Microbiological Reduction of Molybdenum to Molybdenum Blue as a Sustainable Remediation Tool for Molybdenum: A Comprehensive Review
Abstract
1. Introduction
2. Molybdenum (Mo)
2.1. Molybdenum Entry Routes in Animals
2.2. Molybdenum Toxicity
2.2.1. Toxicity to Spermatogenesis and Oogenesis
2.2.2. Mechanism of Molybdenum Toxicity in Ruminants
2.3. Bioremediation
2.3.1. Molybdenum Pollution
2.3.2. Molybdenum Bioremediation
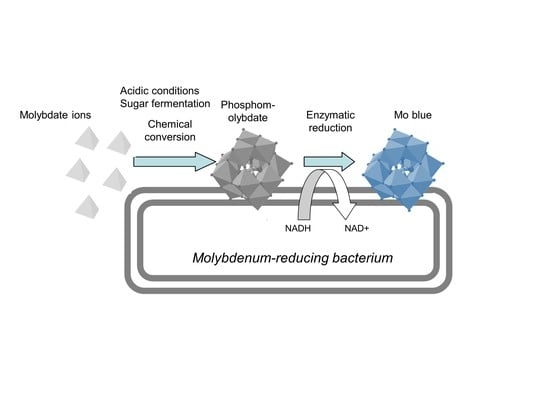
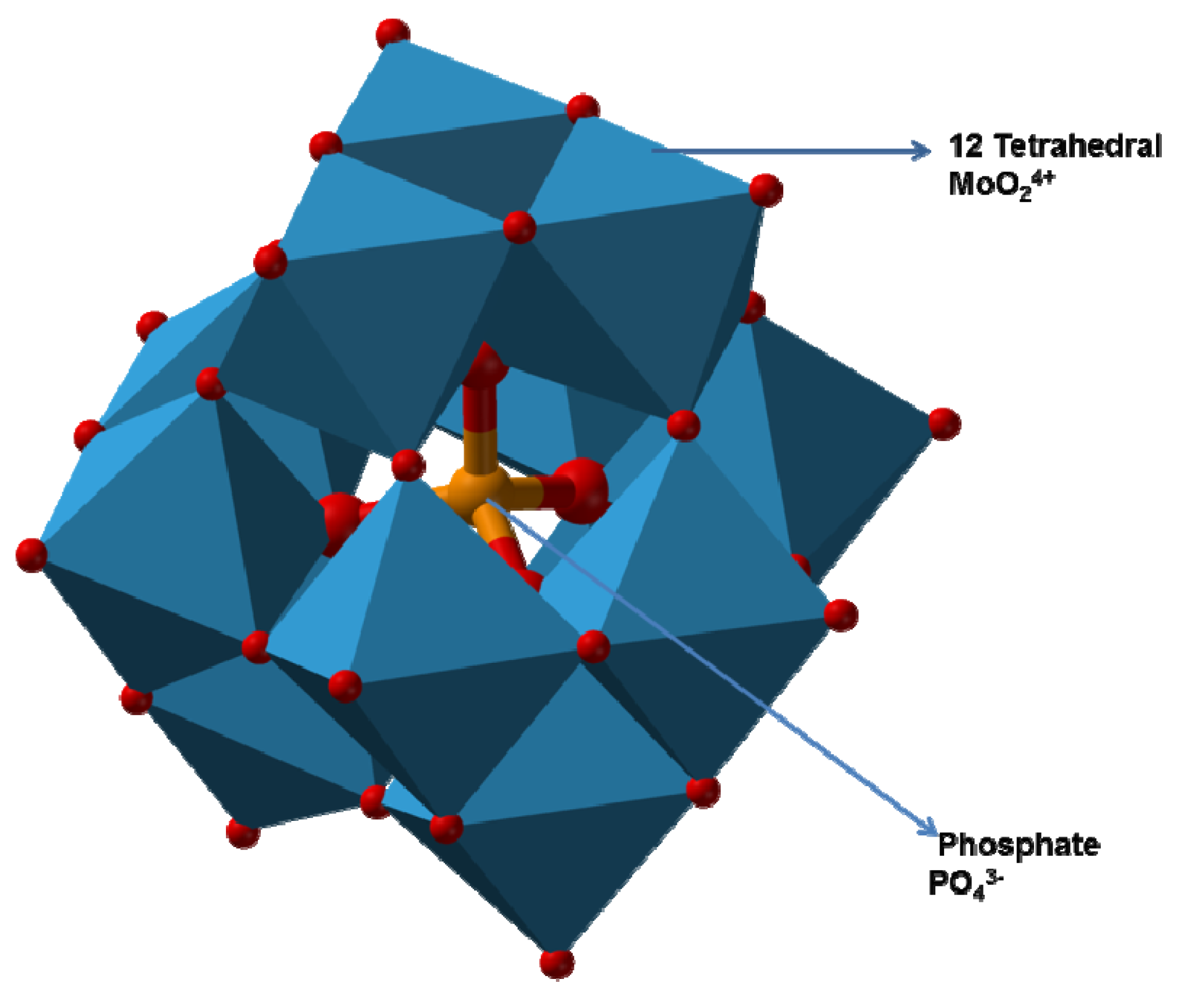
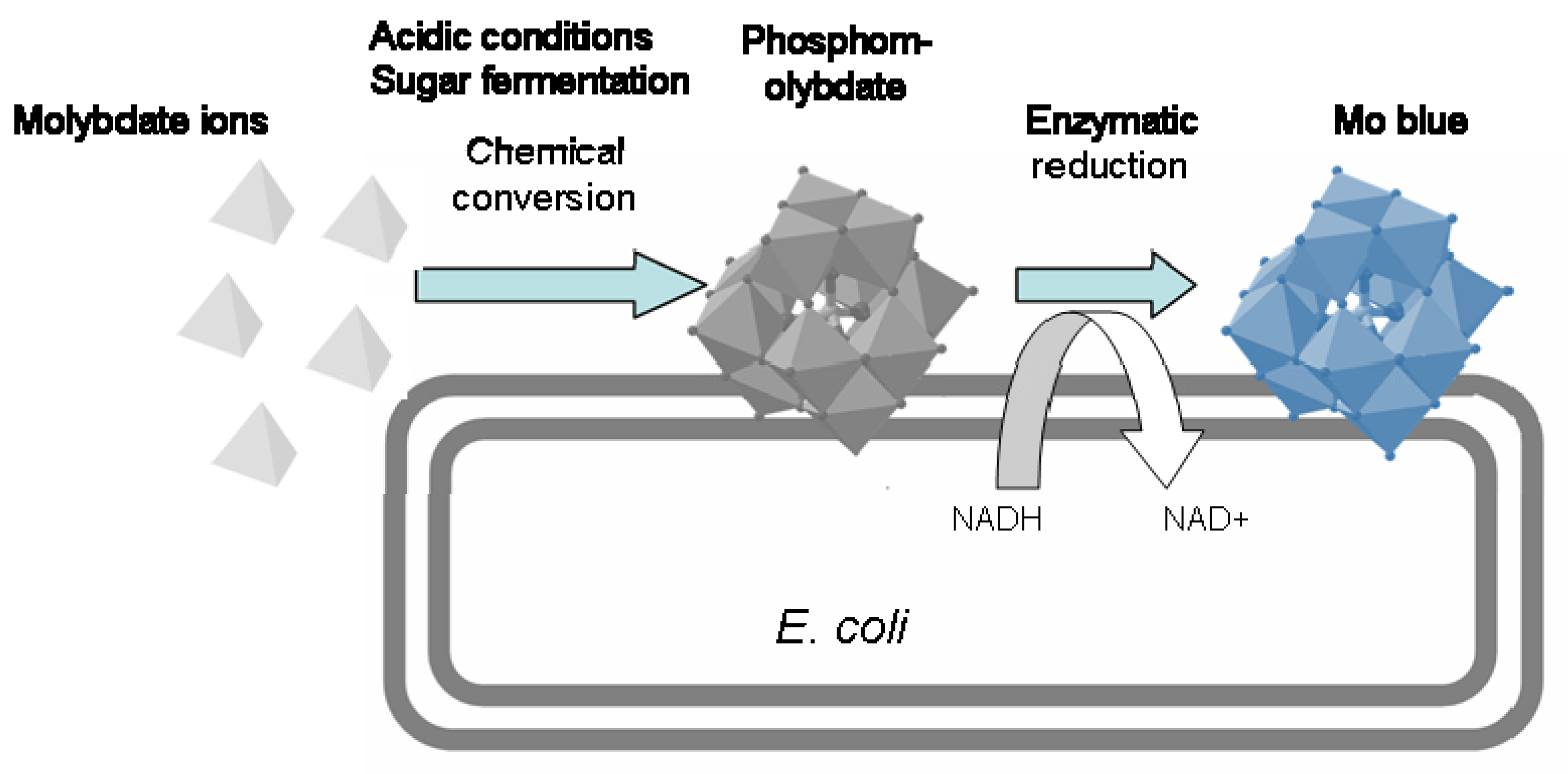
2.3.3. Mechanism of Molybdenum Reduction to Mo-Blue
2.4. Characteristics of Previously Isolated Molybdenum-Reducing Bacteria
| Bacteria | Specialization of the Bacteria | Optimal pH and Temperature | Preferred Carbon Source | MoO4 (mM) | PoO4 (mM) | Heavy Metal Inhibition | 1° Model and Kinetics of Reduction | Optimization Method | Author |
|---|---|---|---|---|---|---|---|---|---|
| Bacillus amyloliquefaciens strain Neni-9 (Indonesia) | Mo reduction Growth on carbaryl and carbofuran | pH 6.3 and 6.5, 30–37 °C | glucose | 20–30 | 5.0–7.5 | Ag+, Cr6+, Cu2+ Hg2+ | 1° model Mo reduction model using modified Gompertz | OFAT | [80] |
| Pseudomonas sp. (Nigeria) | Mo reduction | pH 6.5–7.5 37 °C | glucose | 100 | 3.5–7.5 | n.a. | n.a. | OFAT | [81] |
| Pantoea sp. strain HMY-P4 (Nigeria) | Mo reduction | pH 6.0–8.0 35–40 °C | glucose | 20–40 | 5.0 | n.a. | Aiba; qmax, Ks, and Ki of 0.89 μmol Mo-blue per h, 5.84 mM, and 32.23 mM, respectively | OFAT | [76,110] |
| Enterobacter cloacae (Nigeria) | Mo reduction | pH 6.5–7.0 35–40 °C | glucose | 80–100 | 5.0–7.5 | n.a. | Monod; qmax and Ks of 2.77 μmole Mo-blue h−1, and 12.42 mM, respectively | OFAT | [77,111] |
| Morganella sp. (Nigeria) | Mo reduction | pH 6.0–7.5 35 °C | glucose | 40 | 3.5 | n.a. | Teissier-Edward; qmax, Ks, and Ki of 7.77 mmole Mo-blue h−1, 26.63 mM, and 51.39 mM, respectively. | OFAT | [78] |
| Pseudomonas. strain Dr. Y Kertih(Malaysia) | Mo reduction Growth on phenol, acrylamide, nicotinamide, acetamide, iodoacetamide, propionamide, acetamide, sodium dodecyl sulfate (SDS), and diesel | pH 6.0–6.3 25–40 °C. | glucose | 20 | 5.0 | Ag+, Pb2+, As5+ Hg2+ | n.a. | OFAT | [112] |
| Clostridium pasteurianum BC1 (USA) | metallic (Mo0) nanoparticles 5–20 nm in size Degradation of methyl orange | pH 6.8 n.a. | peptone | 20.67 | 1.74 | OFAT | [113] | ||
| microbial electrolysis cells consortium (China) | Mo reduction, Tungsten reduction and acetate biodegradation Hydrogen production | pH 3.0 22 °C | acetate | 1 | n.a. | n.a. | n.a. | OFAT | [75] |
| Raoultella ornithinolytica strain Mo1 (Egypt) | Mo reduction | pH 6, 30 °C | glucose | 20 | n.a. | n.a. | OFAT | [114] | |
| Raoultella planticola strain MoI (Iraq) | Mo reduction | pH 6 30 °C | glucose | 20 | n.a. | n.a. | OFAT | [114] | |
| Bacillus sonorensis strain Pharon3 (MK078035) (Egypt) | Mo reduction Thermophilic bacterium | pH 7.07 52.2 °C | glucose | 10 | 4.0 | n.a. | RSM (CCD) | [82] | |
| Bacillus tequilensis strain Pharon2 (MK078034) (Egypt) | Mo reduction Thermophilic bacterium | pH 7.02 46.1 °C | sucrose | 10 | 4.0 | n.a. | RSM (CCD) | [82] | |
| Bacillus sp. strain Neni-12 (Indonesia) | Mo reduction Growth on coumaphos | pH 6.3 25–37 °C | glucose | 15–20 | 5.0 | Ag+, Cr6+, Hg2+ | 1° model, coumaphos growth model using modified Gompertz | OFAT | [79] |
| Pseudomonas sp. (Nigeria) | Mo reduction | pH 6.5–7.0 35– 40 °C | glucose | 40–60 | 3.5 | n.a. | n.a. | OFAT | [115] |
| Burkholderia vietnamiensis AQ5-12 (Malaysia) | Mo reduction Glyphosate degradation | pH 6.25–8.0 30–40 °C | glucose | 40–60 | 5.0 | n.a. | n.a. | OFAT | [116] |
| Burkholderia sp. AQ5-13 (Malaysia) | Mo reduction Glyphosate degradation | pH 6.25–8.0 35–40 °C | glucose | 40–50 | 5.0 | n.a. | n.a. | OFAT | [116] |
| Serratia marcescens strain KIK-1 (Nigeria) | Mo reduction Decolorization of various azo and triphenyl methane dyes | pH 5.8–6.5 34–37 °C | glucose | 10–25 | 5.0 | Ag+, Cr6+, Hg2+, Cu2+ | n.a. | OFAT | [117] |
| Pseudomonas putida strain Egypt-15 (Egypt) | Mo reduction Growth on PEG 4000 | pH 6.5 34 °C | glucose | 20 | 5.0 | n.a. | 1° model, PEG 4000 growth model using modified Gompertz | OFAT | [74] |
| Bacillus amyloliquefaciens (Malaysia) | SDS degradation | pH 5.8–6.3 25–34 °C | glucose | 30–50 | 5.0–7.5 | Hg2+, Cu2+, Ag+ | [118] | ||
| Serratia sp. strain HMY1 (Nigeria) | Mo reduction Cyanide degradation Q10 value of 2.038 and a theta value of 1.08 | pH 6.5–7.0 30–35 °C | sucrose | 55 | 3.95 | n.a. | 1° model, Mo reduction model using modified Gompertz | RSM (CCD) | [119,120,121] |
| Enterobacter sp. Strain Saw-2 | Mo reduction Growth on phenol and catechol | pH 6.3–6.8 34–37 °C | glucose | 15–30 | 5.0 | n.a. | n.a. | OFAT | [122] |
| Serratia sp. strain HMY3 (Nigeria) | Mo reduction Cyanide degradation | pH 6.5 35 °C | sucrose | 55–57.5 | 3.95 | As3+, Cr6+, Hg2+, Cu2 | Luong; qmax, Ks, Sm, and n were 25.32 h−1, 113.4 mM, 55.43 mM, and 1.42, respectively. | OFAT | [123] |
| Bacillus sp. strain Neni-10 (Indonesia) | Mo reduction Decolourisation of the dye Metanil Yellow | pH 6.3 34 °C | glucose | 20 | 2.5–7.5 | Ag+, Cu2+, Cr6+, Hg2+ | 1° model Mo reduction best model using Baranyi–Roberts | OFAT | [124,125] |
| Pseudomonas sp. strain 135 (Malaysia) | Mo reduction, Growth on acrylamide, acetamide, and propionamide acrylamide can support Mo reduction | pH 6.0–6.3 25–40 °C | glucose | 15–25 | 5.0–7.5 | Ag+, Cu2+, Cd2+, Hg2+ | n.a. | OFAT | [126] |
| Serratia marcescens strain DR.Y10 (Malaysia) | Mo reduction Growth on acrylamide, propionamide, and acetamide | pH 6.0–6.5 30–37 °C | glucose | 10–30 | 5.0 | Ag+, Cu2+, Cr6+, Hg2+ | n.a. | OFAT | [127] |
| Pseudomonas aeruginosa strain KIK-11 (Malaysia) | Growth on diesel and sodium dodecyl sulphate | pH 5.8–6.0 25–34 °C | glucose | 30–40 | 5.0–7.5 | Ag+, Cu2+, Hg2+ | n.a. | OFAT | [128] |
| Serratia sp. strain MIE2 (Malaysia) | Mo reduction | pH 6.0 27 to 35oC | sucrose | 20 | 3.95 | Hg2+, Zn2+, Cu2 | Teissier-Edward’s qmax, and Ks and Ki 0.89 mmole Mo-blue h−1, 5.84 mM, and 32.23 mM, respectively | RSM (Box-Behnken and CCD) | [129,130] |
| Bacillus sp. strain khayat (Malaysia) | Mo reduction SDS Diesel-degrading | pH 5.8–6.8 34 °C | glucose | 10–20 | 5–7.5 | Ag+, As3+, Pb2+, Hg2+, Cu2+ | n.a. | OFAT | [98] |
| Burkholderia sp. strain Neni-11 (Indonesia) | Mo reduction Amide-degrading | pH 6.0–6.3 30–37 °C | glucose | 15 | 5 | Ag+, Cr6+, Hg2+ | 1° model, Mo reduction model using modified Gompertz | OFAT | [131] |
| Enterobacter sp. strain Aft-3 (Pakistan) | Mo reduction Azo dye-decolorizing | pH 5.8–6.5 37 °C | glucose | 20–25 | 5 | Ag+, Cu2, Hg2+ | n.a. | OFAT | [132] |
| Klebsiella oxytoca strain Saw-5 (Malaysia) | Mo reduction Glyphosate degradation | pH 6.3–6.8 34 °C | glucose | 20–30 | 5 | Ag+, Cd2+, Cr6+, Hg2+, Cu2+ | n.a. | OFAT | [99] |
| P. aeruginosa strain Amr-11 (Egypt) | Mo reduction Phenol degradation | pH 6.3–6.8 34 °C | glucose | 20–30 | 2.5–7.5 | Ag+, As3+, Pb2+, Cd2+, Cr6+, Hg2+, Cu2+ | n.a. | OFAT | [104] |
| Klebsiella oxytoca strain Aft-7 (Pakistan) | Mo reduction SDS degradation | pH 5.8–6.3 25–34 °C | glucose | 5–20 | 5–7.5 | Ag+, As3+, Pb2+, Cd2+, Cr6+, Hg2+, Cu2+ | n.a. | OFAT | [97] |
| Enterobacter sp. strain Zeid-6 (Sudan) | Mo reduction Orange G decolorization | pH 5.5–8.0 30–37 °C | glucose | 20 | 5 | Ag+, Pb2+, Hg2+, Cu2+, | n.a. | OFAT | [92] |
| Pseudomonas putida strain Amr-12 (Egypt) | Mo reduction, Phenol and catechol degradation | pH 6.0–7.0 20–30 °C | glucose | 20–30 | 5.0–7.5 | Ag+, Cr6+, Hg2+ | n.a. | OFAT | [30] |
| Enterobacter sp. Strain Neni-13 | Mo reduction Growth on SDS | pH 6.0–6.5 37 °C | glucose | 15 | 2.5–5.0 | Ag+, Cd2+, Hg2+, Cu2+ | 1° model, SDS growth model using modified Gompertz | OFAT | [133] |
| Bacillus sp. strain Zeid 14 | Mo reduction Growth on amides, acetonitrile, and acrylamide can support Mo reduction | pH 6.0–6.8 25–34 °C | glucose | 10–20 | 5.0–7.5 | Ag+, Cd2+, Cr6+, Hg2+, Cu2+ | 1° model, Mo reduction model using modified Gompertz | OFAT | [134] |
| Klebsiella oxytoca strain DRY14 (Malaysia) | Mo reduction SDS degradation | pH 7.0 25 °C | glucose | 25–30 | 5 | Ag+, Pb2+, Cd2+, Cr6+, Hg2+, Cu2+ | n.a. | OFAT | [67] |
| Bacillus pumilus strain lbna (Malaysia) | Mo reduction | pH 7.0–8.0 37 °C | glucose | 40 | 2.5–5 | As3+, Pb2+, Zn2+, Cd2+, Cr6+, Hg2+, Cu2+ | Luong, qmax, Ks, Sm, and n values of 27.3 μmol Mo-blue h−1, 115.8 mM, 57.83 mM, and 1.405, respectively | OFAT | [135] |
| Bacillus sp. strain A.rzi (Malaysia) | Mo reduction | pH 7.3 28–30 °C | glucose | 50 | 4 | Cd2+, Cr6+, Cu2+, Ag+, Pb2+, Hg2+, Co2+, Zn2+ | Luong, qmax, Ks, Sm, and n values of 5.88 mole Mo-blue h−1, 70.36 mM, 108.22 mM, and 0.74, respectively | OFAT | [68] |
| Pseudomonas sp. strain DRY1 (Antarctica) | Mo reduction | pH 6.5–7.5 15–20 °C | glucose | 30–50 | 5 | Cd2+, Cr6+, Cu2+, Ag+, Pb2+, Hg2+ | n.a. | OFAT | [93] |
| Klebsiella oxytoca strain Hkeem (Malaysia) | Mo reduction | pH 7.3 30 °C | fructose | 80 | 4.5 | Cu2+, Ag+, Hg2+ | n.a. | [102] | |
| Pseudomonas sp. strain DRY2 (Malaysia) | Mo reduction | pH 6.0 40 °C | glucose | 15–20 | 5 | Cr6+, Cu2+, Pb2+, Hg2+ | n.a. | OFAT | [94] |
| Acinetobacter calcoaceticus strain Dr.Y12 (Malaysia) | Mo reduction | pH 6.5 37 °C | glucose | 20 | 5 | Cd2+, Cr6+, Cu2+, Pb2+, Hg2+ | n.a. | OFAT | [96] |
| Enterobacter sp. strain Dr.Y13 (Malaysia) | Mo reduction | pH 6.5 37 °C | glucose | 25–50 | 5 | Cr6+, Cd2+, Cu2+, Ag+, Hg2+ | n.a. | OFAT | [66] |
| S. marcescens strain Dr.Y9 (Malaysia) | Mo reduction | pH 7.0 37 °C | sucrose | 20 | 5 | Cr6+, Cu2+, Ag+, Hg2+ | n.a. | OFAT | [101] |
| Serratia sp. strain Dr.Y8 (Malaysia) | Mo reduction | pH 6.0 37 °C | sucrose | 50 | 5 | Cr6+, Cu2+, Ag+, Hg2+ | n.a. | OFAT | [90] |
| Serratia sp. strain Dr.Y5 (Malaysia) | Mo reduction Purification of 1st Mo-reducing enzyme | pH 7.0 37 °C | sucrose | 30 | 5 | Cu2+ | 1° model Mo reduction best model using Huang model | OFAT | [100,136,137,138,139] |
| Serratia marcescens strain DRY6 (Malaysia) | Mo reduction | pH 7.0 35 °C | sucrose | 15–25 | 5 | Cr6+, Cu2+, Hg2+ | n.a. | OFAT | [95] |
| Enterobacter cloacae strain 48 (Malaysia) | Mo reduction | pH 7.0 30 °C | sucrose | 20 | 2.9 | Cr6+, Cu2+ | n.a. | OFAT | [72] |
| Escherichia coli K12 | Mo reduction | pH 7.0 30–36 °C | glucose | 80 | 5 | Cr6+ | n.a. | OFAT | [70] |
2.5. Statistical-Based Optimization Compared to One-at-a-Time Approach
2.6. Mathematical Modelling of Molybdenum Reduction Profile and Kinetics
2.7. Characteristic of the Partially Purified Molybdenum-Reducing Enzyme from other Bacterial Sources
2.8. Use of Electron Transport Chain Inhibitors in Probing the Location of Molybdenum Reduction
2.9. Identification of the Mo-Reducing Enzyme
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aftab, A.; Memon, A.R.; Shah, S.F.A.; Memon, H.R.; Dahot, M.U. Determination of hydrocarbons and isolation of inherent bacteria from crude oil-contaminated soil for ex-situ bioremediation. Sindh Univ. Res. J. 2013, 45, 476–481. [Google Scholar]
- Aftab, A.; Shah, S.F.A.; Aziz, S.; Memon, H.U.R.; Dahot, M.U. Bioaugmentation of hydrocarbons using bacterial species extracted from oil contaminated site. Sindh Univ. Res. J. 2016, 48, 455–460. [Google Scholar]
- Ilyin, I.; Rozovskaya, O.; Travnikov, M.; Varygina, M. Heavy Metals: Analysis of Long-Term Trends, Country-Specific Research and Progress in Mercury Regional and Global Modelling; MSC-E: Moscow, Russia; CCC: Kjeller, Norway, 2015; Volume 2. [Google Scholar]
- Meyer, M.; Pesch, R.; Schröder, W.; Steinnes, E.; Uggerud, H.T. Spatial patterns and temporal trends of heavy metal concentrations in moss and surface soil specimens collected in Norway between 1990 and 2010. Environ. Sci. Eur. 2014, 26, 1–18. [Google Scholar] [CrossRef]
- Kaplan, D. Absorption and adsorption of heavy metals by microalgae. In Handbook of Microalgal Culture: Applied Phycology and Biotechnology; Richmond, A., Hu, Q., Eds.; Wiley: Hoboken, NJ, USA, 2013; pp. 602–611. [Google Scholar] [CrossRef]
- Soares, E.V.; Soares, H.M.V.M. Bioremediation of industrial effluents containing heavy metals using brewing cells of Saccharomyces cerevisiae as a green technology: A review. Environ. Sci. Pollut. Res. 2012, 19, 1066–1083. [Google Scholar] [CrossRef]
- Dixit, R.; Malaviya, D.; Pandiyan, K.; Singh, U.B.; Sahu, A.; Shukla, R.; Singh, B.P.; Rai, J.P.; Sharma, P.K.; Lade, H.; et al. Bioremediation of heavy metals from soil and aquatic environment: An Overview of principles and criteria of fundamental processes. Sustainability 2015, 7, 2189–2212. [Google Scholar] [CrossRef]
- Rasheed, A.; Ghous, T.; Khan, M.; Ullah, R.S. A simple flow injection pH indicator method for the determination of metal ions by urease inhibition study. J. Chem. Soc. Pak. 2012, 34, 317–320. [Google Scholar]
- Sulaiman, M.R. Update of mercury in fish with a focus on its current status in Malaysia. J. Environ. Bioremediation Toxicol. 2013, 1, 14–19. [Google Scholar]
- Abdullah, A.; Hamzah, Z.; Saat, A.; Wood, A.K.; Alias, M. Accumulation of mercury (Hg) and methyl mercury (MeHg) concentrations in selected marine biota from Manjung coastal area. Malays. J. Anal. Sci. 2015, 19, 669–678. [Google Scholar]
- Samadi, N.; Ansari, R.; Khodavirdilo, B. A suitable method for removing of heavy metal ions from aqueous solutions using proper copolymer and its derivations. Eurasian J. Anal. Chem. 2018, 13, em07. [Google Scholar] [CrossRef]
- Francisco, R.; Alpoim, M.C.; Morais, P.V. Diversity of chromium-resistant and -reducing bacteria in a chromium-contaminated activated sludge. J. Appl. Microbiol. 2002, 92, 837–843. [Google Scholar] [CrossRef]
- Retamal-Morales, G.; Mehnert, M.; Schwabe, R.; Tischler, D.; Zapata, C.; Chávez, R.; Schlömann, M.; Levicán, G. Detection of arsenic-binding siderophores in arsenic-tolerating actinobacteria by a modified CAS Assay. Ecotoxicol. Environ. Saf. 2018, 157, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.; Heine, T.; Malik, L.; Hofmann, S.; Joffroy, K.; Senges, C.H.R.; Bandow, J.E.; Tischler, D. Screening for microbial metal-chelating siderophores for the removal of metal ions from solutions. Microorganisms 2021, 9, 111. [Google Scholar] [CrossRef]
- Sarubbo, L.A.; Rocha, R.B.; Luna, J.M.; Rufino, R.D.; Santos, V.A.; Banat, I.M. Some aspects of heavy metals contamination remediation and role of biosurfactants. Chem. Ecol. 2015, 7540, 707–723. [Google Scholar] [CrossRef]
- Li, P.; Tao, H. Cell surface engineering of microorganisms towards adsorption of heavy metals. Crit. Rev. Microbiol. 2013, 41, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Prasad, S.M. Reduction of heavy metal load in food chain: Technology assessment. Rev. Environ. Sci. Biotechnol. 2011, 10, 199–214. [Google Scholar] [CrossRef]
- Zhai, X.-W.; Zhang, Y.-L.; Qi, Q.; Bai, Y.; Chen, X.-L.; Jin, L.-J.; Ma, X.-G.; Shu, R.-Z.; Yang, Z.-J.; Liu, F.-J. Effects of molybdenum on sperm quality and testis oxidative stress. Syst. Biol. Reprod. Med. 2013, 59, 251–255. [Google Scholar] [CrossRef]
- Wu, S.; Hu, C.; Tan, Q.; Nie, Z.; Sun, X. Effects of molybdenum on water utilization, antioxidative defense system and osmotic-adjustment ability in winter wheat (Triticum aestivum) under drought stress. Plant Physiol. Biochem. 2014, 83, 365–374. [Google Scholar] [CrossRef]
- Pandey, R.; Singh, S.P. Effects of molybdenum on fertility of male rats. BioMetals 2002, 15, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Geng, C.; Gao, Y.; Li, D.; Jian, X.; Hu, Q. Contamination investigation and risk assessment of molybdenum on an industrial site in China. J. Geochem. Explor. 2014, 144, 273–281. [Google Scholar] [CrossRef]
- Rahman, M.A.; Ahmad, S.A.; Salvam, S.; Halmi, M.I.E.; Yusof, M.T.; Shukor, M.Y.; Shamaan, N.A.; Syed, M.A. Dialysis Tubing experiment showed that molybdenum reduction in S. marcescens strain DrY6 is mediated by enzymatic action. J. Environ. Bioremediation Toxicol. 2013, 1, 25–27. [Google Scholar]
- Kosaka, H.; Wakita, K. Some geologic features of the Mamut porphyry copper deposit, Sabah, Malaysia. Econ. Geol. 1978, 73, 618–627. [Google Scholar] [CrossRef]
- Nasernejad, B.; Kaghazchi, T.; Edrisi, M.; Sohrabi, M. Bioleaching of molybdenum from low-grade copper ore. Process Biochem. 1999, 35, 437–440. [Google Scholar] [CrossRef]
- Battogtokh, B.; Lee, J.M.; Woo, N. Contamination of water and soil by the Erdenet copper-molybdenum mine in Mongolia. Environ. Earth Sci. 2014, 71, 3363–3374. [Google Scholar] [CrossRef]
- Barceloux, D.G.; Barceloux, D. Molybdenum. J. Toxicol. Clin. Toxicol. 1999, 37, 231–237. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO/SDE/WSH/03.04/11. Molybdenum in Drinking-Water: Background Document for Development of WHO Guidelines for Drinking-Water Quality; WHO: Geneva, Switzerland, 2011. [Google Scholar]
- Sebenik, R.F.; Burkin, A.R.; Dorfler, R.R. Molybdenum and Molybdenum Compounds. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH: Weinheim, Germany, 2000; pp. 1–63. [Google Scholar]
- Shukor, M.Y.; Rahman, M.F.A.; Shamaan, N.A.; Lee, C.H.; Karim, M.I.A.; Syed, M.A. An improved enzyme assay for molybdenum-reducing activity in bacteria. Appl. Biochem. Biotechnol. 2008, 144, 293–300. [Google Scholar] [CrossRef]
- AbdEl-Mongy, M.A.; Shukor, M.S.; Hussein, S.; Ling, A.P.K.; Shamaan, N.A.; Shukor, M.Y. Isolation and Characterization of a molybdenum-reducing, phenol- and catechol-degrading Pseudomonas putida strain Amr-12 in soils from Egypt. Sci. Study Res. Chem. Chem. Eng. Biotechnol. Food Ind. 2015, 16, 353–369. [Google Scholar]
- Wuana, R.A.; Okieimen, F.E. Heavy metals in contaminated soils: A review of sources, chemistry, risks and best available strategies for remediation. Int. Sch. Res. Not. 2011, 2011, 402647. [Google Scholar] [CrossRef]
- Olaniran, A.O.; Balgobind, A.; Pillay, B. Bioavailability of heavy metals in soil: Impact on microbial biodegradation of organic compounds and possible improvement strategies. Int. J. Mol. Sci. 2013, 14, 10197–10228. [Google Scholar] [CrossRef]
- Stafford, J.M.; Lambert, C.E.; Zyskowski, J.A.; Engfehr, C.L.; Fletcher, O.J.; Clark, S.L.; Tiwary, A.; Gulde, C.M.; Sample, B.E. Dietary toxicity of soluble and insoluble molybdenum to Northern Bobwhite quail (Colinus Virginianus). Ecotoxicology 2016, 25, 291–301. [Google Scholar] [CrossRef]
- Miller, J.K.; Moss, B.R.; Bell, M.C.; Sneed, N.N. Comparison of 99Mo metabolism in young cattle and swine. J. Anim. Sci. 1972, 34, 846–850. [Google Scholar] [CrossRef]
- Robinson, M.F.; McKenzie, J.M.; Tomson, C.D.; van Rij, A.L. Metabolic balance of zinc, copper, cadmium, iron, molybdenum and selenium in young New Zealand women. Br. J. Nutr. 1973, 30, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, H.A.; Frost, D.V.; Balass, J.J. Essential metals in Man: Selenium. J. Chronic Dis. 1970, 23, 227–243. [Google Scholar] [CrossRef]
- Pitt, M.A. Review molybdenum toxicity: Interactions between copper, molybdenum and sulphate. Agents Actions 1976, 6, 758–769. [Google Scholar] [CrossRef]
- Jeter, M.A.; Davis, G.K. The effect of dietary molybdenum upon growth, hemoglobin, reproduction and lactation of rats. J. Nutr. 1954, 54, 215–220. [Google Scholar] [CrossRef]
- Ratnasooriya, W.D.; Wadsworth, R.M. Effects of prazosin on fertility of male rats. Reproduction 1984, 70, 643–647. [Google Scholar] [CrossRef]
- Lyubimov, A.V.; Smith, J.A.; Rousselle, S.D.; Mercieca, M.D.; Tomaszewski, J.E.; Smith, A.C.; Levine, B.S. The effects of tetrathiomolybdate (TTM, NSC-714598) and copper supplementation on fertility and early embryonic development in rats. Reprod. Toxicol. 2004, 19, 223–233. [Google Scholar] [CrossRef]
- Bersényi, A.; Berta, E.; Kádár, I.; Glávits, R.; Szilágyi, M.; Fekete, S.G. Effects of high dietary molybdenum in rabbits. Acta Vet. Hung. 2008, 56, 41–55. [Google Scholar] [CrossRef][Green Version]
- Yamaguchi, S.; Miura, C.; Ito, A.; Agusa, T.; Iwata, H.; Tanabe, S.; Tuyen, B.C.; Miura, T. Effects of lead, molybdenum, rubidium, arsenic and organochlorines on spermatogenesis in fish: Monitoring at Mekong Delta Area and In Vitro Experiment. Aquat. Toxicol. 2007, 83, 43–51. [Google Scholar] [CrossRef]
- Rajagopalan, K.V. Molybdenum: An essential trace element in human nutrition. Annu. Rev. Nutr. 1988, 8, 401–427. [Google Scholar] [CrossRef]
- Silver, S.; Phung, L.T. Bacterial heavy metal resistance: New surprises. Annu. Rev. Microbiol. 1996, 50, 753–789. [Google Scholar] [CrossRef] [PubMed]
- Majak, W.; Steinke, D.; Mcgillivray, J.; Lysyk, T. Clinical signs in cattle grazing high molybdenum forage. Rangel. Ecol. Manag. 2004, 57, 269–274. [Google Scholar] [CrossRef]
- Haywood, S.; Dincer, Z.; Jasani, B.; Loughram, M.J. Molybdenum-associated pituitary endocrinopathy in sheep treated with ammonium tetrathiomolybdate. J. Comp. Pathol. 2004, 130, 21–31. [Google Scholar] [CrossRef]
- Ward, G.M. Molybdenum toxicity and hypocuprosis in ruminants: A Review. J. Anim. Sci. 1978, 46, 1078–1085. [Google Scholar] [CrossRef] [PubMed]
- Parimoolam, S.; Dahalan, F.A. Physicochemical Optimization of Granular Sludge in Rubber Industrial Wastewater. J. Biochem. Microbiol. Biotechnol. 2013, 1, 7–10. [Google Scholar]
- Zahaba, M. Luminescent bacterial testing for monitoring hydrocarbon bioremediation—A Review. J. Biochem. Microbiol. Biotechnol. 2015, 3, 13–20. [Google Scholar]
- Sabullah, M.K.; Marbawi, H.; Faik, A.A.M.; Abdullah, R.; Sani, S.A.; Japanis, F.G.J.; Jaganathan, J.N.; Julius, M.W.; Saini, N.; Roland, R. Bioremediation of hydrocarbon: A mini review. J. Biochem. Microbiol. Biotechnol. 2018, 6, 1–6. [Google Scholar]
- Elekwachi, C.O.; Andresen, J.; Hodgman, T.C. Global use of bioremediation technologies for decontamination of ecosystems. J. Bioremediation Biodegrad. 2014, 5, 225. [Google Scholar] [CrossRef]
- Davis, G.K. Molybdenum. In Metals and Their Compounds in the Environment: Occurrence, Analysis and Biological Relevance; Merian, E., Anke, M., Ihnat, M., Stoeppler, M., Eds.; Wiley-VCH: Weinheim, Germany, 1991; pp. 1089–1100. [Google Scholar]
- Neunhäuserer, C.; Berreck, M.; Insam, H. Remediation of soils contaminated with molybdenum using soil amendments and phytoremediation. Water Air Soil Pollut. 2001, 128, 85–96. [Google Scholar] [CrossRef]
- Qadir, M.A.; Zaidi, J.H.; Ahmad, S.A.; Gulzar, A.; Yaseen, M.; Atta, S.; Tufail, A. Evaluation of trace elemental composition of aerosols in the atmosphere of Rawalpindi and Islamabad using radio analytical methods. Appl. Radiat. Isot. 2012, 70, 906–910. [Google Scholar] [CrossRef]
- Al Kuisi, M.; Al-Hwaiti, M.; Mashal, K.; Abed, A.M. Spatial distribution patterns of molybdenum (Mo) concentrations in potable groundwater in Northern Jordan. Environ. Monit. Assess. 2015, 187, 148. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.A.; Testa, S.M. Acid drainage and sulfide oxidation: Introduction. In Acid Mine Drainage, Rock Drainage, and Acid Sulfate Soils: Causes, Assessment, Prediction, Prevention, and Remediation; Jacobs, J.A., Lehr, J.H., Testa, S.M., Eds.; Wiley: Hoboken, NJ, USA, 2014; pp. 1–8. [Google Scholar] [CrossRef]
- Kargar, M.; Khorasani, N.; Karami, M.; Rafiee, G.; Naseh, R. Study of aluminum, copper and molybdenum pollution in groundwater sources surrounding (Miduk) Shahr-E-Babak copper complex tailings dam. World Acad. Sci. Eng. Technol. 2011, 52, 278–282. [Google Scholar]
- Yu, C.; Xu, S.; Gang, M.; Chen, G.; Zhou, L. Molybdenum Pollution and Speciation in Nver River Sediments Impacted with Mo Mining Activities in Western Liaoning, Northeast China. Int. J. Environ. Res. 2011, 5, 205–212. [Google Scholar]
- Simeonov, L.I.; Kochubovski, M.V.; Simeonova, B.G. Environmental Heavy Metal Pollution and Effects on Child Mental Development; (NATO Science for Peace and Security Series C: Environmental Security); Springer: Dordrecht, The Netherlands, 2011; Volume 1. [Google Scholar]
- LeGendre, G.R.; Runnells, D.D. Removal of dissolved molybdenum from wastewaters by precipitates of ferric iron. Environ. Sci. Technol. 1975, 9, 744–749. [Google Scholar] [CrossRef]
- Runnells, D.D.; Kaback, D.S.; Thurman, E.M. Geochemistry and sampling of molybdenum in sediments, soils, and plants in Colorado. In Molybdenum in the Environment; Chappel, W.R., Peterson, K.K., Eds.; Marcel and Dekker, Inc.: New York, NY, USA, 1976. [Google Scholar]
- Furber, D. Is Molybdenum Lurking In Your Forages? Canadian Cattlemen 2009. Available online: https://www.canadiancattlemen.ca/features/is-molybdenum-lurking-in-your-forages (accessed on 25 May 2020).
- Yong, F.S. Mamut copper mine—The untold story. In Proceedings of the Minerals: Underpinning Yesterday’s Needs, Today’s Development and Tomorrow’s Growth, Kota Kinabalu, Sabah, Malaysia, 22–24 June 2000. [Google Scholar]
- Yakasai, H.M.; Rahman, M.F.; Yasid, N.A.; Ahmad, S.A.; Halmi, M.I.E.; Shukor, M.Y. Elevated molybdenum concentrations in soils contaminated with spent oil lubricant. J. Environ. Microbiol. Toxicol. 2017, 5, 1–3. [Google Scholar]
- Lloyd, J.R. Microbial reduction of metals and radionuclides. FEMS Microbiol. Rev. 2003, 27, 411–425. [Google Scholar] [CrossRef]
- Shukor, M.Y.; Rahman, M.F.; Shamaan, N.A.; Syed, M.S. Reduction of molybdate to molybdenum blue by Enterobacter sp. strain Dr.Y13. J. Basic Microbiol. 2009, 49, S43–S54. [Google Scholar] [CrossRef] [PubMed]
- Halmi, M.I.E.; Zuhainis, S.W.; Yusof, M.T.; Shaharuddin, N.A.; Helmi, W.; Shukor, Y.; Syed, M.A.; Ahmad, S.A. Hexavalent molybdenum reduction to Mo-blue by a sodium-dodecyl-sulfate-degrading Klebsiella oxytoca strain DRY14. BioMed Res. Int. 2013, 2013, 384541. [Google Scholar] [CrossRef] [PubMed]
- Othman, A.R.; Bakar, N.A.; Halmi, M.I.E.; Johari, W.L.W.; Ahmad, S.A.; Jirangon, H.; Syed, M.A.; Shukor, M.Y. Kinetics of molybdenum reduction to molybdenum blue by Bacillus sp. strain A.Rzi. BioMed Res. Int. 2013, 2013, 371058. [Google Scholar] [CrossRef]
- Levine, V.E. The Reducing Properties of microorganisms with special reference to selenium compounds. J. Bacteriol. 1925, 10, 217–263. [Google Scholar] [CrossRef]
- Campbell, A.M.; Del Campillo-Campbell, A.; Villaret, D.B. Molybdate reduction by Escherichia coli K-12 and its Chl mutants. Proc. Natl. Acad. Sci. USA 1985, 82, 227–231. [Google Scholar] [CrossRef]
- Sugio, T.; Tsujita, Y.; Katagiri, T.; Inagaki, K.; Tano, T. Reduction of Mo6+ with elemental sulfur by Thiobacillus ferrooxidans. J. Bacteriol. 1988, 170, 5956–5959. [Google Scholar] [CrossRef] [PubMed]
- Ghani, B.; Takai, M.; Hisham, N.Z.; Kishimoto, N.; Ismail, A.K.M.; Tano, T.; Sugio, T. Isolation and characterization of a Mo6+-reducing bacterium. Appl. Environ. Microbiol. 1993, 59, 1176–1180. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.D.; Barton, L.L.; Thomson, B.M. Reduction and immobilization of molybdenum by Desulfovibrio desulfuricans. J. Environ. Qual. 1997, 26, 1146–1152. [Google Scholar] [CrossRef]
- AbdEl-Mongy, M.A.; Aqlima, S.A.; Shukor, M.S.; Hussein, S.; Ling, A.P.K.; Shukor, M.Y. A PEG 4000-degrading and hexavalent molybdenum-reducing Pseudomonas putida strain Egypt-15. J. Natl. Sci. Found. Sri Lanka 2018, 46, 431–442. [Google Scholar] [CrossRef]
- Huang, L.; Tian, F.; Pan, Y.; Shan, L.; Shi, Y.; Logan, B.E. Mutual benefits of acetate and mixed tungsten and molybdenum for their efficient removal in 40 L microbial electrolysis cells. Water Res. 2019, 162, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Idris, D.; Gafasa, M.A.; Ibrahim, S.S.; Babandi, A.; Shehu, D.; Ya’u, M.; Babagana, K.; Mashi, J.A.; Yakasai, H.M. Pantoea sp. strain HMY-P4 reduced toxic hexavalent molybdenum to insoluble molybdenum blue. J. Biochem. Microbiol. Biotechnol. 2019, 7, 31–37. [Google Scholar]
- Kabir, Z.M.; Gafasa, M.A.; Kabara, H.T.; Ibrahim, S.S.; Babandi, A.; Ya’u, M.; Shehu, D.; Abubakar, S.M.; Babagana, K.; Mashi, J.A.; et al. Isolation and characterization of molybdate-reducing Enterobacter cloacae from agricultural soil in Gwale LGA Kano State, Nigeria. J. Environ. Microbiol. Toxicol. 2019, 7, 1–6. [Google Scholar]
- Mohammed, S.; Gafasa, M.A.; Kabara, H.T.; Babandi, A.; Shehu, D.; Ya’u, M.; Abubakar, S.M.; Babagana, K.; Mashi, J.A.; Yakasai, H.M. Soluble molybdenum reduction by Morganella sp. locally-isolated from agricultural land in Kano. Bioremediation Sci. Technol. Res. 2019, 7, 1–7. [Google Scholar]
- Rusnam; Gusmanizar, N. Isolation and characterization of a molybdenum-reducing and coumaphos-degrading Bacillus sp. strain Neni-12 in soils from West Sumatera, Indonesia. J. Environ. Microbiol. Toxicol. 2019, 7, 20–25. [Google Scholar]
- Rusnam; Gusmanizar, N. Isolation and Characterization of a molybdenum-reducing and carbamate-degrading Bacillus amyloliquefaciens strain Neni-9 in soils from West Sumatera, Indonesia. Bioremediation Sci. Technol. Res. 2020, 8, 17–22. [Google Scholar]
- Alhassan, A.Y.; Babandi, A.; Uba, G.; Yakasai, H.M. Isolation and characterization of molybdenum-reducing Pseudomonas sp. from agricultural land in Northwest-Nigeria. J. Biochem. Microbiol. Biotechnol. 2020, 8, 23–28. [Google Scholar]
- Saeed, A.M.; Sayed, H.A.E.; El-Shatoury, E.H. Optimizing the reduction of molybdate by two novel thermophilic bacilli isolated from Sinai, Egypt. Curr. Microbiol. 2020, 77, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Ariff, A.B.; Rosfarizan, M.; Ghani, B.; Sugio, T.; Karim, M.I.A. Molybdenum reductase in Enterobacter cloacae. World J. Microbiol. Biotecnol. 1997, 13, 643–647. [Google Scholar] [CrossRef]
- Shukor, M.Y.; Lee, C.H.; Omar, I.; Karim, M.I.A.; Syed, M.A.; Shamaan, N.A. Isolation and characterization of a molybdenum-reducing enzyme in Enterobacter cloacae strain 48. Pertanika J. Sci. Technol. 2003, 11, 261–272. [Google Scholar]
- Lee, J.D. Concise Inorganic Chemistry; Van Nostrand Reinhold Company: New York, NY, USA, 1977. [Google Scholar]
- Shukor, M.Y.; Adam, H.; Ithnin, K.; Yunus, I.; Shamaan, N.A.; Syed, A. Molybdate reduction to molybdenum blue in microbe proceeds via a phosphomolybdate intermediate. J. Biol. Sci. 2007, 7, 1448–1452. [Google Scholar] [CrossRef][Green Version]
- Sims, R.P.A. Formation of heteropoly blue by some reduction procedures used in the micro-determination of phosphorus. Analyst 1961, 86, 584–590. [Google Scholar] [CrossRef]
- Kazansky, L.P.; Fedotov, M.A. Phosphorus-31 and Oxygen-17 N.M.R. Evidence of trapped electrons in reduced 18-molybdodiphosphate(V), P2Mo18O628−. J. Chem. Soc. Chem. Commun. 1980, 644–646. [Google Scholar] [CrossRef]
- Chae, H.K.; Klemperer, W.G.; Marquart, T.A. High-nuclearity oxomolybdenum(V) complexes. Coord. Chem. Rev. 1993, 128, 209–224. [Google Scholar] [CrossRef]
- Shukor, M.Y.; Rahman, M.F.; Suhaili, Z.; Shamaan, N.A.; Syed, M.A. Bacterial reduction of hexavalent molybdenum to molybdenum blue. World J. Microbiol. Biotechnol. 2009, 25, 1225–1234. [Google Scholar] [CrossRef]
- You, X.-Y.; Wang, H.; Ren, G.-Y.; Li, J.-J.; Duan, X.; Zheng, H.-J.; Jiang, Z.-Q. Complete genome sequence of the molybdenum-resistant bacterium Bacillus subtilis strain LM 4–2. Stand. Genom. Sci. 2015, 10, 127. [Google Scholar] [CrossRef]
- Othman, A.R.; Abu Zeid, I.M.; Rahman, M.F.; Ariffin, F.; Shukor, M.Y. Isolation and characterization of a molybdenum-reducing and Orange G-decolorizing Enterobacter sp. strain Zeid-6 in soils from Sudan. Bioremediation Sci. Technol. Res. 2015, 3, 13–19. [Google Scholar]
- Ahmad, S.A.; Shukor, M.Y.; Shamaan, N.A.; Mac, C.; Syed, M.A. Molybdate reduction to molybdenum blue by an Antarctic bacterium. BioMed Res. Int. 2013, 2013, 871941. [Google Scholar] [CrossRef] [PubMed]
- Shukor, M.Y.; Ahmad, S.A.; Nadzir, M.M.M.; Abdullah, M.P.; Shamaan, N.A.; Syed, M.A. Molybdate reduction by Pseudomonas sp. strain DRY2. J. Appl. Microbiol. 2010, 108, 2050–2058. [Google Scholar] [CrossRef]
- Shukor, M.Y.; Habib, S.H.M.; Rahman, M.F.A.; Jirangon, H.; Abdullah, M.P.A.; Shamaan, N.A.; Syed, M.A. Hexavalent molybdenum reduction to molybdenum blue by S. marcescens strain Dr. Y6. Appl. Biochem. Biotechnol. 2008, 149, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Shukor, M.Y.; Rahman, M.F.; Suhaili, Z.; Shamaan, N.A.; Syed, M.A. Hexavalent molybdenum reduction to Mo-blue by Acinetobacter calcoaceticus. Folia Microbiol. 2010, 55, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Masdor, N.; Abd Shukor, M.S.; Khan, A.; Bin Halmi, M.I.E.; Abdullah, S.R.S.; Shamaan, N.A.; Shukor, M.Y. Isolation and characterization of a molybdenum-reducing and SDS- degrading Klebsiella oxytoca strain Aft-7 and its bioremediation application in the environment. Biodiversitas 2015, 16, 238–246. [Google Scholar] [CrossRef]
- Khayat, M.E.; Rahman, M.F.A.; Shukor, M.S.; Ahmad, S.A.; Shamaan, N.A.; Shukor, M.Y. Characterization of a molybdenum-reducing Bacillus sp. strain Khayat with the ability to grow on SDS and diesel. Rend. Fis. Acc. Lincei 2016, 27, 547–556. [Google Scholar] [CrossRef]
- Sabullah, M.K.; Rahman, M.F.; Ahmad, S.A.; Sulaiman, M.R.; Shukor, M.S.; Shamaan, N.A.; Shukor, M.Y. Isolation and characterization of a molybdenum-reducing and glyphosate-degrading Klebsiella oxytoca strain Saw-5 in Soils from Sarawak. Agrivita 2016, 38, 1–13. [Google Scholar] [CrossRef]
- Rahman, M.F.A.; Shukor, M.Y.; Suhaili, Z.; Mustafa, S.; Shamaan, N.A.; Syed, M.A. Reduction of Mo(VI) by the Bacterium Serratia Sp. Strain DRY5. J. Environ. Biol. 2009, 30, 65–72. [Google Scholar]
- Yunus, S.M.; Hamim, H.M.; Anas, O.M.; Aripin, S.N.; Arif, S.M. Mo(VI) Reduction to molybdenum blue by Serratia marcescens strain Dr. Y 9. Pol. J. Microbiol. 2009, 58, 141–147. [Google Scholar]
- Lim, H.K.; Syed, M.A.; Shukor, M.Y. Reduction of molybdate to molybdenum blue by Klebsiella sp. strain Hkeem. J. Basic Microbiol. 2012, 52, 296–305. [Google Scholar] [CrossRef]
- Abo-Shakeer, L.K.A.; Ahmad, S.A.; Shukor, M.Y.; Shamaan, N.A.; Syed, M.A. Isolation and characterization of a molybdenum-reducing Bacillus pumilus strain Lbna. J. Environ. Microbiol. Toxicol. 2013, 1, 9–14. [Google Scholar]
- Ibrahim, Y.; Abdel-Mongy, M.; Shukor, M.S.; Hussein, S.; Ling, A.P.K.; Shukor, M.Y. Characterization of a Molybdenum-reducing bacterium with the ability to degrade phenol, isolated in soils from Egypt. Biotechnol. J. Biotechnol. Comput. Biol. Bionanotechnol. 2015, 96, 234–245. [Google Scholar] [CrossRef]
- Elangovan, R.; Abhipsa, S.; Rohit, B.; Ligy, P.; Chandraraj, K. Reduction of Cr(VI) by a Bacillus Sp. Biotechnol. Lett. 2006, 28, 247–252. [Google Scholar] [CrossRef]
- Rege, M.A.; Petersen, J.N.; Johnstone, D.L.; Turick, C.E.; Yonge, D.R.; Apel, W.A. Bacterial reduction of hexavalent chromium by Enterobacter cloacae strain H01 grown on sucrose. Biotechnol. Lett. 1997, 19, 691–694. [Google Scholar] [CrossRef]
- Hettiarachchi, G.M.; Pierzynski, G.M.; Ransom, M.D. In situ stabilization of soil lead using phosphorus and manganese oxide. Environ. Sci. Technol. 2000, 34, 4614–4619. [Google Scholar] [CrossRef]
- Deeb, B.E.; Altalhi, A.D. Degradative plasmid and heavy metal resistance plasmid naturally coexist in phenol and cyanide assimilating bacteria. Am. J. Biochem. Biotechnol. 2009, 5, 84–93. [Google Scholar] [CrossRef]
- Halmi, M.I.E.; Wasoh, H.; Sukor, S.; Ahmad, S.A.; Yusof, M.T.; Shukor, M.Y. Bioremoval of molybdenum from aqueous solution. Int. J. Agric. Biol. 2014, 16, 848–850. [Google Scholar]
- Yakasai, H.M.; Babandi, A.; Uba, G. Inhibition kinetics study of molybdenum reduction by Pantoea sp. strain HMY-P4. J. Environ. Microbiol. Toxicol. 2020, 8, 24–29. [Google Scholar]
- Yakasai, H.M.; Babandi, A.; Ibrahim, S. Modelling the inhibition kinetics of molybdenum reduction by the molybdate-reducing Enterobacter cloacae. Bull. Environ. Sci. Sustain. Manag. 2020, 4, 11–17. [Google Scholar]
- Kesavan, V.; Mansur, A.; Suhaili, Z.; Salihan, M.S.R.; Rahman, M.F.A.; Shukor, M.Y. Isolation and characterization of a heavy metal-reducing Pseudomonas sp. strain Dr.Y Kertih with the ability to assimilate phenol and diesel. Bioremediation Sci. Technol. Res. 2018, 6, 14–22. [Google Scholar]
- Nordmeier, A.; Woolford, J.; Celeste, L.; Chidambaram, D. Sustainable batch production of biosynthesized nanoparticles. Mater. Lett. 2017, 191, 53–56. [Google Scholar] [CrossRef]
- Saeed, A.M.; El Shatoury, E.; Hadid, R. Production of molybdenum blue by two novel molybdate-reducing bacteria belonging to the genus Raoultella isolated from Egypt and Iraq. J. Appl. Microbiol. 2019, 126, 1722–1728. [Google Scholar] [CrossRef] [PubMed]
- Gafasa, M.A.; Ibrahim, S.S.; Babandi, A.; Abdullahi, N.; Shehu, D.; Ya’u, M.; Babagana, K.; Mashi, J.A.; Yakasai, H.M. Characterizing the molybdenum-reducing properties of Pseudomonas sp. locally isolated from agricultural soil in Kano Metropolis Nigeria. Bioremediation Sci. Technol. Res. 2019, 7, 34–40. [Google Scholar]
- Manogaran, M.; Ahmad, S.A.; Yasid, N.A.; Yakasai, H.M.; Shukor, M.Y. Characterisation of the simultaneous molybdenum reduction and glyphosate degradation by Burkholderia vietnamiensis AQ5-12 and Burkholderia sp. AQ5-13. 3 Biotech. 2018, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Karamba, I.K.; Yakasai, H. Isolation and characterization of a molybdenum-reducing and methylene blue-decolorizing Serratia marcescens strain KIK-1 in soils from Nigeria. Bioremediation Sci. Technol. Res. 2018, 6, 1–8. [Google Scholar]
- Maarof, M.Z.; Shukor, M.Y.; Mohamad, O.; Karamba, K.I.; Halmi, M.I.E.; Rahman, M.F.A.; Yakasai, H.M. Isolation and characterization of a molybdenum-reducing Bacillus amyloliquefaciens strain KIK-12 in soils from Nigeria with the ability to grow on SDS. J. Environ. Microbiol. Toxicol. 2018, 6, 13–20. [Google Scholar]
- Yakasai, M.H.; Ibrahim, K.K.; Yasid, N.A.; Halmi, M.I.E.; Rahman, M.F.A.; Shukor, M.Y. Mathematical modelling of molybdenum reduction to Mo-blue by a cyanide-degrading bacterium. Bioremediation Sci. Technol. Res. 2016, 4, 1–5. [Google Scholar]
- Yakasai, H.M.; Yasid, N.A.; Shukor, M.Y. Temperature coefficient and Q10 value estimation for the growth of molybdenum-reducing Serratia sp. strain HMY1. Bioremediation Sci. Technol. Res. 2018, 6, 22–24. [Google Scholar]
- Yakasai, H.; Karamba, K.; Yasid, N.; Halmi, M.I.E.; Rahman, M.F.; Ahmad, S.A.; Shukor, M. Response surface-based optimization of a novel molybdenum-reducing and cyanide-degrading Serratia sp. strain HMY1. Desalination Water Treat. 2019, 145, 220–231. [Google Scholar] [CrossRef]
- Sabullah, M.K.; Rahman, M.F.; Ahmad, S.A.; Sulaiman, M.R.; Shukor, M.S.; Gansau, A.J.; Shamaan, N.A.; Shukor, M.Y. Isolation and characterization of a molybdenum-reducing and phenolic- and catechol-degrading Enterobacter sp. strain Saw-2. Biotropia Southeast Asian J. Trop. Biol. 2017, 24, 47–58. [Google Scholar] [CrossRef]
- Yakasai, M.H. Bioreduction of Hexavalent Molybdenum to Molybdenum Blue by Serratia sp. Strain Mie2 and Purification of the Molybdenum Reducing Enzyme. Ph.D. Thesis, Department of Biochemistry, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Seri Kembangan, Malaysia, 2017. [Google Scholar]
- Mansur, R.; Gusmanizar, N.; Roslan, M.A.H.; Ahmad, S.A.; Shukor, M.Y. Isolation and characterisation of a molybdenum-reducing and metanil yellow dye-decolourising Bacillus sp. strain Neni-10 in soils from West Sumatera, Indonesia. Trop. Life Sci. Res. 2017, 28, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Yakasai, M.H.; Manogaran, M. Kinetic modelling of molybdenum-blue production by Bacillus sp. strain Neni-10. J. Environ. Microbiol. Toxicol. 2020, 8, 5–10. [Google Scholar]
- Yakasai, M.H.; Rahman, M.F.A.; Rahim, M.B.H.A.; Khayat, M.E.; Shamaan, N.A.; Shukor, M.Y. Isolation and characterization of a metal-reducing Pseudomonas sp. strain 135 with amide-degrading capability. Bioremediation Sci. Technol. Res. 2017, 5, 32–38. [Google Scholar]
- Chee, H.-S.; Manogaran, M.; Suhaili, Z.; Yakasai, M.H.; Rahman, M.F.A.; Shamaan, N.A.; Yasid, N.A.; Othman, A.R. Isolation and characterisation of a Mo-reducing bacterium from Malaysian Soil. Bioremediation Sci. Technol. Res. 2017, 5, 17–24. [Google Scholar]
- Mohamad, O.; Yakasai, H.M.; Karamba, K.I.; Halmi, M.I.E.; Rahman, M.F.; Shukor, M.Y. Reduction of molybdenum by Pseudomonas aeruginosa strain KIK-11 isolated from a metal-contaminated soil with ability to grow on diesel and sodium dodecyl sulphate. J. Environ. Microbiol. Toxicol. 2017, 5, 19–26. [Google Scholar]
- Halmi, M.I.E.; Abdullah, S.R.S.; Johari, W.L.W.; Ali, M.S.M.; Shaharuddin, N.A.; Khalid, A.; Shukor, M.Y. Modelling the kinetics of hexavalent molybdenum (Mo6+) reduction by the Serratia sp. strain MIE2 in batch culture. Rend. Fis. Acc. Lincei 2016, 27, 653–663. [Google Scholar] [CrossRef]
- Aziz, N.F.; Halmi, M.I.E.; Johari, W.L.W. Statistical optimization of hexavalent molybdenum reduction by Serratia sp. strain MIE2 using Central Composite Design (CCD). J. Biochem. Microbiol. Biotechnol. 2017, 5, 8–11. [Google Scholar]
- Mansur, R.; Gusmanizar, N.; Dahalan, F.A.; Masdor, N.A.; Ahmad, S.A.; Shukor, M.S.; Roslan, M.A.H.; Shukor, M.Y. Isolation and characterization of a molybdenum-reducing and amide-degrading Burkholderia cepacia strain Neni-11 in soils from West Sumatera, Indonesia. IIOAB 2016, 7, 28–40. [Google Scholar]
- Shukor, M.S.; Khan, A.; Masdor, N.; Halmi, M.I.E.; Abdullah, S.R.S.; Shukor, M.Y. Isolation of a novel molybdenum-reducing and azo dye decolorizing Enterobacter sp. strain Aft-3 from Pakistan. Chiang Mai Univ. J. Nat. Sci. 2016, 15, 95–114. [Google Scholar] [CrossRef]
- Rahman, M.F.; Rusnam, M.; Gusmanizar, N.; Masdor, N.A.; Lee, C.H.; Shukor, M.S.; Roslan, M.A.H.; Shukor, M.Y. Molybdate-Reducing and SDS-Degrading Enterobacter sp. strain Neni-13. Nova Biotechnol. Chim. 2016, 15, 166–181. [Google Scholar] [CrossRef][Green Version]
- Mohd Adnan, A.S.; Abu Zeid, I.M.; Ahmad, S.A.; Halmi, M.I.E.; Abdullah, S.R.S.; Masdor, M.A.; Shukor, M.S.; Shukor, M.Y. A Molybdenum-reducing Bacillus sp. strain Zeid 14 in soils from Sudan that could grow on amides and acetonitrile. Malays. J. Soil Sci. 2016, 20, 111–134. [Google Scholar]
- Abo-Shakeer, L.K.A.; Rahman, M.F.A.; Yakasai, M.H.; Bakar, N.A.; Othman, A.R.; Syed, M.A.; Abdullah, N.; Shukor, M.Y. Kinetic studies of the partially purified molybdenum-reducing enzyme from Bacillus pumilus strain Lbna. Bioremediation Sci. Technol. Res. 2017, 5, 18–23. [Google Scholar]
- Shukor, M.Y.; Bakar, N.A.; Othman, A.R.; Yunus, I.; Shamaan, N.A.; Syed, M.A. Development of an inhibitive enzyme assay for copper. J. Environ. Biol. 2009, 30, 39–44. [Google Scholar] [PubMed]
- Shukor, M.Y.; Halmi, M.I.E.; Rahman, M.F.A.; Shamaan, N.A.; Syed, M.A. Molybdenum Reduction to Molybdenum Blue in Serratia sp. strain DRY5 is catalyzed by a novel molybdenum-reducing enzyme. BioMed Res. Int. 2014, 2014, 853084. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Halmi, M.I.E.; Ahmad, S.A.; Yusof, M.T.; Shukor, M.Y.; Syed, M.A. Entrapment of Mo-reducing bacterium increase its resistance towards heavy metals. Bull. Environ. Sci. Manag. 2013, 1, 11–13. [Google Scholar]
- Syed, M.A.; Shamaan, N.A.; Shukor, M.Y. Mathematical modeling of the molybdenum blue production from Serratia sp. strain DRY5. J. Environ. Microbiol. Toxicol. 2020, 8, 12–17. [Google Scholar]
- Box, G.E.P.; Wilson, K.B. On the experimental attainment of optimum conditions. J. R. Stat. Soc. 1951, 13, 1–45. [Google Scholar] [CrossRef]
- Montgomery, D.C.; Runger, G.C. Applied Statistics and Probability for Engineers; John Wiley and Sons: Chichester, UK, 1994. [Google Scholar]
- Sharma, Y.C.; Uma; Upadhyay, S.N. Removal of a cationic dye from wastewaters by adsorption on activated carbon developed from coconut coir. Energy Fuels 2009, 23, 2983–2988. [Google Scholar] [CrossRef]
- Zin, K.M.; Halmi, M.I.E.; Abd Gani, S.S.; Zaidan, U.H.; Samsuri, A.W.; Abd Shukor, M.Y. Microbial Decolorization of Triazo dye, Direct Blue 71: An Optimization Approach Using Response Surface Methodology (RSM) and Artificial Neural Network (ANN). BioMed Res. Int. 2020, 2020, 2734135. Available online: https://www.hindawi.com/journals/bmri/2020/2734135/ (accessed on 10 September 2020). [CrossRef] [PubMed]
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; Van’t Riet, K. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef] [PubMed]
- Baranyi, J.; Roberts, T.A. A Dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Buchanan, R.L.; Whiting, R.C.; Damert, W.C. When is simple good enough: A comparison of the gompertz, baranyi, and three-phase linear models for fitting bacterial growth curves. Food Microbiol. 1997, 14, 313–326. [Google Scholar] [CrossRef]
- Fujikawa, H. Development of a new logistic model for microbial growth in foods. Biocontrol Sci. 2010, 15, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, A.R.; Binning, P.J.; Aamand, J.; Badawi, N.; Rosenbom, A.E. The Gompertz Function can coherently describe microbial mineralization of growth-sustaining pesticides. Environ. Sci. Technol. 2013, 47, 8508–8514. [Google Scholar] [CrossRef] [PubMed]
- Shukor, M.S.; Shukor, M.Y. Bioremoval of toxic molybdenum using dialysis tubing. Chem. Eng. Res. Bull. 2015, 18, 6–11. [Google Scholar] [CrossRef]
- Gusmanizar, N.; Halmi, M.; Rusnam, M.; Rahman, M.; Shukor, M.; Azmi, N.; Shukor, M.Y. Isolation and characterization of a molybdenum-reducing and azo-dye decolorizing Serratia marcescens strain Neni-1 from Indonesian soil. J. Urban Environ. Eng. 2016, 10, 113–123. [Google Scholar] [CrossRef]
- Halmi, M.I.E.; Ahmad, S.A.; Syed, M.A.; Shamaan, N.A.; Shukor, M.Y. Mathematical modelling of the molybdenum reduction kinetics in Bacillus pumilus strain Lbna. Bull. Environ. Sci. Manag. 2014, 2, 24–29. [Google Scholar]
- Ricker, F.J. 11 Growth rates and models. In Bioenergetics and Growth; Hoar, W.S., Randall, D.J., Brett, J.R., Eds.; Academic Press: Cambridge, MA, USA, 1979; Volume 8, pp. 677–743. [Google Scholar]
- Gompertz, B. On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies. Philos. Trans. R. Soc. 1825, 115, 513–585. [Google Scholar]
- Richards, F.J. A flexible growth function for empirical use. J. Exp. Bot. 1959, 10, 290–300. [Google Scholar] [CrossRef]
- Baranyi, J. Mathematics of predictive food microbiology. Int. J. Food Microbiol. 1995, 26, 199–218. [Google Scholar] [CrossRef]
- Babák, L.; Šupinová, P.; Burdychová, R. Growth models of thermus aquaticus and Thermus scotoductus. Acta Univ. Agric. Silvic. Mendel. Brun. 2012, 60, 19–26. [Google Scholar] [CrossRef]
- López, S.; Prieto, M.; Dijkstra, J.; Dhanoa, M.S.; France, J. Statistical evaluation of mathematical models for microbial growth. Int. J. Food Microbiol. 2004, 96, 289–300. [Google Scholar] [CrossRef]
- Buchanan, R.L. Predictive food microbiology. Trends Food Sci. and Technol. 1993, 4, 6–11. [Google Scholar] [CrossRef]
- Huang, L. Optimization of a new mathematical model for bacterial growth. Food Control 2013, 32, 283–288. [Google Scholar] [CrossRef]
- Glusczak, L.; dos Santos Miron, D.; Crestani, M.; Braga da Fonseca, M.; de Araújo Pedron, F.; Duarte, M.F.; Vieira, V.L.P. Effect of glyphosate herbicide on acetylcholinesterase activity and metabolic and hematological parameters in piava (Leporinus obtusidens). Ecotoxicol. Environ. Saf. 2006, 65, 237–241. [Google Scholar] [CrossRef]
- Soda, S.O.; Yamamura, S.; Zhou, H.; Ike, M.; Fujita, M. Reduction Kinetics of As (V) to As (III) by a Dissimilatory arsenate-reducing bacterium, Bacillus sp. SF-1. Biotechnol. Bioeng. 2006, 93, 812–815. [Google Scholar] [CrossRef]
- Sukumar, M. Reduction of hexavalent chromium by Rhizopus oryzae. Afr. J. Environ. Sci. Technol. 2010, 4, 412–418. [Google Scholar]
- Truex, M.J.; Peyton, B.M.; Valentine, N.B.; Gorby, Y.A. Kinetics of U(VI) reduction by a dissimilatory Fe(III)-reducing bacterium under non-growth conditions. Biotechnol. Bioeng. 1997, 55, 490–496. [Google Scholar] [CrossRef]
- King, R.B.; Long, G.M.; Sheldon, J.K. Practical Environmental Bioremediation: The Field Guide; CRC Press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef]
- Boon, B.; Laudelout, H. Kinetics of nitrite oxidation by Nitrobacter winogradskyi. Biochem. J. 1962, 85, 440–447. [Google Scholar] [CrossRef]
- Teissier, G. Growth of bacterial populations and the available substrate concentration. Rev. Sci. Instrum. 1942, 3208, 209–214. [Google Scholar]
- Aiba, S.; Shoda, M.; Nagatani, M. Kinetics of product inhibition in alcohol fermentation. Biotechnol. Bioeng. 1968, 10, 845–864. [Google Scholar] [CrossRef]
- Yano, T.; Koga, S. Dynamic behavior of the chemostat subject to substrate inhibition. Biotechnol. Bioeng. 1969, 11, 139–153. [Google Scholar] [CrossRef]
- Han, K.; Levenspiel, O. Extended Monod kinetics for substrate, product, and cell inhibition. Biotechnol. Bioeng. 1988, 32, 430–437. [Google Scholar] [CrossRef]
- Luong, J.H.T. Generalization of Monod kinetics for analysis of growth data with substrate inhibition. Biotechnol. Bioeng. 1987, 29, 242–248. [Google Scholar] [CrossRef]
- Halmi, M.I.E. Bioreduction of Hexavalent Molybdenum to Molybdenum Blue by Serratia sp. Strain MIE2 and Purification of the Molybdenum Reducing Enzyme. Ph.D. Thesis, Department of Biochemistry, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Seri Kembangan, Malaysia, 2012. [Google Scholar]
- Opperman, D.J.; Piater, L.A.; van Heerden, E. A novel chromate reductase from Thermus scotoductus SA-01 related to old yellow enzyme. J. Bacteriol. 2008, 190, 3076–3082. [Google Scholar] [CrossRef]
- Freedman, Z.; Zhu, C.; Barkay, T. Mercury resistance and mercuric reductase activities and expression among chemotrophic thermophilic aquificae. Appl. Environ. Microbiol. 2012, 78, 6568–6575. [Google Scholar] [CrossRef]
- Schröder, I.; Rech, S.; Krafft, T.; Macy, J.M. Purification and characterization of the selenate reductase from Thauera selenatis. J. Biol. Chem. 1997, 272, 23765–23768. [Google Scholar] [CrossRef]
- Shukor, M.Y. Revisiting the role of the electron transport chain in molybdate reduction by Enterobacter cloacae Strain 48. Indian J. Biotechnol. 2014, 13, 404–407. [Google Scholar]
- Shukor, M.Y.; Syed, M.A.; Lee, C.H.; Karim, M.I.A.; Shamaan, N.A. A method to distinguish between chemical and enzymatic reduction of molybdenum in Enterobacter cloacae Strain 48. Malays. J. Biochem. 2002, 7, 71–72. [Google Scholar]
| Model | p | Equation | Best Model for Mo-Reducing Bacterium | Reference |
|---|---|---|---|---|
| Modified Logistic | 3 | nil | ||
| Modified Gompertz | 3 | Bacillus amyloliquefaciens strain Neni-9 Bacillus sp. strain Neni-12 Serratia sp. strain HMY1 Burkholderia sp. strain Neni-11 Bacillus sp. strain Zeid 14 | [79,80,119,131,134] | |
| Modified Richards | 4 | nil | ||
| Modified Schnute | 4 | nil | ||
| Baranyi–Roberts | 4 | nil | ||
| Von Bertalanffy | 3 | nil | ||
| Huang | 4 | Serratia sp. strain Dr.Y5 | [139] | |
| Buchanan Three-phase linear model | 3 | y = A, if x < lag y = A + k(x − λ), if λ ≤ x ≤ xmax y = ymax, if x ≥ xmax | nil |
| Author | p | First Reported by | Reduction Rate | Best Model for Mo-Reducing Bacterium | Reference |
|---|---|---|---|---|---|
| Monod | 2 | [165] | Enterobacter cloacae | [111] | |
| Haldane | 3 | [166] | Nil | ||
| Teissier-Edward | 3 | [167] | Morganella sp. | [78] | |
| Aiba | 4 | [168] | Pantoea sp. strain HMY-P4 | [110] | |
| Yano and Koga | 4 | [169] | Nil | ||
| Han and Levenspiel | 5 | [170] | Nil | ||
| Luong | 4 | [171] | Bacillus pumilus strain lbna Bacillus sp. strain A.rzi Serratia sp. strain HMY3 | [68,103,123] |
| Bacteria | Molecular Weight | pH | Temp °C | Km NADH (mM) | Vmax NADH mU mg−1 | Km PM (mM) | Vmax PM mU mg−1 | Kcat/Km M−1s−1 | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Enterobacter cloacae strain 48 | n.p but 80, 90, and 100 bands observed | 6.5 | 25 | 1.38 | 102.6 | 2.56 | 99.4 | n.a. | [29] |
| Pseudomonas sp. strain DRY1 | n.p. | 6.0 | 20 | 4.68 | 26.98 | 3.52 | 23.48 | n.a. | [93] |
| Serratia sp. strain DRY5 | 105 | 6.0 | 25 to 35 | 0.79 | 12 | 3.87 | 12.05 | 5.47 | [137] |
| Bacillus pumilus strain Lbna | n.p | 5.5 | 25 to 35 | 6.646 | 0.057 | 3.399 | 0.106 | n.a. | [135] |
| Serratia sp. strain MIE2 | 100 | 5.0 | 35 | 0.859 | 16.11 | 6.02 | 6.89 | 7.89 | [172] |
| Serratia sp. strain HMY1 | 100 | 5.5 | 25 to 35 | 1.81 | 21.2 | 4.53 | 21.66 | 5.35 | [123] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yakasai, H.M.; Rahman, M.F.; Manogaran, M.; Yasid, N.A.; Syed, M.A.; Shamaan, N.A.; Shukor, M.Y. Microbiological Reduction of Molybdenum to Molybdenum Blue as a Sustainable Remediation Tool for Molybdenum: A Comprehensive Review. Int. J. Environ. Res. Public Health 2021, 18, 5731. https://doi.org/10.3390/ijerph18115731
Yakasai HM, Rahman MF, Manogaran M, Yasid NA, Syed MA, Shamaan NA, Shukor MY. Microbiological Reduction of Molybdenum to Molybdenum Blue as a Sustainable Remediation Tool for Molybdenum: A Comprehensive Review. International Journal of Environmental Research and Public Health. 2021; 18(11):5731. https://doi.org/10.3390/ijerph18115731
Chicago/Turabian StyleYakasai, Hafeez Muhammad, Mohd Fadhil Rahman, Motharasan Manogaran, Nur Adeela Yasid, Mohd Arif Syed, Nor Aripin Shamaan, and Mohd Yunus Shukor. 2021. "Microbiological Reduction of Molybdenum to Molybdenum Blue as a Sustainable Remediation Tool for Molybdenum: A Comprehensive Review" International Journal of Environmental Research and Public Health 18, no. 11: 5731. https://doi.org/10.3390/ijerph18115731
APA StyleYakasai, H. M., Rahman, M. F., Manogaran, M., Yasid, N. A., Syed, M. A., Shamaan, N. A., & Shukor, M. Y. (2021). Microbiological Reduction of Molybdenum to Molybdenum Blue as a Sustainable Remediation Tool for Molybdenum: A Comprehensive Review. International Journal of Environmental Research and Public Health, 18(11), 5731. https://doi.org/10.3390/ijerph18115731