Identification of a New Heavy-Metal-Resistant Strain of Geobacillus stearothermophilus Isolated from a Hydrothermally Active Volcanic Area in Southern Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
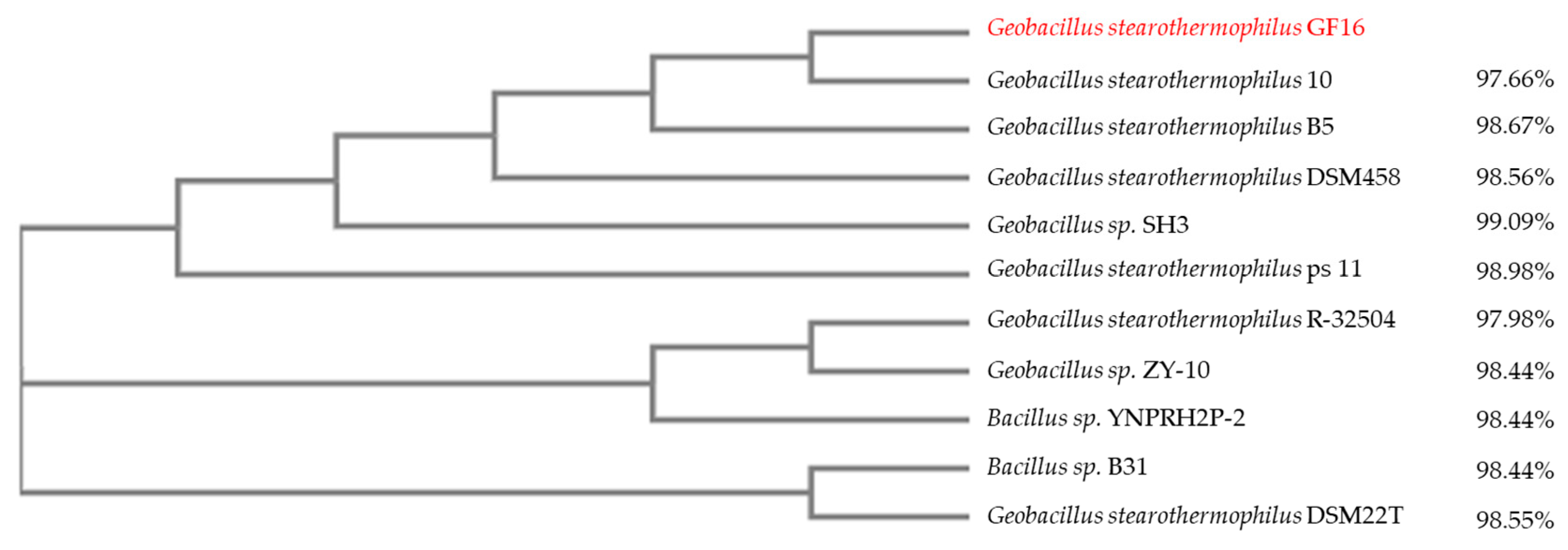
2.2. Strain Isolation and Molecular Identification
Neighbor-Joining Tree Development
2.3. Geobacillus stearothermophilus Physiological Analyses
2.3.1. Determination of Optimal Growth Conditions
2.3.2. Antibiotic Susceptibility
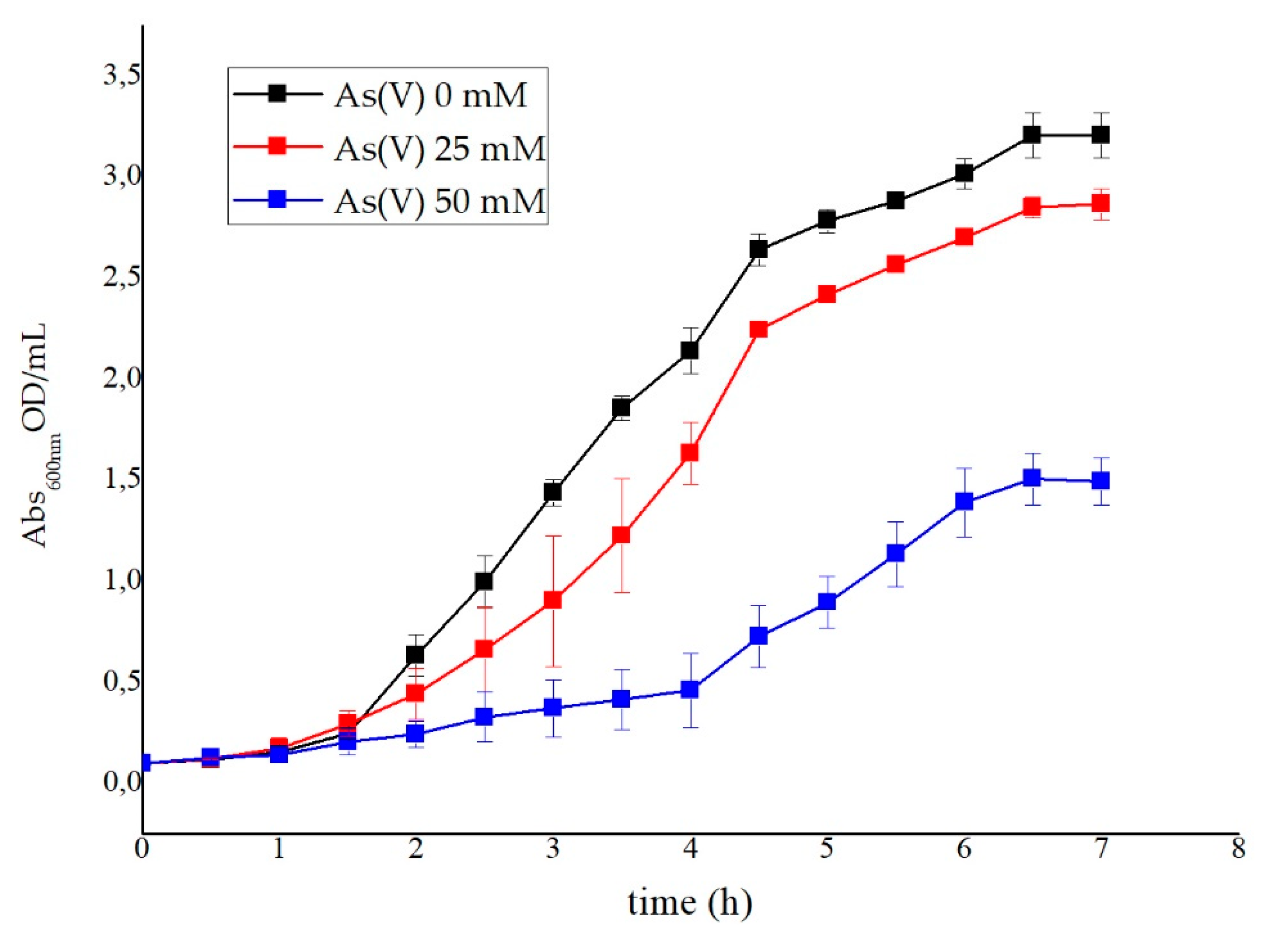
2.3.3. Heavy-Metal Resistance
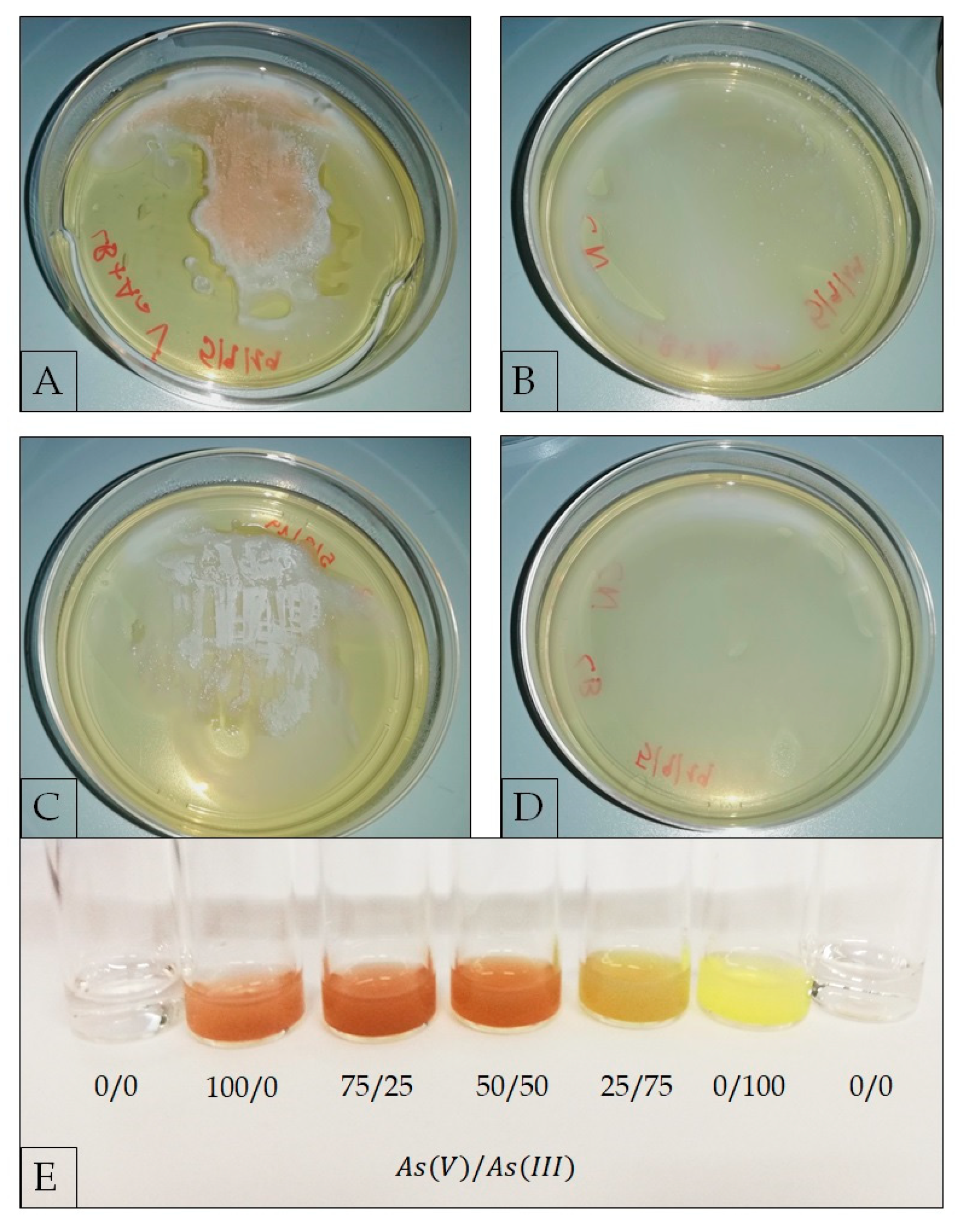
2.3.4. Evaluation of As(V) Biotransformation
2.3.5. Bioinformatic Analysis
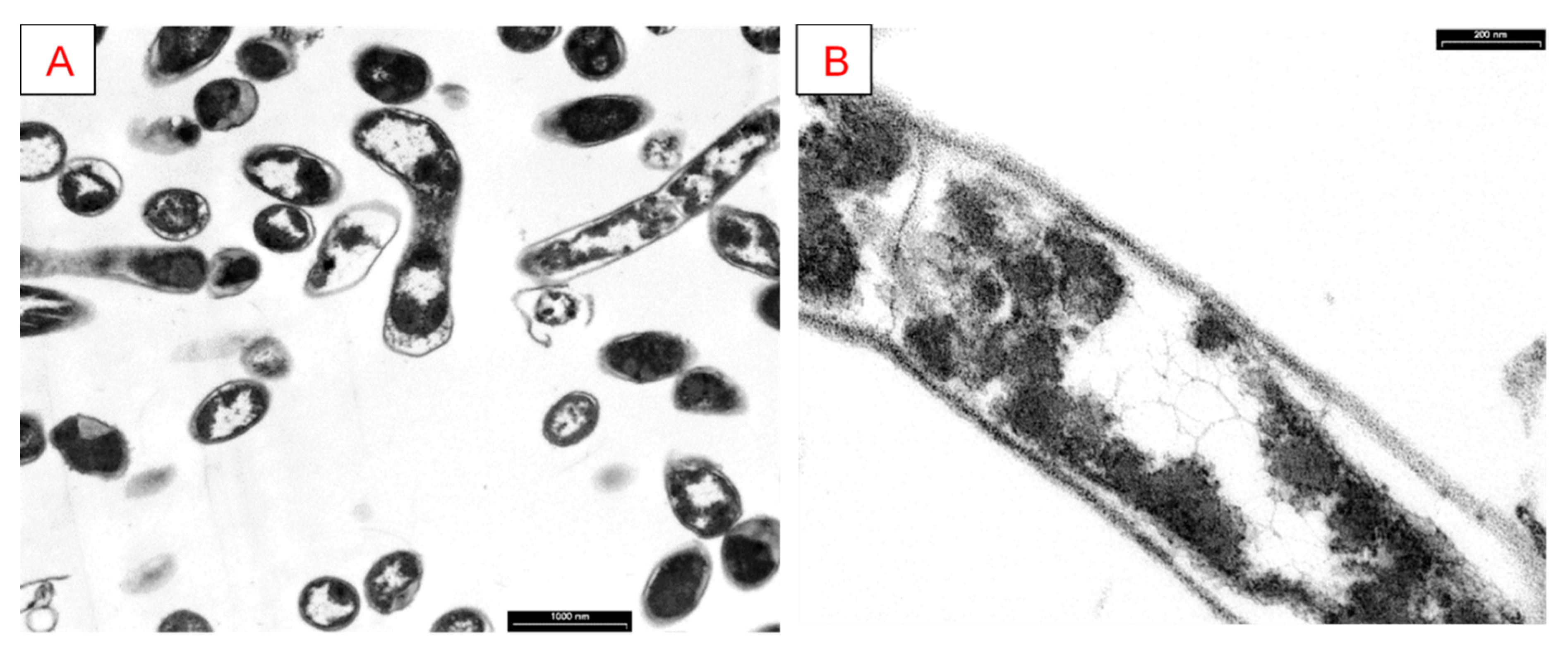
2.3.6. Transmission Electron Microscopy (TEM)
3. Results and Discussion
3.1. Geochemical Characterization of the Sampling Site
3.2. Isolation and Identification of Geobacillus stearothermophilus GF16
3.3. Metal Ion Resistance and Antibiotic Susceptibility in G. stearothermophilus GF16
3.4. Bionformatic Analyses
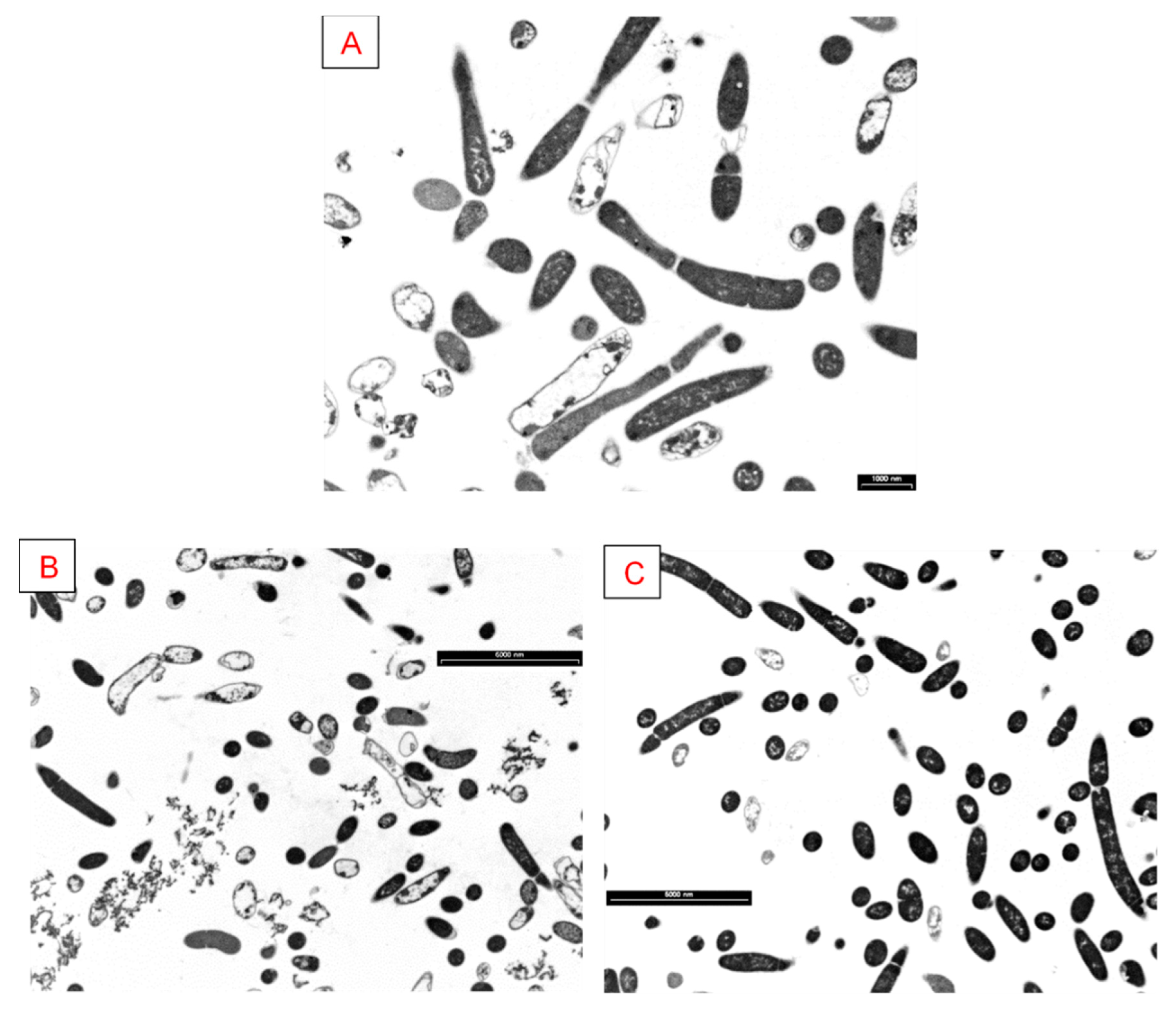
3.5. Analysis of Cellular Morphology
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mathew, B.B.; Singh, H.; Biju, G.; Krishnamurthy, N.B. Classification, Source, and Effect of Environmental Pollutants and Their Biodegradation. J. Environ. Pathol. Toxicol. Oncol. 2017, 36, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Rensing, C.; Rosen, B.P. Heavy Metals Cycle (Arsenic, Mercury, Selenium, others). In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: San Diego, CA, USA, 2009; Volume 3, pp. 205–219. ISBN 9780123739445. [Google Scholar] [CrossRef]
- Kadirvelu, K.; Thamaraiselvi, K.; Namasivayam, C. Removal of heavy metals from industrial wastewaters by adsorption onto activated carbon prepared from an agricultural solid waste. Bioresour. Technol. 2001, 76, 63–65. [Google Scholar] [CrossRef]
- Cole, M.; Lindeque, P.; Halsband, C.; Galloway, T.S. Microplastics as contaminants in the marine environment: A review. Mar. Pollut. Bull. 2011, 62, 2588–2597. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Human Health Risk Assessment Toolkit: Chemical Hazards. 2010. Available online: https://apps.who.int/iris/handle/10665/44458 (accessed on 14 April 2020).
- International Agency for Research on Cancer. International Agency for Research on Cancer Iarc Monographs on the Evaluation of Carcinogenic Risks to Humans. In Iarc Monographs on the Evaluation of Carcinogenic Risks to Humans; 2002; Available online: https://monographs.iarc.fr/list-of-classifications (accessed on 14 April 2020).
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Arsenic, metals, fibres, and dusts. In A Review of Human Carcinogens; The International Agency for Research on Cancer: Lyon, France, 2012; ISBN 978-92-832-1320-8. [Google Scholar]
- Mamtani, R.; Stern, P.; Dawood, I.; Cheema, S. Metals and disease: A global primary health care perspective. J. Toxicol. 2011, 2011, 1–11. [Google Scholar] [CrossRef]
- Gillispie, E.C.; Sowers, T.D.; Duckworth, O.W.; Polizzotto, M.L. Soil Pollution Due to Irrigation with Arsenic-Contaminated Groundwater: Current State of Science. Curr. Pollut. Rep. 2015, 1, 1–12. [Google Scholar] [CrossRef]
- Waseem, A.; Arshad, J.; Iqbal, F.; Sajjad, A.; Mehmood, Z.; Murtaza, G. Pollution Status of Pakistan: A Retrospective Review on Heavy Metal Contamination of Water, Soil, and Vegetables. BioMed Res. Int. 2014, 2014, 1–29. [Google Scholar] [CrossRef]
- Gunduz, O.; Simsek, C.; Hasozbek, A. Arsenic pollution in the groundwater of Simav Plain, Turkey: Its impact on water quality and human health. Water Air Soil Pollut. 2010, 205, 43–62. [Google Scholar] [CrossRef]
- Bartolucci, S.; Contursi, P.; Fiorentino, G.; Limauro, D.; Pedone, E. Responding to toxic compounds: A genomic and functional overview of Archaea. Front. Biosci. 2013, 18, 165–189. [Google Scholar] [CrossRef][Green Version]
- Oremland, R.S.; Stolz, J.F. The ecology of arsenic. Science 2003, 300, 939–944. [Google Scholar] [CrossRef]
- Kulp, T.R.; Hoeft, S.E.; Asao, M.; Madigan, M.T.; Hollibaugh, J.T.; Fisher, J.C.; Stolz, J.F.; Culbertson, C.W.; Miller, L.G.; Oremland, R.S. Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science 2008, 321, 967–970. [Google Scholar] [CrossRef]
- Pedone, E.; Bartolucci, S.; Fiorentino, G. Sensing and adapting to environmental stress: The archaeal tactic. Front. Biosci. A J. Virtual Libr. 2004, 9, 2909–2926. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, R.; Rosen, B.P.; Phung, L.T.; Silver, S. Microbial arsenic: From geocycles to genes and enzymes. FEMS Microbiol. Rev. 2002, 26, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.P. Bacterial resistance to heavy metals and metalloids. J. Biol. Inorg. Chem. 1996, 1, 273–277. [Google Scholar] [CrossRef]
- Saltikov, C.W.; Olson, B.H. Homology of Escherichia coli R773 arsA, arsB, and arsC genes in arsenic-resistant bacteria isolated from raw sewage and arsenic-enriched creek waters. Appl. Environ. Microbiol. 2002, 68, 280–288. [Google Scholar] [CrossRef]
- Ji, G.; Silver, S. Regulation and expression of the arsenic resistance operon from Staphylococcus aureus plasmid pI258. J. Bacteriol. 1992, 164, 3684–3694. [Google Scholar] [CrossRef] [PubMed]
- Antonucci, I.; Gallo, G.; Limauro, D.; Contursi, P.; Ribeiro, A.L.; Blesa, A.; Berenguer, J.; Bartolucci, S.; Fiorentino, G. An ArsR/SmtB family member regulates arsenic resistance genes unusually arranged in Thermus thermophilus HB27. Microb. Biotechnol. 2017, 10, 1690–1701. [Google Scholar] [CrossRef] [PubMed]
- Gallo, G.; Antonucci, I.; Pirone, L.; Amoresano, A.; Contursi, P.; Limauro, D.; Pedone, E.; Bartolucci, S.; Fiorentino, G. A physicochemical investigation on the metal binding properties of TtSmtB, a thermophilic member of the ArsR/SmtB transcription factor family. Int. J. Biol. Macromol. 2019, 138, 1056–1063. [Google Scholar] [CrossRef]
- Fernández, M.; Morel, B.; Ramos, J.L.; Krell, T. Paralogous regulators ArsR1 and ArsR2 of Pseudomonas putida KT2440 as a basis for arsenic biosensor development. Appl. Environ. Microbiol. 2016, 82, 4133–4144. [Google Scholar] [CrossRef]
- Patel, P.C.; Goulhen, F.; Boothman, C.; Gault, A.G.; Charnock, J.M.; Kalia, K.; Lloyd, J.R. Arsenate detoxification in a Pseudomonad hypertolerant to arsenic. Arch. Microbiol. 2007, 187, 171–183. [Google Scholar] [CrossRef]
- Del Giudice, I.; Limauro, D.; Pedone, E.; Bartolucci, S.; Fiorentino, G. A novel arsenate reductase from the bacterium Thermus thermophilus HB27: Its role in arsenic detoxification. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 2071–2079. [Google Scholar] [CrossRef]
- Politi, J.; Spadavecchia, J.; Fiorentino, G.; Antonucci, I.; Casale, S.; De Stefano, L. Interaction of Thermus thermophilus ArsC enzyme and gold nanoparticles naked-eye assays speciation between As(III) and As(V). Nanotechnology 2015, 26, 435703. [Google Scholar] [CrossRef] [PubMed]
- Politi, J.; Spadavecchia, J.; Fiorentino, G.; Antonucci, I.; De Stefano, L. Arsenate reductase from Thermus thermophilus conjugated to polyethylene glycol-stabilized gold nanospheres allow trace sensing and speciation of arsenic ions. J. R. Soc. Interface 2016, 13, 20160629. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.P.; Bao, P.; Sun, G.X. Identification and catalytic residues of the arsenite methyltransferase from a sulfate-reducing bacterium, Clostridium sp. BXM. FEMS Microbiol. Lett. 2015, 362, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Antonucci, I.; Gallo, G.; Limauro, D.; Contursi, P.; Ribeiro, A.L.; Blesa, A.; Berenguer, J.; Bartolucci, S.; Fiorentino, G. Characterization of a promiscuous cadmium and arsenic resistance mechanism in Thermus thermophilus HB27 and potential application of a novel bioreporter system. Microb. Cell Factories 2018, 17, 1–9. [Google Scholar] [CrossRef]
- Dhankher, O.P.; Shasti, N.A.; Rosen, B.P.; Fuhrmann, M.; Meagher, R.B. Increased cadmium tolerance and accumulation by plants expressing bacterial arsenate reductase. New Phytol. 2003, 159, 431–441. [Google Scholar] [CrossRef]
- Xu, C.; Rosen, B.P. Metalloregulation of Soft Metal Resistance Pumps. In Metals and Genetics; Sarkar, B., Ed.; Springer: Boston, MA, USA, 1999; pp. 5–19. ISBN 978-1-4615-4723-5. [Google Scholar] [CrossRef]
- Abbas, S.Z.; Rafatullah, M.; Hossain, K.; Ismail, N.; Tajarudin, H.A.; Abdul Khalil, H.P.S. A review on mechanism and future perspectives of cadmium-resistant bacteria. Int. J. Environ. Sci. Technol. 2018, 15, 243–262. [Google Scholar] [CrossRef]
- Sabadell, J.E.; Axtmann, R.C. Heavy metal contamination from geothermal sources. Environ. Health Perspect. 1975, 12, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nriagu, J.O. A global assessment of natural sources of atmospheric trace metals. Nature 1989, 338, 47–49. [Google Scholar] [CrossRef]
- Hawkes, R.B.; Franzmann, P.D.; Plumb, J.J. Moderate thermophiles including “Ferroplasma cupricumulans” sp. nov. dominate an industrial-scale chalcocite heap bioleaching operation. Hydrometallurgy 2006, 83, 229–236. [Google Scholar] [CrossRef]
- Özdemir, S.; Kilinc, E.; Poli, A.; Nicolaus, B.; Güven, K. Biosorption of Cd, Cu, Ni, Mn and Zn from aqueous solutions by thermophilic bacteria, Geobacillus toebii sub.sp. decanicus and Geobacillus thermoleovorans sub.sp. stromboliensis: Equilibrium, kinetic and thermodynamic studies. Chem. Eng. J. 2009, 152, 195–206. [Google Scholar] [CrossRef]
- Gallo, G.; Puopolo, R.; Limauro, D.; Bartolucci, S.; Fiorentino, G. Metal-Tolerant Thermophiles: From the Analysis of Resistance Mechanisms to their Biotechnological Exploitation. Open Biochem. J. 2018, 12, 149–160. [Google Scholar] [CrossRef]
- Poli, A.; Salerno, A.; Laezza, G.; di Donato, P.; Dumontet, S.; Nicolaus, B. Heavy metal resistance of some thermophiles: Potential use of α-amylase from Anoxybacillus amylolyticus as a microbial enzymatic bioassay. Res. Microbiol. 2009, 160, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.K.; Bhattacharjee, I.; Chandra, G. Biosorption of heavy metals from industrial waste water by Geobacillus thermodenitrificans. J. Hazard. Mater. 2010, 175, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Piochi, M.; Mormone, A.; Strauss, H.; Balassone, G. The acid sulfate zone and the mineral alteration styles of the Roman Puteoli (Neapolitan area, Italy): Clues on fluid fracturing progression at the Campi Flegrei volcano. Solid Earth 2019, 10, 1809–1831. [Google Scholar] [CrossRef]
- Valentino, G.M.; Cortecci, G.; Franco, E.; Stanzione, D. Chemical and isotopic compositions of minerals and waters from the Campi Flegrei volcanic system, Naples, Italy. J. Volcanol. Geotherm. Res. 1999, 91, 329–344. [Google Scholar] [CrossRef]
- Piochi, M.; Mormone, A.; Balassone, G.; Strauss, H.; Troise, C.; De Natale, G. Native sulfur, sulfates and sulfides from the active Campi Flegrei volcano (southern Italy): Genetic environments and degassing dynamics revealed by mineralogy and isotope geochemistry. J. Volcanol. Geotherm. Res. 2015, 304, 180–193. [Google Scholar] [CrossRef]
- Macwilliams, M.P.; Liao, M. Luria Broth (LB) and Luria Agar (LA) Media and Their Uses Protocol. 2006. Available online: https://www.asm.org/Protocols/Luria-Broth-LB-and-Luria-Agar-LA-Media-and-Their-U (accessed on 14 April 2020).
- Schumann, P.; Pukall, R. The discriminatory power of ribotyping as automatable technique for differentiation of bacteria. Syst. Appl. Microbiol. 2013, 36, 369–375. [Google Scholar] [CrossRef]
- Schumann, P.; Maier, T. MALDI-TOF mass spectrometry applied to classification and identification of bacteria. Methods Microbiol. 2014, 41, 275–306. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Rankin, I.D. MIC Testing. In Manual of Antimicrobial Susceptibility Testing; Coyle, M.B., Ed.; American Society for Microbiology: Washington, DC, USA, 2005; pp. 53–62. ISBN 1-55581-349-6. [Google Scholar]
- Simeonova, D.D.; Lièvremont, D.; Lagarde, F.; Muller, D.A.E.; Groudeva, V.I.; Lett, M.C. Microplate screening assay for the detection of arsenite-oxidizing and arsenate-reducing bacteria. FEMS Microbiol. Lett. 2004, 237, 249–253. [Google Scholar] [CrossRef]
- Pinho, S.C.; Nunes, O.C.; Lobo-da-Cunha, A.; Almeida, M.F. Inactivation of Geobacillus stearothermophilus spores by alkaline hydrolysis applied to medical waste treatment. J. Environ. Manag. 2015, 161, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Giggenbach, W.F. Geothermal solute equilibria. Derivation of Na-K-Mg-Ca geoindicators. Geochim. Cosmochim. Acta 1988, 52, 2749–2765. [Google Scholar] [CrossRef]
- Hedenquist, J.W.; Lowenstern, J.B. The role of magmas in the formation of hydrothermal ore deposits. Nature 1994, 370, 519–527. [Google Scholar] [CrossRef]
- Ellis, A.J.; Mahon, W.A.J. Chemistry and Geothermal Systems; Academic Press: New York, NY, USA; San Francisco, CA, USA; London, UK, 1977; p. 392. ISBN 0122374509. [Google Scholar] [CrossRef]
- Webster, J.G.; Nordstrom, D.K. Chapter 4: Geothermal Arsenic. The source, transport and fate of arsenic in geothermal systems. In Arsenic in Ground Water; Welch, A.H., Stollenwerk, K.G., Eds.; Springer: Boston, MA, USA, 2003; pp. 101–125. ISBN 978-0-306-47956-4. [Google Scholar] [CrossRef]
- Farnfield, H.R.; Marcilla, A.L.; Ward, N.I. Arsenic speciation and trace element analysis of the volcanic río Agrio and the geothermal waters of Copahue, Argentina. Sci. Total. Environ. 2012, 433, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Aiuppa, A.; Avino, R.; Brusca, L.; Caliro, S.; Chiodini, G.; D’Alessandro, W.; Favara, R.; Federico, C.; Ginevra, W.; Inguaggiato, S.; et al. Mineral control of arsenic content in thermal waters from volcano-hosted hydrothermal systems: Insights from island of Ischia and Phlegrean Fields (Campanian Volcanic Province, Italy). Chem. Geol. 2006, 229, 313–330. [Google Scholar] [CrossRef]
- Cardellini, C.; Chiodini, G.; Frondini, F.; Avino, R.; Bagnato, E.; Caliro, S.; Lelli, M.; Rosiello, A. Monitoring diffuse volcanic degassing during volcanic unrests: The case of Campi Flegrei (Italy). Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Valentino, G.M.; Stanzione, D. Source processes of the thermal waters from the Phlegraean Fields (Naples, Italy) by means of the study of selected minor and trace elements distribution. Chem. Geol. 2003, 194, 245–274. [Google Scholar] [CrossRef]
- Nazina, T.N.; Tourova, T.P.; Poltaraus, A.B.; Novikova, E.V.; Grigoryan, A.A.; Ivanova, A.E.; Lysenko, A.M.; Petrunyaka, V.V.; Osipov, G.A.; Belyaev, S.S.; et al. Taxonomic study of aerobic thermophilic bacilli: Descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus th. Int. J. Syst. Evol. Microbiol. 2001, 51, 433–446. [Google Scholar] [CrossRef]
- Kananavičiute, R.; Čitavičius, D. Genetic engineering of Geobacillus spp. J. Microbiol. Methods 2015, 111, 31–39. [Google Scholar] [CrossRef]
- Suzuki, H. Peculiarities and biotechnological potential of environmental adaptation by Geobacillus species. Appl. Microbiol. Biotechnol. 2018, 102, 10425–10437. [Google Scholar] [CrossRef]
- Burgess, S.A.; Flint, S.H.; Lindsay, D.; Cox, M.P.; Biggs, P.J. Insights into the Geobacillus stearothermophilus species based on phylogenomic principles. BMC Microbiol. 2017, 17, 140. [Google Scholar] [CrossRef]
- Kliem, M.; Sauer, S. The essence on mass spectrometry based microbial diagnostics. Curr. Opin. Microbiol. 2012, 15, 397–402. [Google Scholar] [CrossRef]
- Majumder, A.; Bhattacharyya, K.; Bhattacharyya, S.; Kole, S.C. Arsenic-tolerant, arsenite-oxidising bacterial strains in the contaminated soils of West Bengal, India. Sci. Total. Environ. 2013, 463–464, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Jean, J.S.; Kar, S.; Chou, M.L.; Chen, C.Y. Screening of plant growth-promoting traits in arsenic-resistant bacteria isolated from agricultural soil and their potential implication for arsenic bioremediation. J. Hazard. Mater. 2014, 272, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Cuebas, M.; Sannino, D.; Bini, E. Isolation and characterization of arsenic resistant Geobacillus kaustophilus strain from geothermal soils. J. Basic Microbiol. 2011, 51, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.C.; Fu, H.L.; Lin, Y.F.; Rosen, B.P. Pathways of Arsenic Uptake and Efflux. Curr. Top. Membr. 2012, 69, 325–358. [Google Scholar] [CrossRef]
- Cai, L.; Liu, G.; Rensing, C.; Wang, G. Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 2009, 9, 4. [Google Scholar] [CrossRef]
- Lebrun, E.; Brugna, M.; Baymann, F.; Muller, D.; Lièvremont, D.; Lett, M.C.; Nitschke, W. Arsenite oxidase, an ancient bioenergetic enzyme. Mol. Biol. Evol. 2003, 20, 686–693. [Google Scholar] [CrossRef]
- Dangayach, S.; Sharma, P.; Singhai, P.; Gupta, N. Microbial removal of arsenic: Mechanisms and Applications. Asian J. Multidisciplanary Stud. 2014, 2, 159–170. [Google Scholar]
- Rosen, B.P. Families of arsenic transporters. Trends Microbiol. 1999, 7, 207–212. [Google Scholar] [CrossRef]
- Yang, H.C.; Cheng, J.; Finan, T.M.; Rosen, B.P.; Bhattacharjee, H. Novel pathway for arsenic detoxification in the legume symbiont Sinorhizobium meliloti. J. Bacteriol. 2005, 187, 6991–6997. [Google Scholar] [CrossRef] [PubMed]
- Hetzer, A.; Daughney, C.J.; Morgan, H.W. Cadmium ion biosorption by the thermophilic bacteria Geobacillus stearothermophilus and G. thermocatenulatus. Appl. Environ. Microbiol. 2006, 72, 4020–4027. [Google Scholar] [CrossRef] [PubMed]
- Ranawat, P.; Rawat, S. Metal-tolerant thermophiles: Metals as electron donors and acceptors, toxicity, tolerance and industrial applications. Environ. Sci. Pollut. Res. 2017, 25, 4105–4133. [Google Scholar] [CrossRef]
- Özdemir, S.; Klnç, E.; Poli, A.; Nicolaus, B. Biosorption of heavy metals (Cd2+, Cu2+, Co 2+, and Mn2+) by thermophilic bacteria, geobacillus thermantarcticus and anoxybacillus amylolyticus: Equilibrium and kinetic studies. Bioremediation J. 2013, 17, 86–96. [Google Scholar] [CrossRef]
- Wu, D.; Zhang, Z.; Gao, Q.; Ma, Y. Isolation and characterization of aerobic, culturable, arsenic-tolerant bacteria from lead–zinc mine tailing in southern China. World J. Microbiol. Biotechnol. 2018, 34, 177. [Google Scholar] [CrossRef] [PubMed]
- Krumova, K.; Nikolovska, M.; Groudeva, V. Isolation and identification of arsenic-transforming bacteria from arsenic contaminated sites in Bulgaria. Biotechnol. Biotechnol. Equip. 2008, 22, 721–728. [Google Scholar] [CrossRef]
- Krumova, K.; Nikolovska, M.; Groudeva, V. Characterization of arsenic-transforming bacteria from arsenic contaminated sites in Bulgaria. Biotechnol. Biotechnol. Equip 2008, 22, 729–735. [Google Scholar] [CrossRef][Green Version]
- Egan, K.; Kelleher, P.; Field, D.; Rea, M.C.; Paul Ross, R.; Cotter, P.D.; Hilla, C. Genome sequence of Geobacillus stearothermophilus DSM 458, an antimicrobial-producing thermophilic bacterium, isolated from a sugar beet factory. Genome Announc. 2017, 5, 43. [Google Scholar] [CrossRef]
- Koechler, S.; Arsène-Ploetze, F.; Brochier-Armanet, C.; Goulhen-Chollet, F.; Heinrich-Salmeron, A.; Jost, B.; Lièvremont, D.; Philipps, M.; Plewniak, F.; Bertin, P.N.; et al. Constitutive arsenite oxidase expression detected in arsenic-hypertolerant pseudomonas xanthomarina S11. Res. Microbiol. 2015, 166, 205–2014. [Google Scholar] [CrossRef]
- Andres, J.; Bertin, P.N. The microbial genomics of arsenic. FEMS Microbiol. Rev. 2016, 40, 299–322. [Google Scholar] [CrossRef]
- Chang, J.S.; Kim, Y.H.; Kim, K.W. The ars genotype characterization of arsenic-resistant bacteria from arsenic-contaminated gold-silver mines in the Republic of Korea. Appl. Microbiol. Biotechnol. 2008, 80, 155–165. [Google Scholar] [CrossRef] [PubMed]

| Metal Ions | mM | ± |
|---|---|---|
| As(III) | 1.90 | 0.10 |
| As(V) | 117 | 3.00 |
| Cd(II) | 0.90 | 0.10 |
| Co(II) | 2.00 | 0.50 |
| Co(III) | 2.75 | 0.25 |
| Cr(VI) | 0.25 | 0.01 |
| Cu(II) | 4.10 | 0.10 |
| Hg(II) | 0.02 | 0.00 |
| Ni(II) | 1.30 | 0.10 |
| V(V) | 128 | 2.00 |
| Organism | Strain | Origin | Genome Size (Mb) | CG% | Gene | Protein | BioProject |
|---|---|---|---|---|---|---|---|
| Geobacillus stearothermophilus | 10 | Yellowstone thermal spring | 3.67 | 52.61 | 3645 | 3312 | PRJNA252389 |
| Geobacillus stearothermophilus | DSM458 | Austria sugar beet factory | 3.46 | 52.10 | 3683 | 3165 | PRJNA327158 |
| Geobacillus stearothermophilus | B5 | China rice stack | 3.39 | 52.50 | 3426 | 3045 | PRJNA513473 |
| Geobacillus stearothermophilus 10 (PRJNA252389) | Geobacillus stearothermophilus DSM458 (PRJNA327158) | Geobacillus stearothermophilus B5 (PRJNA513473) | |
|---|---|---|---|
| Putative Proteins | Locus | Locus | Locus |
| ArsR | GT50_RS07590 | EPB69_RS07030 | |
| EPB69_RS15665 | |||
| EPB69_RS15730 | |||
| ArsB | GT50_RS07510 | GS458_RS16835 | EPB69_RS15660 |
| ArsC | GT50_RS07505 | GS458_RS16830 | EPB69_RS15655 |
| GT50_RS06280 | GS458_RS15800 | ||
| CadA | GT50_RS12470 | GS458_RS03700 | EPB69_RS03440 |
| CadC | GT50_RS12465 | GS458_RS03695 | EPB69_RS03435 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puopolo, R.; Gallo, G.; Mormone, A.; Limauro, D.; Contursi, P.; Piochi, M.; Bartolucci, S.; Fiorentino, G. Identification of a New Heavy-Metal-Resistant Strain of Geobacillus stearothermophilus Isolated from a Hydrothermally Active Volcanic Area in Southern Italy. Int. J. Environ. Res. Public Health 2020, 17, 2678. https://doi.org/10.3390/ijerph17082678
Puopolo R, Gallo G, Mormone A, Limauro D, Contursi P, Piochi M, Bartolucci S, Fiorentino G. Identification of a New Heavy-Metal-Resistant Strain of Geobacillus stearothermophilus Isolated from a Hydrothermally Active Volcanic Area in Southern Italy. International Journal of Environmental Research and Public Health. 2020; 17(8):2678. https://doi.org/10.3390/ijerph17082678
Chicago/Turabian StylePuopolo, Rosanna, Giovanni Gallo, Angela Mormone, Danila Limauro, Patrizia Contursi, Monica Piochi, Simonetta Bartolucci, and Gabriella Fiorentino. 2020. "Identification of a New Heavy-Metal-Resistant Strain of Geobacillus stearothermophilus Isolated from a Hydrothermally Active Volcanic Area in Southern Italy" International Journal of Environmental Research and Public Health 17, no. 8: 2678. https://doi.org/10.3390/ijerph17082678
APA StylePuopolo, R., Gallo, G., Mormone, A., Limauro, D., Contursi, P., Piochi, M., Bartolucci, S., & Fiorentino, G. (2020). Identification of a New Heavy-Metal-Resistant Strain of Geobacillus stearothermophilus Isolated from a Hydrothermally Active Volcanic Area in Southern Italy. International Journal of Environmental Research and Public Health, 17(8), 2678. https://doi.org/10.3390/ijerph17082678








