Sex and Exposure to Postnatal Chlorpyrifos Influence the Epigenetics of Feeding-Related Genes in a Transgenic APOE Mouse Model: Long-Term Implications on Body Weight after a High-Fat Diet
Abstract
1. Introduction
2. Material and Methods
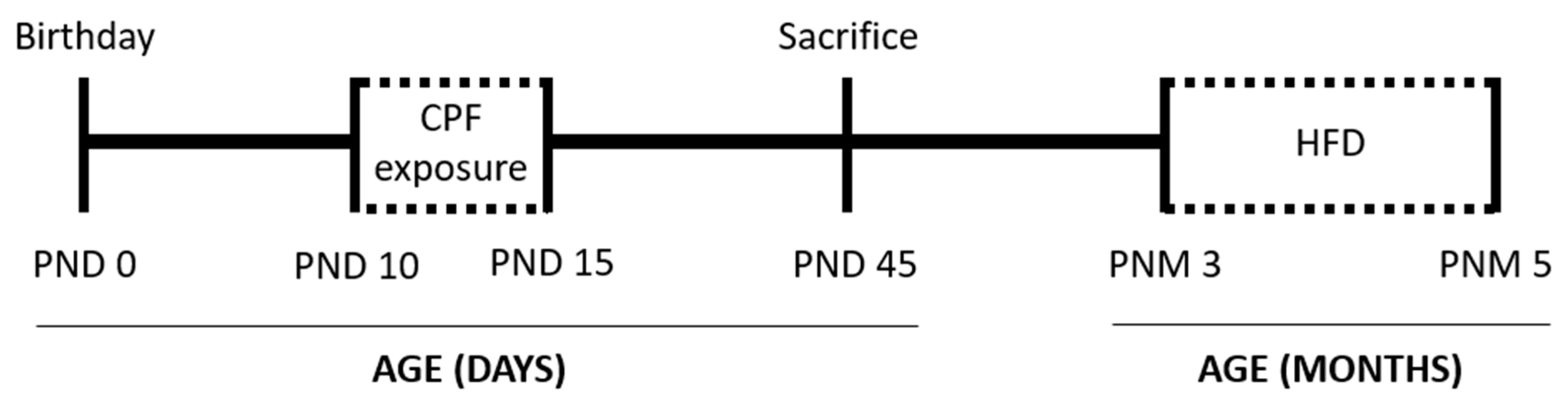
2.1. Animals and Care
2.2. Chemicals and Treatment
2.3. Brain Sampling
2.4. Isolation of DNA and RNA
2.5. Methylation Analysis: Bisulfite Modification and Pyrosequencing
2.6. Gene Expression Analysis
2.7. Dietary Intervention: High-Fat Diet
2.8. Statistical Analysis
3. Results
3.1. DNA Methylation and Gene Expression
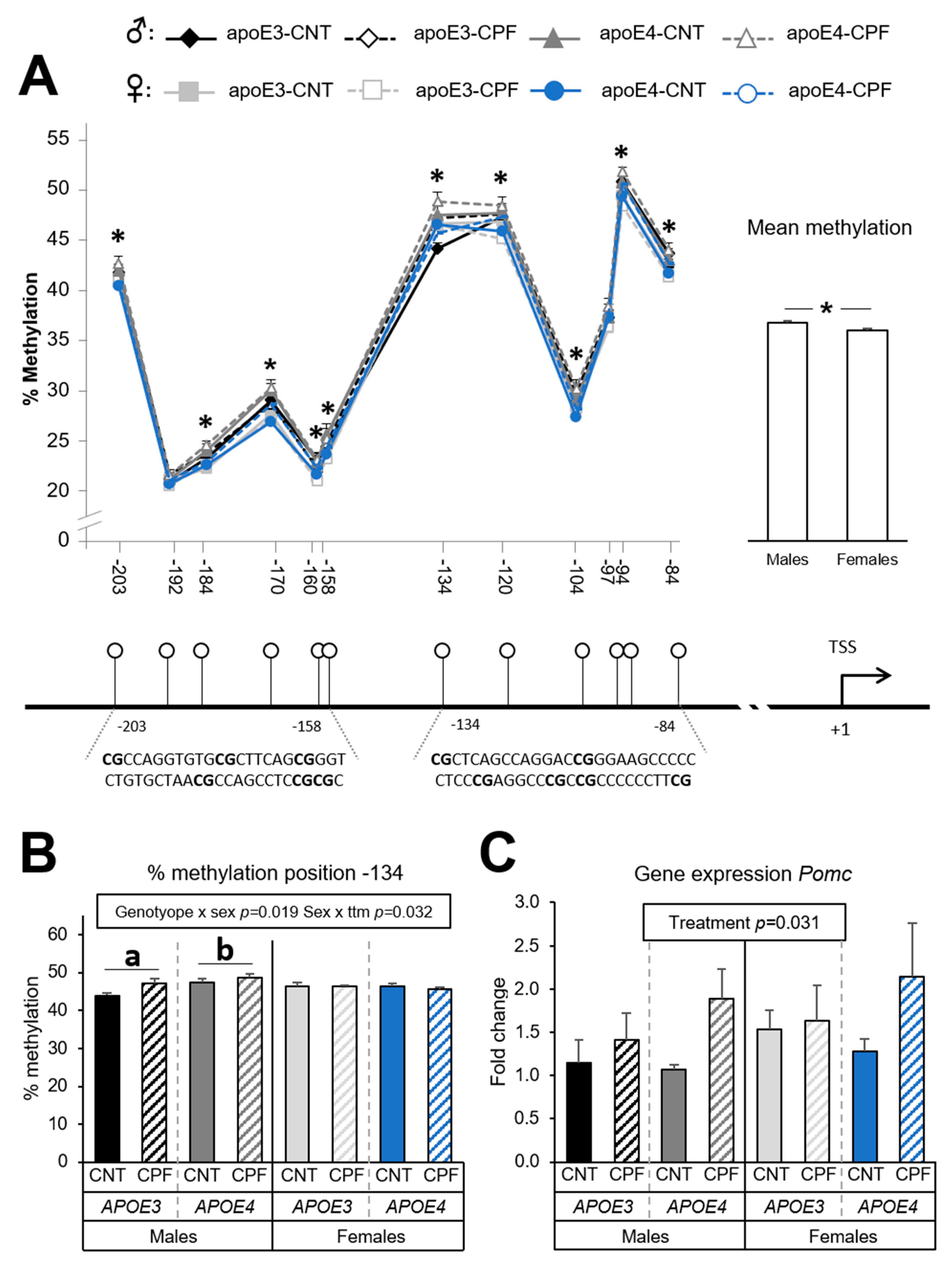
3.1.1. Pomc Methylation is Highly Influenced by Sex while CPF Modulates Gene Expression Levels
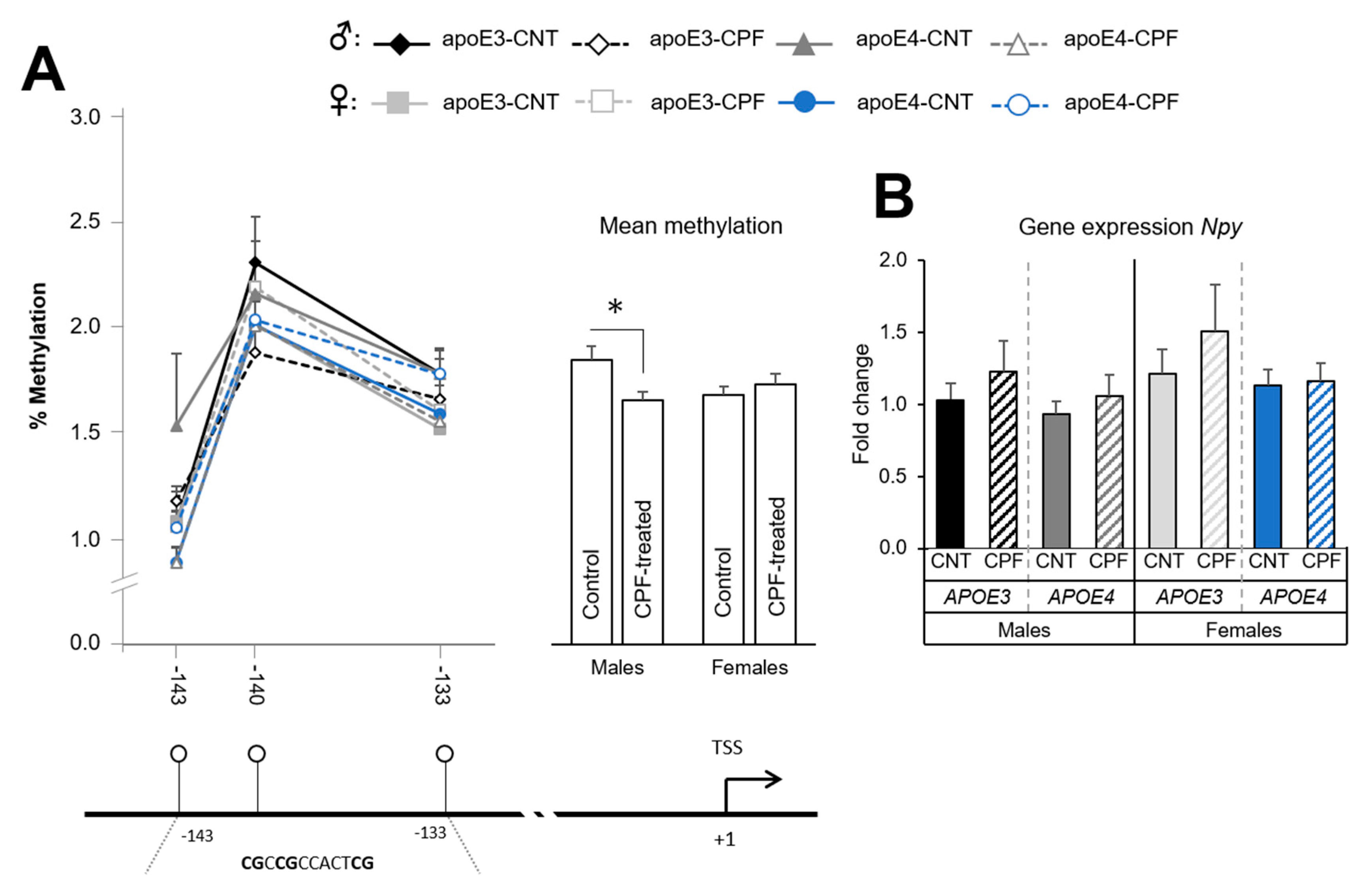
3.1.2. CPF Treatment Modulates Npy Methylation Only in Males
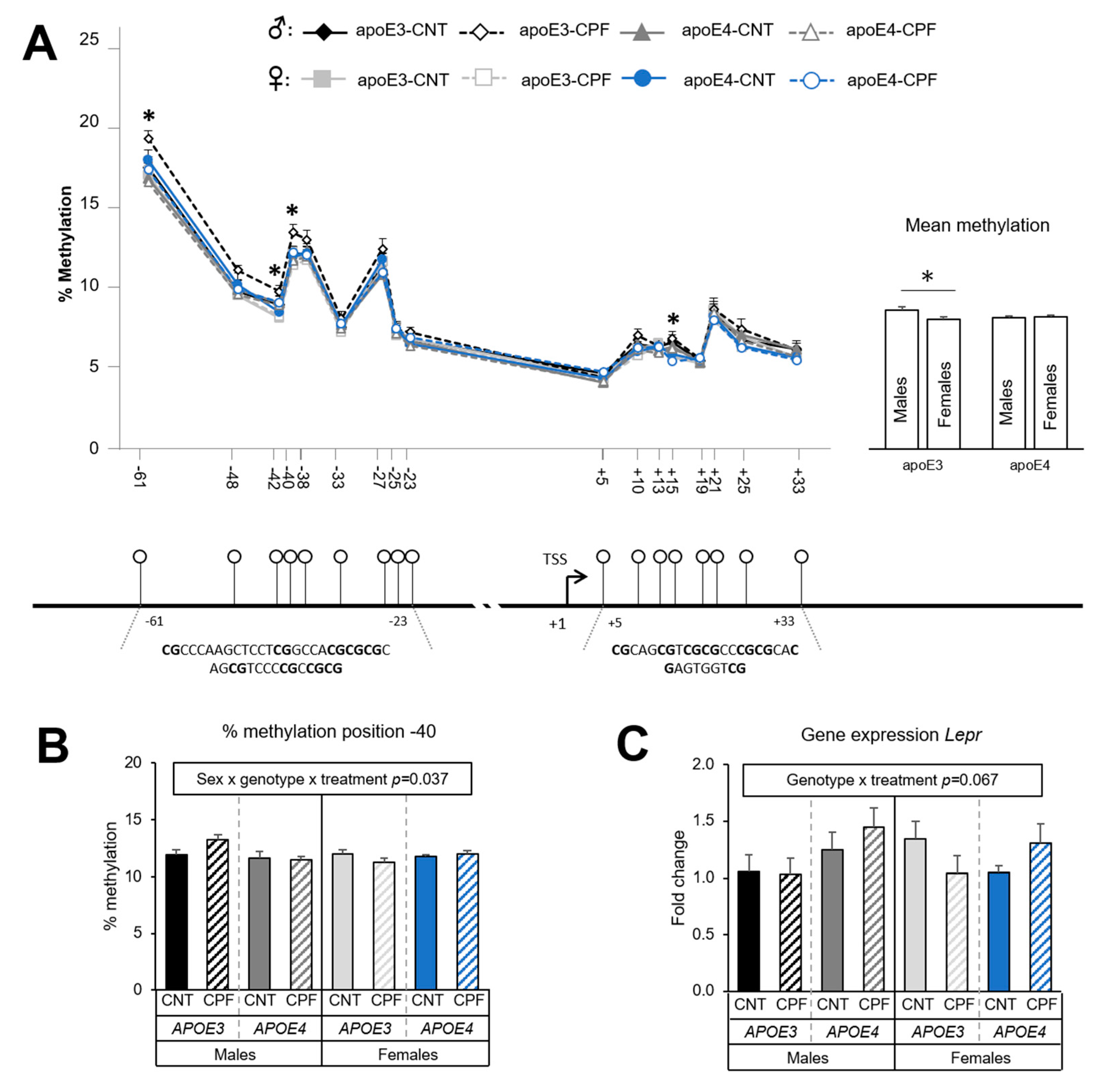
3.1.3. Lepr Presents Sexual-Dimorphic Differences only in the APOE3 Genotype
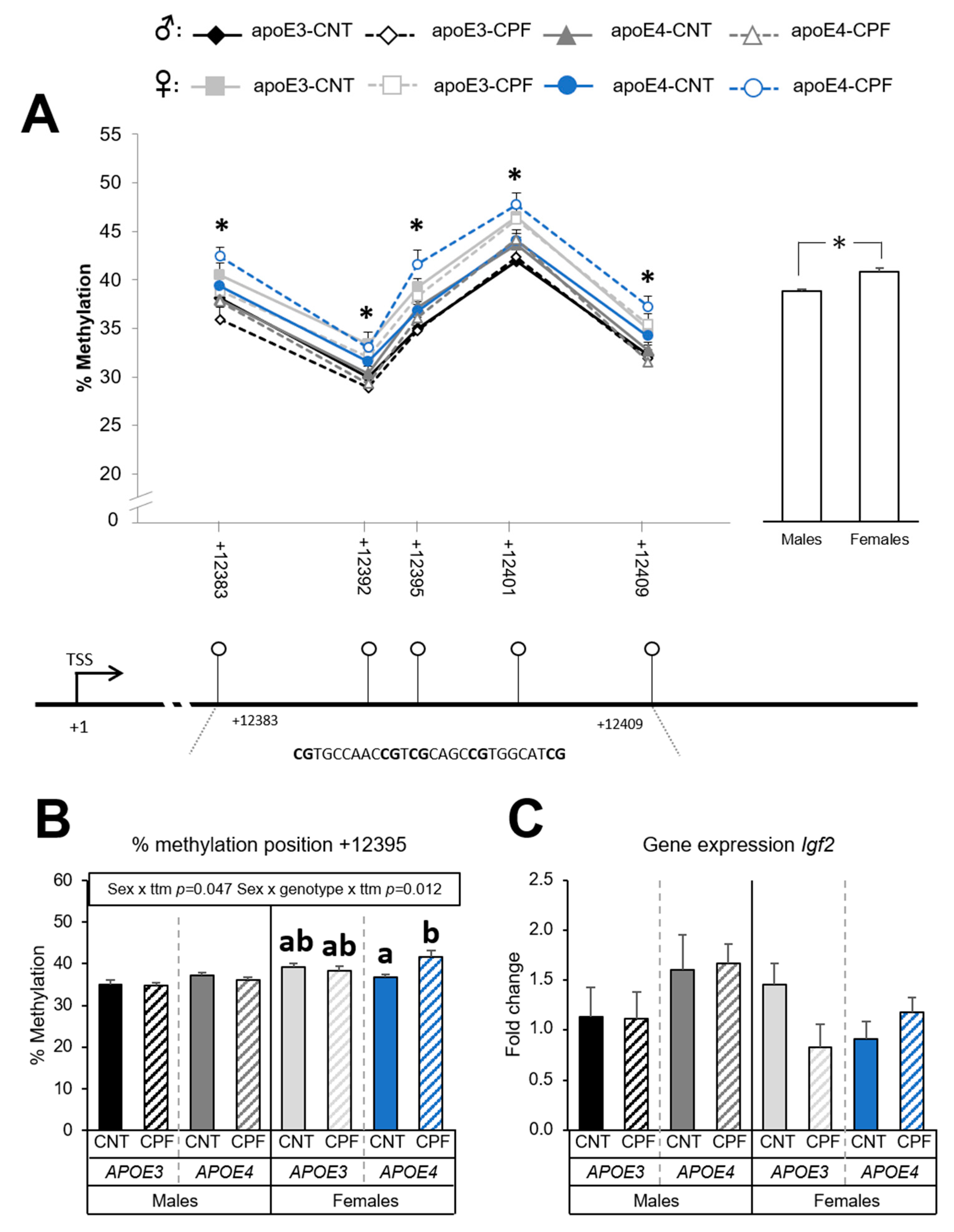
3.1.4. Igf2 Methylation Levels Depend on Sex
3.2. Dietary Challenge: High-Fat Diet
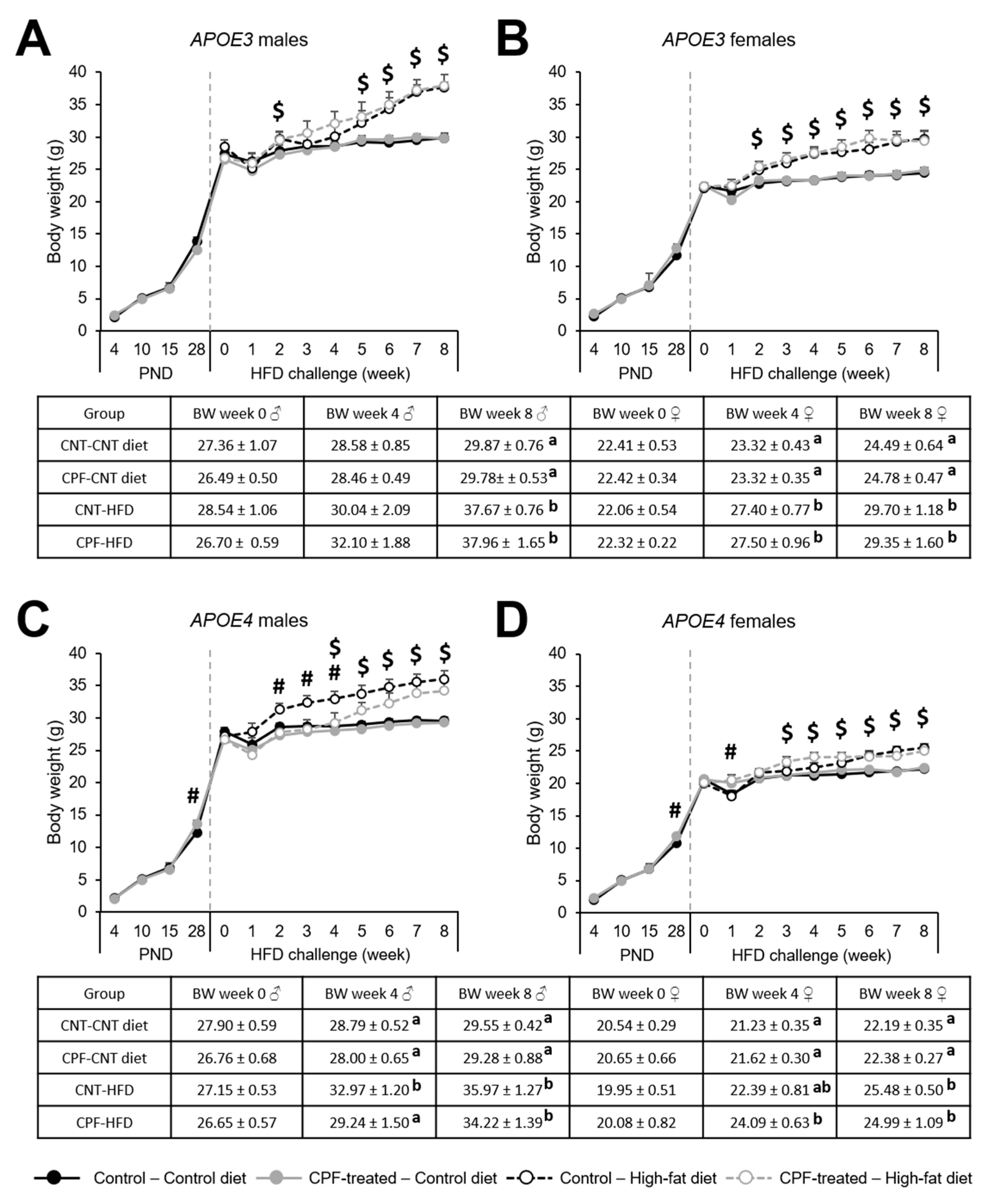
Body Weight Progression is Influenced by CPF Exposure Only for the APOE4 Genotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manna, P.; Jain, S.K. Obesity, Oxidative Stress, Adipose Tissue Dysfunction, and the Associated Health Risks: Causes and Therapeutic Strategies. Metab. Syndr. Relat. Disord. 2015, 13, 423–444. [Google Scholar] [CrossRef] [PubMed]
- Grandjean, P.; Barouki, R.; Bellinger, D.C.; Casteleyn, L.; Chadwick, L.H.; Cordier, S.; Etzel, R.A.; Gray, K.A.; Ha, E.H.; Junien, C.; et al. Life-long implications of developmental exposure to environmental stressors: New perspectives. Endocrinology 2015, 156, 3408–3415. [Google Scholar] [CrossRef] [PubMed]
- Sargis, R.M.; Heindel, J.J.; Padmanabhan, V. Interventions to address environmental metabolism—Disrupting chemicals: Changing the narrative to empower action to restore metabolic health. Front. Endocrinol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Heindel, J.J.; Blumberg, B.; Cave, M.; Machtinger, R.; Mantovani, A.; Mendez, M.A.; Nadal, A.; Palanza, P.; Panzica, G.; Sargis, R.; et al. Metabolism disrupting chemicals and metabolic disorders. Reprod. Toxicol. 2017, 68, 3–33. [Google Scholar] [CrossRef]
- Flaskos, J. The developmental neurotoxicity of organophosphorus insecticides: A direct role for the oxon metabolites. Toxicol. Lett. 2012, 209, 86–93. [Google Scholar] [CrossRef]
- Saunders, M.; Magnanti, B.L.; Correia Carreira, S.; Yang, A.; Alamo-Hernández, U.; Riojas-Rodriguez, H.; Calamandrei, G.; Koppe, J.G.; Krayer von Krauss, M.; Keune, H.; et al. Chlorpyrifos and neurodevelopmental effects: A literature review and expert elicitation on research and policy. Environ. Health 2012, 11, S5. [Google Scholar] [CrossRef]
- Reiss, R.; Chang, E.T.; Richardson, R.J.; Goodman, M. A review of epidemiologic studies of low-level exposures to organophosphorus insecticides in non-occupational populations. Crit. Rev. Toxicol. 2015, 45, 531–641. [Google Scholar] [CrossRef]
- Peris-Sampedro, F.; Cabré, M.; Basaure, P.; Reverte, I.; Domingo, J.L.; Colomina, M.T. Adulthood dietary exposure to a common pesticide leads to an obese-like phenotype and a diabetic profile in apoE3 mice. Environ. Res. 2015, 142, 169–176. [Google Scholar] [CrossRef]
- Peris-Sampedro, F.; Basaure, P.; Reverte, I.; Cabré, M.; Domingo, J.L.; Colomina, M.T. Chronic exposure to chlorpyrifos triggered body weight increase and memory impairment depending on human apoE polymorphisms in a targeted replacement mouse model. Physiol. Behav. 2015, 144, 37–45. [Google Scholar] [CrossRef]
- Blanco, J.; Guardia-Escote, L.; Mulero, M.; Basaure, P.; Biosca-Brull, J.; Cabré, M.; Colomina, M.T.; Domingo, J.L.; Sánchez, D.J. Obesogenic effects of chlorpyrifos and its metabolites during the differentiation of 3T3-L1 preadipocytes. Food Chem. Toxicol. 2020, 137, 111171. [Google Scholar] [CrossRef]
- Huang, Y.; Mahley, R.W. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis. 2014, 72, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Arbones-Mainar, J.M.; Johnson, L.A.; Altenburg, M.K.; Maeda, N. Differential modulation of diet-induced obesity and adipocyte functionality by human apolipoprotein E3 and E4 in mice. Int. J. Obes. 2008, 32, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
- Huebbe, P.; Dose, J.; Schloesser, A.; Campbell, G.; Glüer, C.-C.C.; Gupta, Y.; Ibrahim, S.; Minihane, A.-M.M.; Baines, J.F.; Nebel, A.; et al. Apolipoprotein E (APOE) genotype regulates body weight and fatty acid utilization-Studies in gene-targeted replacement mice. Mol. Nutr. Food Res. 2015, 59, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Segev, Y.; Livne, A.; Mints, M.; Rosenblum, K. Concurrence of High Fat Diet and APOE Gene Induces Allele Specific Metabolic and Mental Stress Changes in a Mouse Model of Alzheimer’s Disease. Front. Behav. Neurosci. 2016, 10, 170. [Google Scholar] [CrossRef]
- Basaure, P.; Guardia-Escote, L.; Cabré, M.; Peris-Sampedro, F.; Sánchez-Santed, F.; Domingo, J.L.; Colomina, M.T. Postnatal chlorpyrifos exposure and apolipoprotein E (APOE) genotype differentially affect cholinergic expression and developmental parameters in transgenic mice. Food Chem. Toxicol. 2018, 118, 42–52. [Google Scholar] [CrossRef]
- Basaure, P.; Guardia-Escote, L.; Cabré, M.; Peris-Sampedro, F.; Sánchez-Santed, F.; Domingo, J.L.; Colomina, M.T. Learning, memory and the expression of cholinergic components in mice are modulated by the pesticide chlorpyrifos depending upon age at exposure and apolipoprotein E (APOE) genotype. Arch. Toxicol. 2019, 93, 693–707. [Google Scholar] [CrossRef]
- Peris-Sampedro, F.; Blanco, J.; Cabré, M.; Basaure, P.; Guardia-Escote, L.; Domingo, J.L.; Sánchez, D.J.; Colomina, M.T. New mechanistic insights on the metabolic-disruptor role of chlorpyrifos in apoE mice: A focus on insulin- and leptin-signalling pathways. Arch. Toxicol. 2018, 92, 1717–1728. [Google Scholar] [CrossRef]
- Slotkin, T.A.; Brown, K.K.; Seidler, F.J. Developmental exposures of rats to chlorpyrifos elicits sex-selective hyperlipidemia and hyperinsulinemia in adulthood. Environ. Health Perspect. 2005, 113, 1291–1294. [Google Scholar] [CrossRef][Green Version]
- Guardia-Escote, L.; Basaure, P.; Peris-Sampedro, F.; Biosca-Brull, J.; Cabré, M.; Sánchez-Santed, F.; Domingo, J.L.; Colomina, M.T. APOE genetic background and sex confer different vulnerabilities to postnatal chlorpyrifos exposure and modulate the response to cholinergic drugs. Behav. Brain Res. 2019, 376, 112195. [Google Scholar] [CrossRef]
- Guardia-Escote, L.; Basaure, P.; Blanco, J.; Cabré, M.; Pérez-Fernández, C.; Sánchez-Santed, F.; Domingo, J.L.; Colomina, M.T. Postnatal exposure to chlorpyrifos produces long-term effects on spatial memory and the cholinergic system in mice in a sex- and APOE genotype-dependent manner. Food Chem. Toxicol. 2018, 122, 1–10. [Google Scholar] [CrossRef]
- Jones, P.A.; Liang, G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009, 10, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Benite-Ribeiro, S.A.; Putt, D.A.; Soares-Filho, M.C.; Santos, J.M. The link between hypothalamic epigenetic modifications and long-term feeding control. Appetite 2016, 107, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Bouret, S.G.; Draper, S.J.; Simerly, R.B. Formation of Projection Pathways from the Arcuate Nucleus of the Hypothalamus to Hypothalamic Regions Implicated in the Neural Control of Feeding Behavior in Mice. J. Neurosci. 2004, 24, 2797–2805. [Google Scholar] [CrossRef] [PubMed]
- Padilla, S.; Carmody, J.; Zeltser, L. Populations in Hypothalamic Feeding Circuits. Nat. Med. 2010, 16, 403–405. [Google Scholar] [CrossRef]
- Jirtle, R.L.; Skinner, M.K. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007, 8, 253–262. [Google Scholar] [CrossRef]
- Schwartz, M.W.; Woods, S.C.; Porte, D.; Seeley, R.J.; Baskin, D.G. Central nervous system control of food intake. Nature 2000, 404, 661–671. [Google Scholar] [CrossRef]
- Morton, G.J.; Cummings, D.E.; Baskin, D.G.; Barsh, G.S.; Schwartz, M.W. Central nervous system control of food intake and body weight. Nature 2006, 443, 289–295. [Google Scholar] [CrossRef]
- Dhillo, W.S. Appetite regulation: An overview. Thyroid 2007, 17, 433–445. [Google Scholar] [CrossRef]
- Kwon, O.; Kim, K.W.; Kim, M.S. Leptin signalling pathways in hypothalamic neurons. Cell. Mol. Life Sci. 2016, 73, 1457–1477. [Google Scholar] [CrossRef]
- Hyland, L.; Park, S.B.; Abdelaziz, Y.; Abizaid, A. Ghrelin infused into the dorsomedial hypothalamus of male mice increases food intake and adiposity. Physiol. Behav. 2020, 220, 112882. [Google Scholar] [CrossRef] [PubMed]
- Sahu, A.; Dube, M.G.; Phelps, C.P.; Sninsky, C.A.; Kalra, P.S.; Kalra, S.P. Insulin and insulin-like growth factor II suppress neuropeptide Y release from the nerve terminals in the paraventricular nucleus: A putative hypothalamic site for energy homeostasis. Endocrinology 1995, 136, 5718–5724. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.M.; Mezdour, H.; Aratanti, Y.; Knouff, C.; Najib, J.; Reddick, R.L.; Quarfordt, S.H.; Maeda, N. Targeted Replacement of the Mouse Apolipoprotein E Gene with the Common Human APOE3 Allele Enhances Diet-induced Hypercholesterolemia and Atherosclerosis. J. Biol. Chem. 1997, 272, 17972–17980. [Google Scholar] [CrossRef] [PubMed]
- Freitag, N.; Zwier, M.V.; Barrientos, G.; Tirado-González, I.; Conrad, M.L.; Rose, M.; Scherjon, S.A.; Plösch, T.; Blois, S.M. Influence of relative NK-DC abundance on placentation and its relation to epigenetic programming in the offspring. Cell Death Dis. 2014, 5, e1392. [Google Scholar] [CrossRef]
- Kabra, D.G.; Pfuhlmann, K.; García-Cáceres, C.; Schriever, S.C.; García, V.C.; Kebede, A.F.; Fuente-Martin, E.; Trivedi, C.; Heppner, K.; Uhlenhaut, N.H.; et al. Hypothalamic leptin action is mediated by histone deacetylase 5. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Herman, A.M.; Ortiz-Guzman, J.; Kochukov, M.; Herman, I.; Quast, K.B.; Patel, J.M.; Tepe, B.; Carlson, J.C.; Ung, K.; Selever, J.; et al. A cholinergic basal forebrain feeding circuit modulates appetite suppression. Nature 2016, 538, 253–256. [Google Scholar] [CrossRef]
- Zhu, W.-L.; Wang, Z.-K. Effects of random food deprivation and refeeding on energy metabolism, behavior and hypothalamic neuropeptide expression in Apodemus chevrieri. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2016, 201, 71–78. [Google Scholar] [CrossRef]
- Phillipps, H.R.; Rand, C.J.; Brown, R.S.E.E.; Kokay, I.C.; Stanton, J.-A.A.; Grattan, D.R. Prolactin regulation of insulin-like growth factor 2 gene expression in the adult mouse choroid plexus. FASEB J. 2019, 33, 6115–6128. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, L.; Liang, Y.; Sui, L.; Guo, L.; Zhou, J.; Fan, K.; Jing, J.; Zhang, Y.; Yao, B. Blastomere removal from cleavage-stage mouse embryos alters placental function, which is associated with placental oxidative stress and inflammation. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Murrell, A.; Heeson, S.; Bowden, L.; Constância, M.; Dean, W.; Kelsey, G.; Reik, W. An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO Rep. 2001, 2, 1101–1106. [Google Scholar] [CrossRef]
- Milagro, F.I.; Mansego, M.L.; De Miguel, C.; Martínez, J.A. Dietary factors, epigenetic modifications and obesity outcomes: Progresses and perspectives. Mol. Asp. Med. 2013, 34, 782–812. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, P.; Anderson, J.T. Obesity: Epigenetic aspects. Biomol. Concepts 2016, 7, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Gali Ramamoorthy, T.; Allen, T.J.; Davies, A.; Harno, E.; Sefton, C.; Murgatroyd, C.; White, A. Maternal overnutrition programs epigenetic changes in the regulatory regions of hypothalamic Pomc in the offspring of rats. Int. J. Obes. 2018, 42, 1431–1444. [Google Scholar] [CrossRef] [PubMed]
- Plagemann, A.; Harder, T.; Brunn, M.; Harder, A.; Roepke, K.; Wittrock-Staar, M.; Ziska, T.; Schellong, K.; Rodekamp, E.; Melchior, K.; et al. Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: An epigenetic model of obesity and the metabolic syndrome. J. Physiol. 2009, 587, 4963–4976. [Google Scholar] [CrossRef]
- Shi, X.; Wang, X.; Li, Q.; Su, M.; Chew, E.; Wong, E.T.; Lacza, Z.; Radda, G.K.; Tergaonkar, V.; Han, W. Nuclear factor κb (NF-κB) suppresses food intake and energy expenditure in mice by directly activating the Pomc promoter. Diabetologia 2013, 56, 925–936. [Google Scholar] [CrossRef]
- Marco, A.; Kisliouk, T.; Weller, A.; Meiri, N. High fat diet induces hypermethylation of the hypothalamic Pomc promoter and obesity in post-weaning rats. Psychoneuroendocrinology 2013, 38, 2844–2853. [Google Scholar] [CrossRef]
- Marco, A.; Kisliouk, T.; Tabachnik, T.; Meiri, N.; Weller, A. Overweight and CpG methylation of the Pomc promoter in offspring of high-fat-diet-fed dams are not “reprogrammed” by regular chow diet in rats. FASEB J. 2014, 28, 4148–4157. [Google Scholar] [CrossRef]
- Schellong, K.; Melchior, K.; Ziska, T.; Henrich, W.; Rancourt, R.C.; Plagemann, A. Sex-specific epigenetic alterations of the hypothalamic Agrp-Pomc system do not explain ‘diabesity’ in the offspring of high-fat diet (HFD) overfed maternal rats. J. Nutr. Biochem. 2020, 75, 108257. [Google Scholar] [CrossRef]
- Unnikrishnan, A.; Jackson, J.; Matyi, S.A.; Hadad, N.; Wronowski, B.; Georgescu, C.; Garrett, K.P.; Wren, J.D.; Freeman, W.M.; Richardson, A. Role of DNA methylation in the dietary restriction mediated cellular memory. GeroScience 2017, 39, 331–345. [Google Scholar] [CrossRef]
- Hua, X.; Xiong, J.W.; Zhang, Y.J.; Cao, X.Y.; Sun, P.; Wu, J.; Chen, L. Exposure of pregnant mice to triclosan causes hyperphagic obesity of offspring via the hypermethylation of proopiomelanocortin promoter. Arch. Toxicol. 2019, 93, 547–558. [Google Scholar] [CrossRef]
- Zhang, X.; Wallace, A.D.; Du, P.; Kibbe, W.A.; Jafari, N.; Xie, H.; Lin, S.; Baccarelli, A.; Soares, M.B.; Hou, L. DNA methylation alterations in response to pesticide exposure in vitro. Environ. Mol. Mutagen. 2012, 53, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Paul, K.C.; Chuang, Y.-H.; Cockburn, M.; Bronstein, J.M.; Horvath, S.; Ritz, B. Organophosphate pesticide exposure and differential genome-wide DNA methylation. Sci. Total Environ. 2018, 645, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Liu, X.; Shen, W.; Deckelbaum, R.J.; Qi, K. The regulation of leptin, leptin receptor and pro-opiomelanocortin expression by N-3 PUFAs in diet-induced obese mice is not related to the methylation of their promoters. Nutr. Metab. 2011, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, R.; Jia, Y.; Cai, D.; Zhou, B.; Qu, X.; Han, H.; Xu, L.; Wang, L.; Yao, Y.; et al. Hypermethylation of Sp1 binding site suppresses hypothalamic POMC in neonates and may contribute to metabolic disorders in adults: Impact of maternal dietary CLAs. Diabetes 2014, 63, 1475–1487. [Google Scholar] [CrossRef] [PubMed]
- Mauvais-Jarvis, F. Sex differences in metabolic homeostasis, diabetes, and obesity. Biol. Sex Differ. 2015, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Lim, C.Y.; Li, C.; Radda, G.K.; Li, C.; Cao, X.; Han, W. FoxO1 inhibits leptin regulation of pro-opiomelanocortin promoter activity by blocking STAT3 interaction with specificity protein 1. J. Biol. Chem. 2009, 284, 3719–3727. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lacza, Z.; Sun, Y.E.; Han, W. Leptin resistance and obesity in mice with deletion of methyl-CpG-binding protein 2 (MeCP2) in hypothalamic pro-opiomelanocortin (POMC) neurons. Diabetologia 2014, 57, 236–245. [Google Scholar] [CrossRef]
- Cifani, C.; Micioni Di Bonaventura, M.V.; Pucci, M.; Giusepponi, M.E.; Romano, A.; Di Francesco, A.; Maccarrone, M.; D’Addario, C. Regulation of hypothalamic neuropeptides gene expression in diet induced obesity resistant rats: Possible targets for obesity prediction? Front. Neurosci. 2015, 9, 187. [Google Scholar] [CrossRef]
- Palou, M.; Picó, C.; McKay, J.A.; Sánchez, J.; Priego, T.; Mathers, J.C.; Palou, A. Protective effects of leptin during the suckling period against later obesity may be associated with changes in promoter methylation of the hypothalamic pro-opiomelanocortin gene. Br. J. Nutr. 2011, 106, 769–778. [Google Scholar] [CrossRef]
- Bo, E.; Farinetti, A.; Marraudino, M.; Sterchele, D.; Eva, C.; Gotti, S.; Panzica, G. Adult exposure to tributyltin affects hypothalamic neuropeptide Y, Y1 receptor distribution, and circulating leptin in mice. Andrology 2016, 4, 723–734. [Google Scholar] [CrossRef]
- Chen, M.; Wang, X.; Hu, Z.; Zhou, H.; Xu, Y.; Qiu, L.; Qin, X.; Zhang, Y.; Ying, Z. Programming of mouse obesity by maternal exposure to concentrated ambient fine particles. Part. Fibre Toxicol. 2017, 14, 20. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Tso, P.; Woods, S.C.; Clegg, D.J.; Barber, K.L.; Carey, K.; Liu, M. Brain apolipoprotein E: An important regulator of food intake in rats. Diabetes 2008, 57, 2092–2098. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shen, L.; Tso, P.; Wang, D.Q.H.; Woods, S.C.; Davidson, W.S.; Sakai, R.; Liu, M. Up-regulation of apolipoprotein E by leptin in the hypothalamus of mice and rats. Physiol. Behav. 2009, 98, 223–228. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Agba, O.B.; Lausser, L.; Huse, K.; Bergmeier, C.; Jahn, N.; Groth, M.; Bens, M.; Sahm, A.; Gall, M.; Witte, O.W.; et al. Tissue-, sex-, and age-specific DNA methylation of rat glucocorticoid receptor gene promoter and insulin-like growth factor 2 imprinting control region. Physiol. Genom. 2017, 49, 690–702. [Google Scholar] [CrossRef]
- Do, E.K.; Zucker, N.L.; Huang, Z.Y.; Schechter, J.C.; Kollins, S.H.; Maguire, R.L.; Murphy, S.K.; Hoyo, C.; Fuemmeler, B.F. Associations between imprinted gene differentially methylated regions, appetitive traits and body mass index in children. Pediatr. Obes. 2019, 14. [Google Scholar] [CrossRef]
- Baskin, D.G.; Wilcox, B.J.; Figlewicz, D.P.; Dorsa, D.M. Insulin and insulin-like growth factors in the CNS. Trends Neurosci. 1988, 11, 107–111. [Google Scholar] [CrossRef]
- Lauterio, T.J.; Marson, L.; Daughaday, W.H.; Baile, C.A. Evidence for the role of insulin-like growth factor II (IGF-II) in the control of food intake. Physiol. Behav. 1987, 40, 755–758. [Google Scholar] [CrossRef]
- Panzica, G.C.; Melcangi, R.C. Structural and molecular brain sexual differences: A tool to understand sex differences in health and disease. Neurosci. Biobehav. Rev. 2016, 67, 2–8. [Google Scholar] [CrossRef]
- Chowen, J.A.; Freire-Regatillo, A.; Argente, J. Neurobiological characteristics underlying metabolic differences between males and females. Prog. Neurobiol. 2019, 176, 18–32. [Google Scholar] [CrossRef]
- El-Maarri, O.; Becker, T.; Junen, J.; Manzoor, S.S.; Diaz-Lacava, A.; Schwaab, R.; Wienker, T.; Oldenburg, J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: A tendency toward higher methylation levels in males. Hum. Genet. 2007, 122, 505–514. [Google Scholar] [CrossRef]
- Liu, J.; Morgan, M.; Hutchison, K.; Calhoun, V.D. A study of the influence of sex on genome wide methylation. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, N.S.; Melton, P.E.; Cadby, G.; Yazar, S.; Franchina, M.; Moses, E.K.; Mackey, D.A.; Hewitt, A.W. Meta-analysis of human methylation data for evidence of sex-specific autosomal patterns. BMC Genom. 2014, 15, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.S.; Watson, K.Q.; Rebeck, G.W. Metabolic disturbances of a high-fat diet are dependent on APOE genotype and sex. eNeuro 2019, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Peris-Sampedro, F.; Guardia-Escote, L.; Basaure, P.; Cabré, M.; Colomina, M.T.; Teresa, M. Improvement of APOE4-dependent non-cognitive behavioural traits by postnatal cholinergic stimulation in female mice. Behav. Brain Res. 2020, 384, 112552. [Google Scholar] [CrossRef] [PubMed]
- Lassiter, T.L.; Ryde, I.T.; Levin, E.D.; Seidler, F.J.; Slotkin, T.A. Neonatal exposure to parathion alters lipid metabolism in adulthood: Interactions with dietary fat intake and implications for neurodevelopmental deficits. Brain Res. Bull. 2010, 81, 85–91. [Google Scholar] [CrossRef]
- Lassiter, T.L.; Brimijoin, S. Rats gain excess weight after developmental exposure to the organophosphorothionate pesticide, chlorpyrifos. Neurotoxicol. Teratol. 2008, 30, 125–130. [Google Scholar] [CrossRef]
- Lassiter, T.L.; Ryde, I.T.; MacKillop, E.A.; Brown, K.K.; Levin, E.D.; Seidler, F.J.; Slotkin, T.A. Exposure of neonatal rats to parathion elicits sex-selective reprogramming of metabolism and alters the response to a high-fat diet in adulthood. Environ. Health Perspect. 2008, 116, 1456–1462. [Google Scholar] [CrossRef][Green Version]
- Ahima, R.S.; Prabakaran, D.; Flier, J.S. Postnatal leptin surge and regulation of circadian rhythm of leptin by feeding. Implications for energy homeostasis and neuroendocrine function. J. Clin. Investig. 1998, 101, 1020–1027. [Google Scholar] [CrossRef]
- Granado, M.; Fuente-Martín, E.; García-Cáceres, C.; Argente, J.; Chowen, J.A. Leptin in early life: A key factor for the development of the adult metabolic profile. Obes. Facts 2012, 5, 138–150. [Google Scholar] [CrossRef]
- Attig, L.; Solomon, G.; Ferezou, J.; Abdennebi-Najar, L.; Taouis, M.; Gertler, A.; Djiane, J. Early postnatal leptin blockage leads to a long-term leptin resistance and susceptibility to diet-induced obesity in rats. Int. J. Obes. 2008, 32, 1153–1160. [Google Scholar] [CrossRef]
- Maioli, S.; Lodeiro, M.; Merino-Serrais, P.; Falahati, F.; Khan, W.; Puerta, E.; Codita, A.; Rimondini, R.; Ramirez, M.J.; Simmons, A.; et al. Alterations in brain leptin signalling in spite of unchanged CSF leptin levels in Alzheimer’s disease. Aging Cell. 2015, 14, 122–129. [Google Scholar] [CrossRef] [PubMed]
| Gene | Sequences 5′–3′ |
|---|---|
| LepR | F: Biotin-GTTGTAAAGGTTAGAGAGGATAGAAT |
| R: AACTAAAAAAATATCCCACCTATATCC | |
| Seq1: ACACTAACCACTCAAAT | |
| Seq2: ACTCTATACTACTAACTCAAAA | |
| Pomc | F: Biotin-TAGGGTTGGGTGGGTGAG |
| R: CACAAAAACCCTAAACCTCTATCCAATTCT | |
| Seq1: CTTTCCAAACAAATATACCTT | |
| Seq2: ATTAAATTCTTCCTAACCAC | |
| Igf2 DMR2 | F: GGGTTGGGGTGGTTATTTTAATGG |
| R: gacgggacaccgctgatcgtttaATCTCTTTATCTCACCCCATAATTC | |
| Universal biotinylated primer: Biotin-gggacaccgctgatcgttta | |
| Seq1: GAATTTTTAGGTAGGTTTTTAAG | |
| Npy | F: GGGTAGTTTAGAATTGGGGTGTGG |
| R: ATAACTCCCACAACCACTTC-Biotin | |
| Seq1: AATTGGGGTGTGGGT |
| Gene | Forward Sequence | Reverse Sequence | Reference |
|---|---|---|---|
| LepR | CGTGGTGAAGCATCGTACTG | GGGCCATGAGAAGGTAAGGT | [35] |
| Pomc | AGAACGCCATCATCAAGAAC | AAGAGGCTAGAGGTCATCAG | [36] |
| Npy * | TGGACTGACCCTCGCTCTAT | GTGTCTCAGGGCTGGATCTC | [37] |
| Igf2 | ACACGCTTCAGTTTGTCTGTTC | GGGGGTGGCACAGTATGTC | [38] |
| Gapdh | ACAACTTTGGCATTGTGGAA | GATGCAGGGATGATGTTCTG | [39] |
| Genotype | Treatment | Males | Females |
|---|---|---|---|
| APOE3 | CNT—CNT | 8 | 8 |
| CNT—HFD | 8 | 8 | |
| CPF—CNT | 8 | 8 | |
| CPF—HFD | 8 | 8 | |
| APOE4 | CNT—CNT | 8 | 8 |
| CNT—HFD | 8 | 8 | |
| CPF—CNT | 12 | 8 | |
| CPF—HFD | 7 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guardia-Escote, L.; Blanco, J.; Basaure, P.; Biosca-Brull, J.; Verkaik-Schakel, R.N.; Cabré, M.; Peris-Sampedro, F.; Pérez-Fernández, C.; Sánchez-Santed, F.; Plösch, T.; et al. Sex and Exposure to Postnatal Chlorpyrifos Influence the Epigenetics of Feeding-Related Genes in a Transgenic APOE Mouse Model: Long-Term Implications on Body Weight after a High-Fat Diet. Int. J. Environ. Res. Public Health 2021, 18, 184. https://doi.org/10.3390/ijerph18010184
Guardia-Escote L, Blanco J, Basaure P, Biosca-Brull J, Verkaik-Schakel RN, Cabré M, Peris-Sampedro F, Pérez-Fernández C, Sánchez-Santed F, Plösch T, et al. Sex and Exposure to Postnatal Chlorpyrifos Influence the Epigenetics of Feeding-Related Genes in a Transgenic APOE Mouse Model: Long-Term Implications on Body Weight after a High-Fat Diet. International Journal of Environmental Research and Public Health. 2021; 18(1):184. https://doi.org/10.3390/ijerph18010184
Chicago/Turabian StyleGuardia-Escote, Laia, Jordi Blanco, Pia Basaure, Judit Biosca-Brull, Rikst Nynke Verkaik-Schakel, Maria Cabré, Fiona Peris-Sampedro, Cristian Pérez-Fernández, Fernando Sánchez-Santed, Torsten Plösch, and et al. 2021. "Sex and Exposure to Postnatal Chlorpyrifos Influence the Epigenetics of Feeding-Related Genes in a Transgenic APOE Mouse Model: Long-Term Implications on Body Weight after a High-Fat Diet" International Journal of Environmental Research and Public Health 18, no. 1: 184. https://doi.org/10.3390/ijerph18010184
APA StyleGuardia-Escote, L., Blanco, J., Basaure, P., Biosca-Brull, J., Verkaik-Schakel, R. N., Cabré, M., Peris-Sampedro, F., Pérez-Fernández, C., Sánchez-Santed, F., Plösch, T., Domingo, J. L., & Colomina, M. T. (2021). Sex and Exposure to Postnatal Chlorpyrifos Influence the Epigenetics of Feeding-Related Genes in a Transgenic APOE Mouse Model: Long-Term Implications on Body Weight after a High-Fat Diet. International Journal of Environmental Research and Public Health, 18(1), 184. https://doi.org/10.3390/ijerph18010184









