D.L. Ixtoc 1 oil spill: Flaking of surface mousse in the Gulf of Mexico. Nat. Cell Biol. 1981, 290, 235–238. [Google Scholar] [CrossRef]
- Suja, L.D.; Summers, S.; Gutierrez, T. Role of EPS, Dispersant and Nutrients on the Microbial Response and MOS Formation in the Subarctic Northeast Atlantic. Front. Microbiol. 2017, 8, 676. [Google Scholar] [CrossRef]
- Romero, I.C.; Schwing, P.T.; Brooks, G.R.; Larson, R.A.; Hastings, D.W.; Ellis, G.F.R.; Goddard, E.A.; Hollander, D.J. Hydrocarbons in Deep-Sea Sediments following the 2010 Deepwater Horizon Blowout in the Northeast Gulf of Mexico. PLoS ONE 2015, 10, e0128371. [Google Scholar] [CrossRef]
- Scoma, A.; Yakimov, M.M.; Boon, N. Challenging Oil Bioremediation at Deep-Sea Hydrostatic Pressure. Front. Microbiol. 2016, 7, 1203. [Google Scholar] [CrossRef]
- Wang, W.; Cai, B.; Shao, Z. Oil degradation and biosurfactant production by the deep sea bacterium Dietzia maris As-13-3. Front. Microbiol. 2014, 16, 711. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.K.; Krishnan, K.; Kurian, P.J. Complete genome sequence and comparative genome analysis of Alcanivorax sp. IO_7, a marine alkane-degrading bacterium isolated from hydrothermally-influenced deep seawater of southwest Indian ridge. Genomics 2021, 113, 884–891. [Google Scholar] [CrossRef]
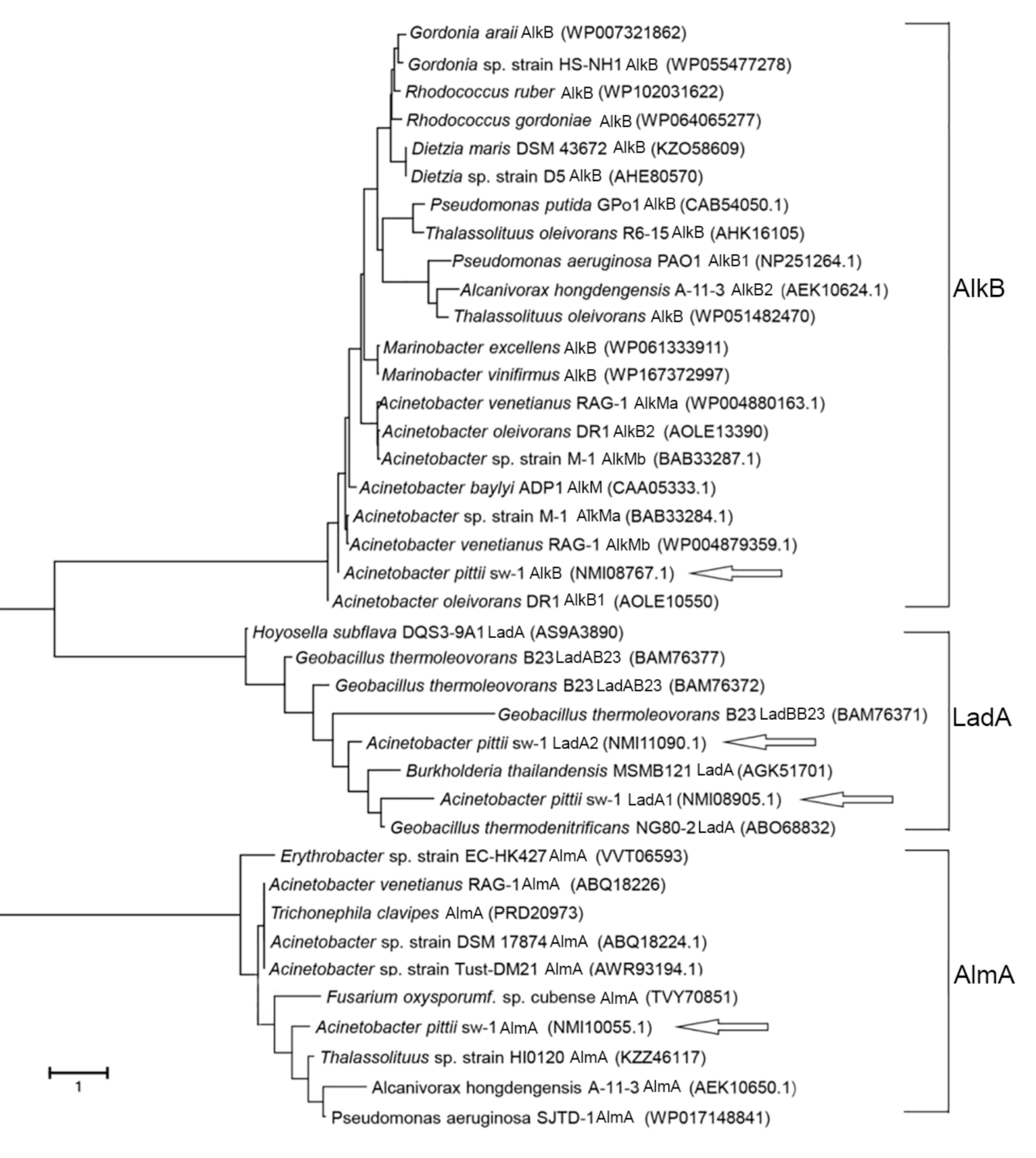
| Strain Name | Gene Name | Responsible for Range of Alkanes | Amino Acid Identity (%) | References |
|---|---|---|---|---|
| Acinetobacter M-1 | alkMa | C22–C30 | 86.27 % | [33] |
| Acinetobacter M-1 | alkMb | C16–C22 | 60.79 % | [33] |
| Acinetobacter DR1 | alkB1 | C24–C26 | 95.58 % | [17] |
| Acinetobacter DR1 | alkB2 | C12–C16 | 61.04 % | [17] |
| Acinetobacter ADP1 | alkM | C12–C18 | 83.09 % | [11] |
| Acinetobacter RAG-1 | alkMa | C12 | 61.04 % | [34] |
| Acinetobacter RAG-1 | alkMb | C12 | 86.42 % | [34] |
| Acinetobacter DSM 17,874 | almA | C32 and longer | 81.49 % | [18] |
| Alcanivorax Hongdengensis A-11-3 | almA | C18–C36 | 50.20 % | [35] |
| P. aeruginosa SJTD-1 | almA | C18–C24 | 48.50 % | [20] |
| G. thermodenitrificans NG80-2 | ladA | C15–C36 | 45.44% (with ladA1) 49.78% (with ladA2) | [19] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, W.; Zhao, C.; Gao, X.; Wang, L.; Tian, Q.; Liu, Y.; Xue, S.; Han, Z.; Chen, F.; Wang, S. Characterization and Transcriptome Analysis of a Long-Chain n-Alkane-Degrading Strain Acinetobacter pittii SW-1. Int. J. Environ. Res. Public Health 2021, 18, 6365. https://doi.org/10.3390/ijerph18126365
Kong W, Zhao C, Gao X, Wang L, Tian Q, Liu Y, Xue S, Han Z, Chen F, Wang S. Characterization and Transcriptome Analysis of a Long-Chain n-Alkane-Degrading Strain Acinetobacter pittii SW-1. International Journal of Environmental Research and Public Health. 2021; 18(12):6365. https://doi.org/10.3390/ijerph18126365
Chicago/Turabian StyleKong, Weina, Cheng Zhao, Xingwang Gao, Liping Wang, Qianqian Tian, Yu Liu, Shuwen Xue, Zhuang Han, Fulin Chen, and Shiwei Wang. 2021. "Characterization and Transcriptome Analysis of a Long-Chain n-Alkane-Degrading Strain Acinetobacter pittii SW-1" International Journal of Environmental Research and Public Health 18, no. 12: 6365. https://doi.org/10.3390/ijerph18126365
APA StyleKong, W., Zhao, C., Gao, X., Wang, L., Tian, Q., Liu, Y., Xue, S., Han, Z., Chen, F., & Wang, S. (2021). Characterization and Transcriptome Analysis of a Long-Chain n-Alkane-Degrading Strain Acinetobacter pittii SW-1. International Journal of Environmental Research and Public Health, 18(12), 6365. https://doi.org/10.3390/ijerph18126365






