Molecular Probes to Evaluate the Synthesis and Production Potential of an Odorous Compound (2-methylisoborneol) in Cyanobacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Cyanobacterial Strains and Field Samples
2.2. Genomic DNA and RNA Extraction
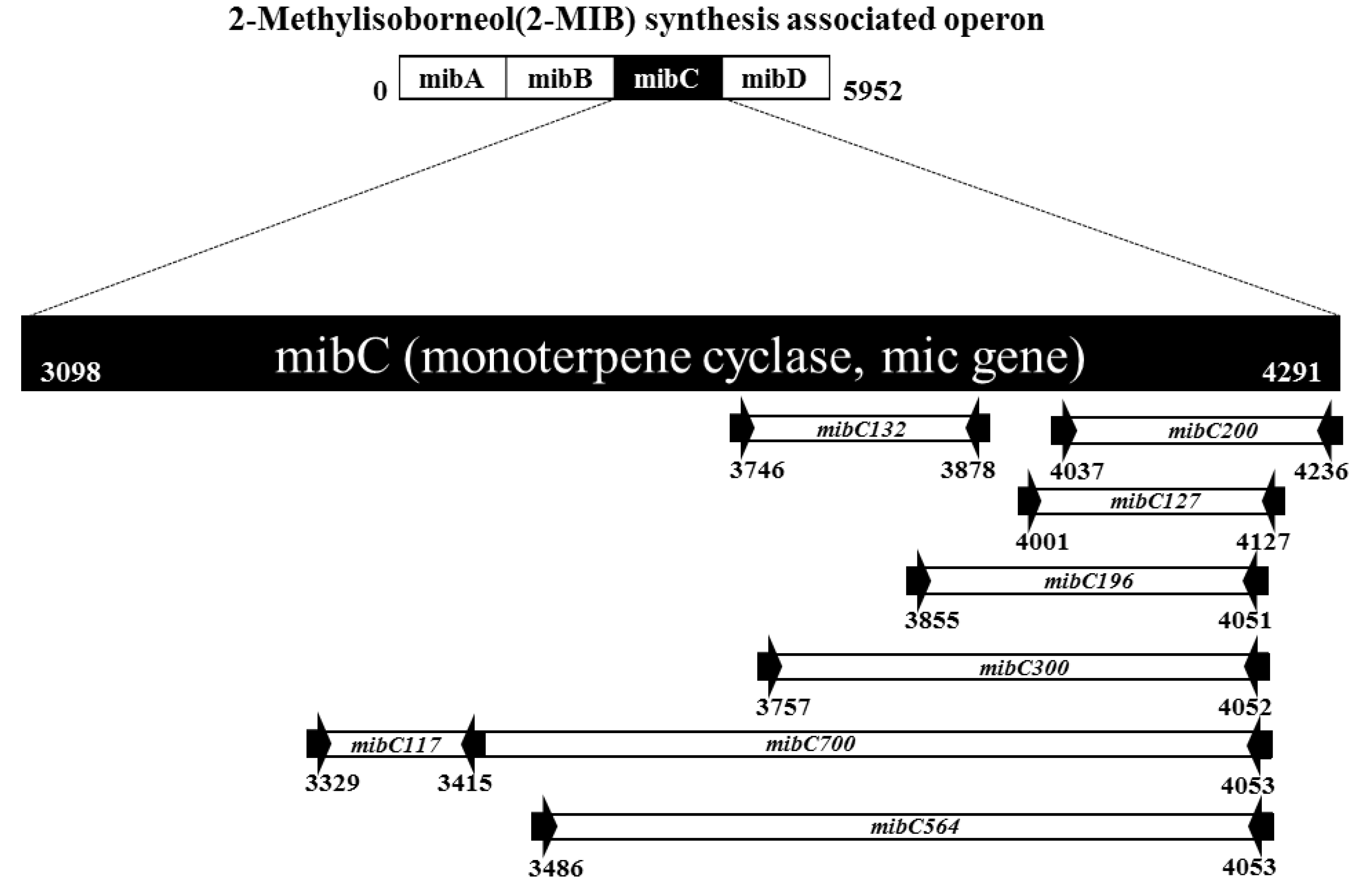
2.3. Sequence Analysis of mibC Genes and Primer Design
2.4. Detection of mibC and Field Application
2.5. Sequence Analysis of mibC
2.6. Phylogenetic Analysis
3. Results
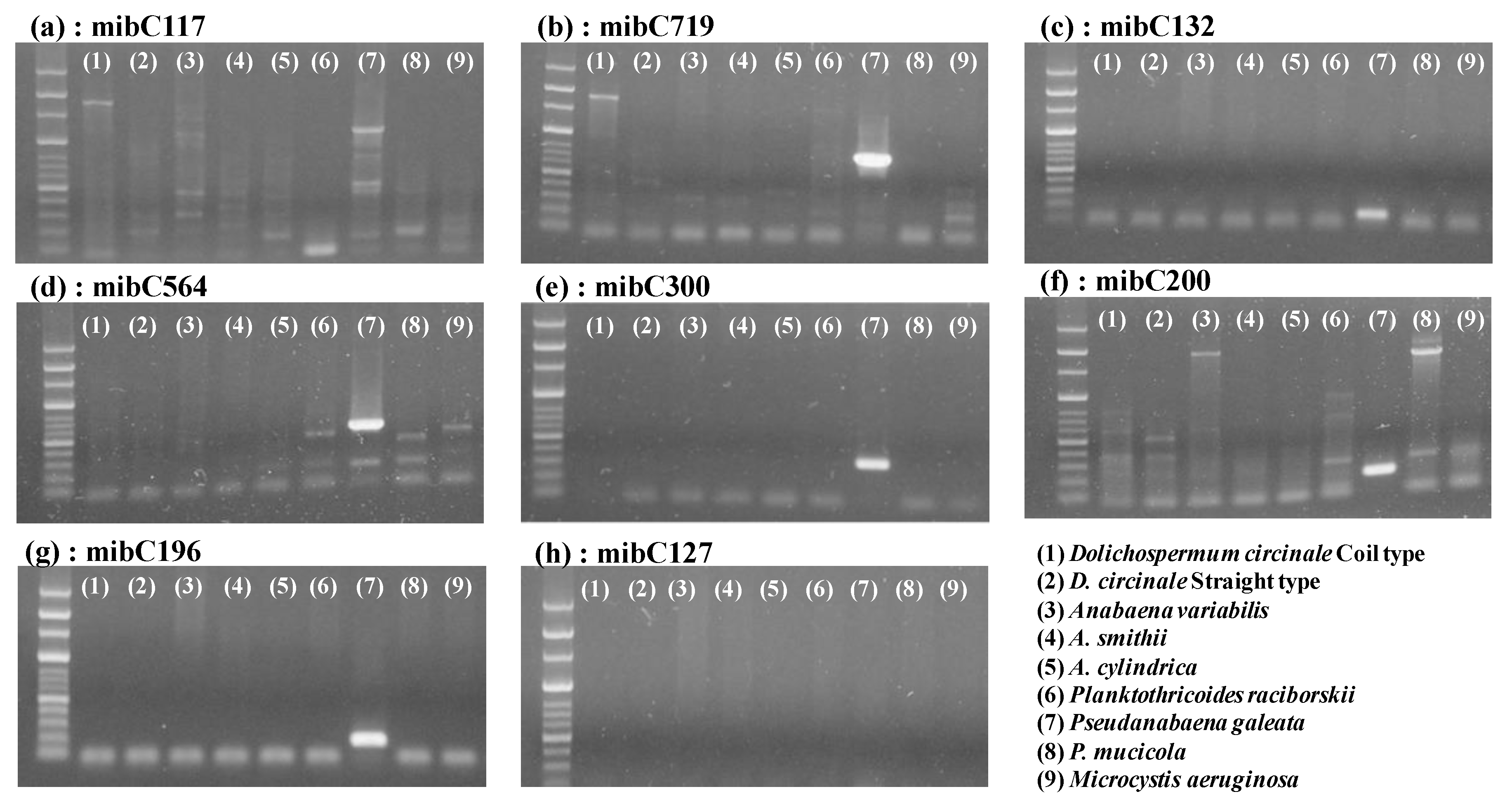
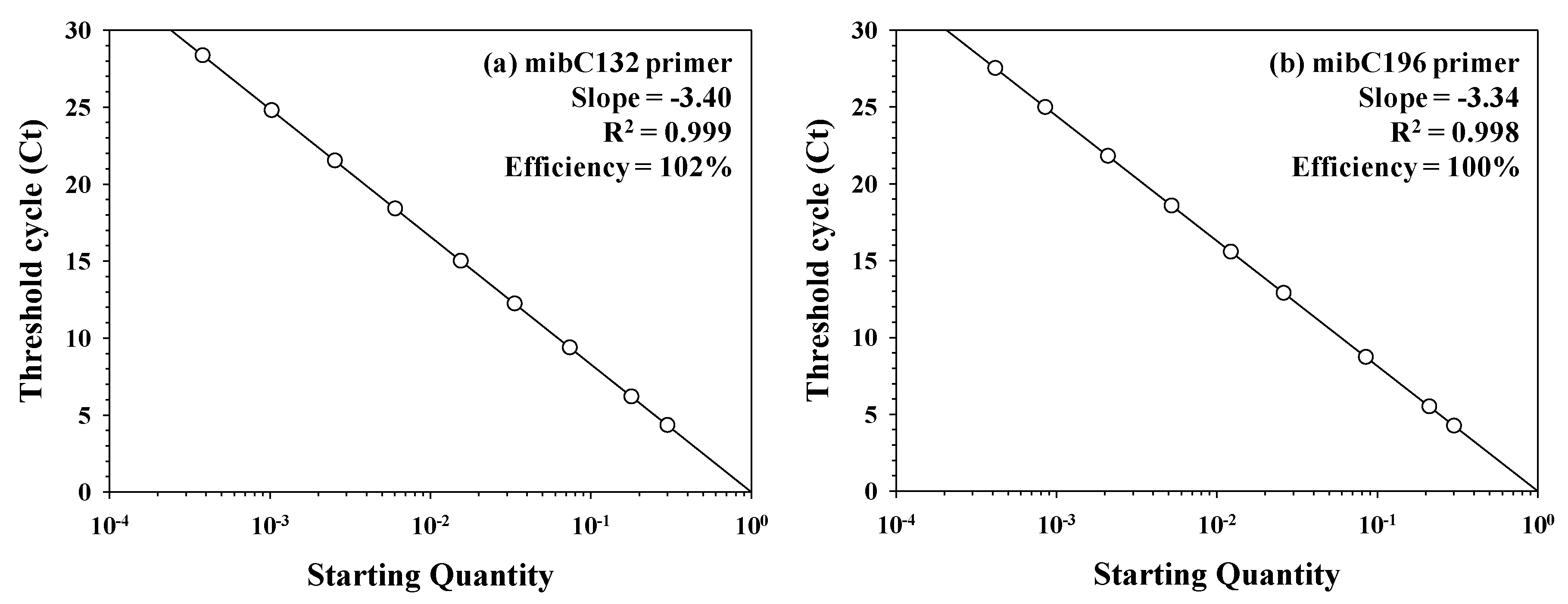
3.1. Applicability of the Designed Primers
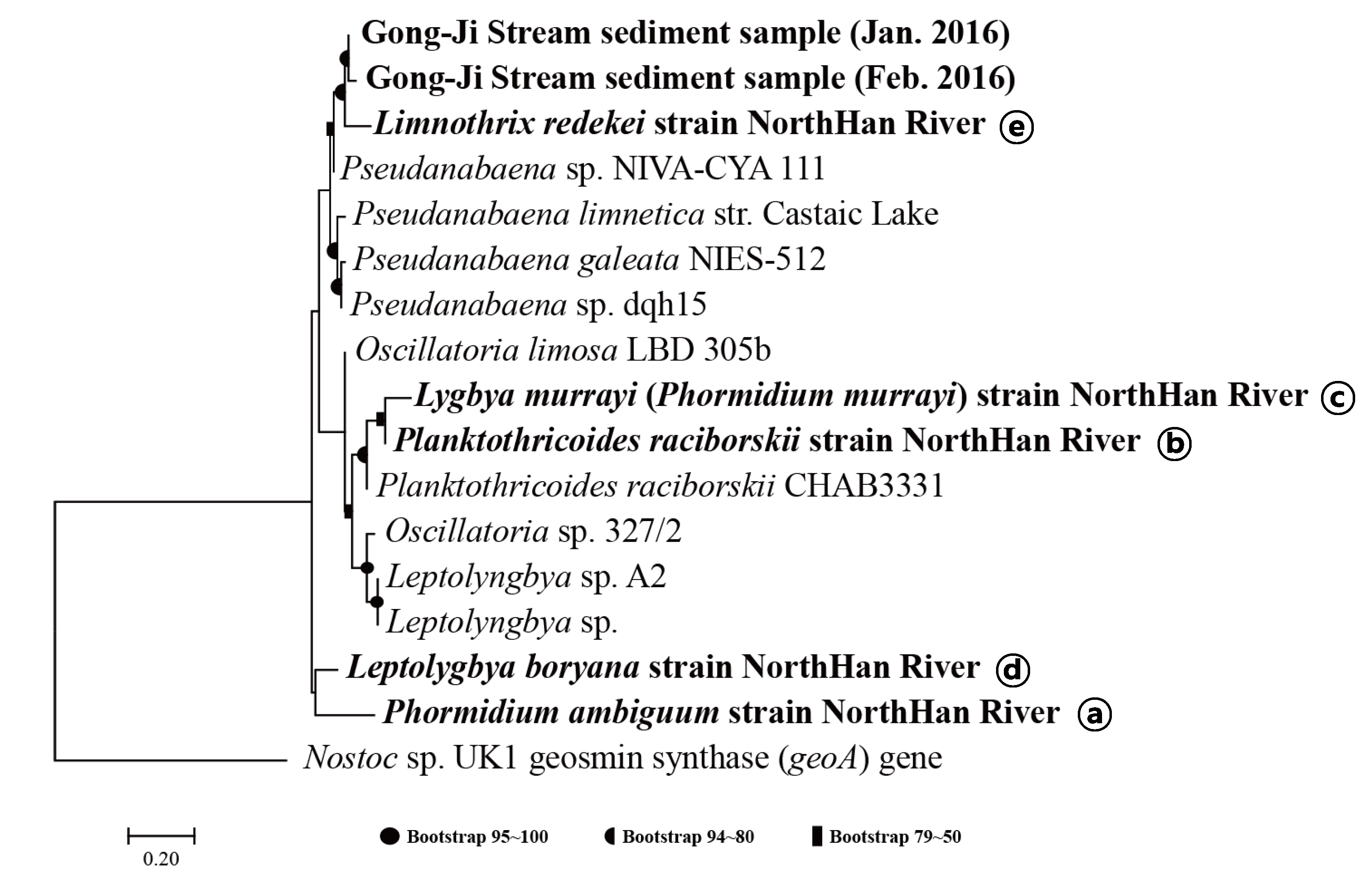
3.2. Phylogenic Analysis of Primer Amplicon Sequences
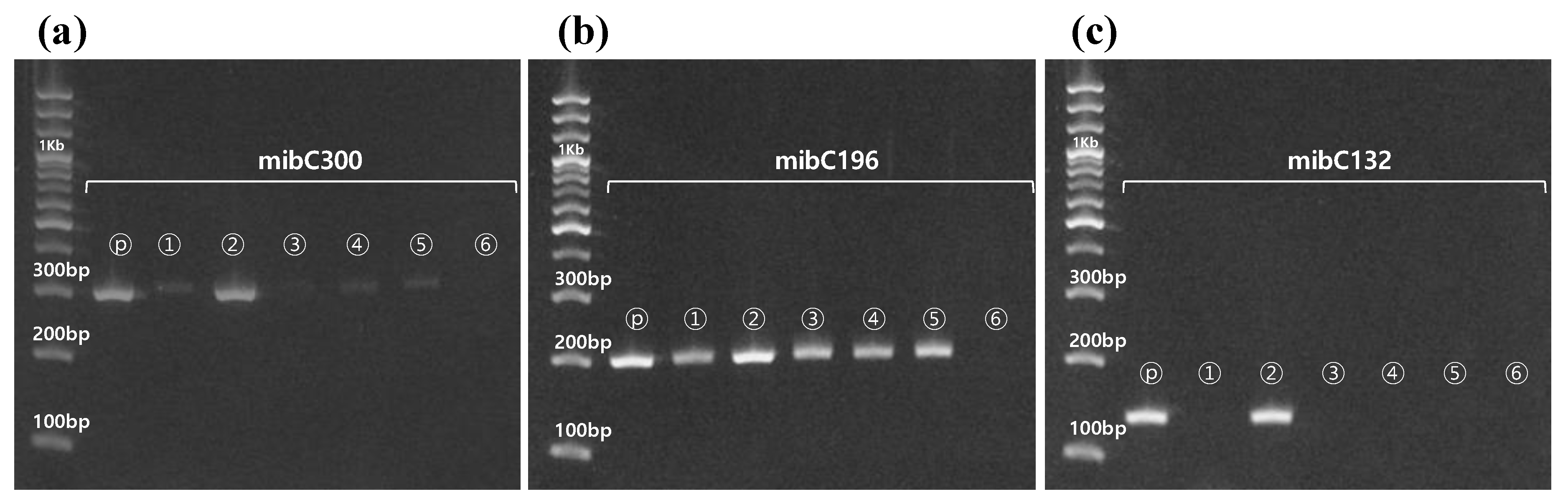
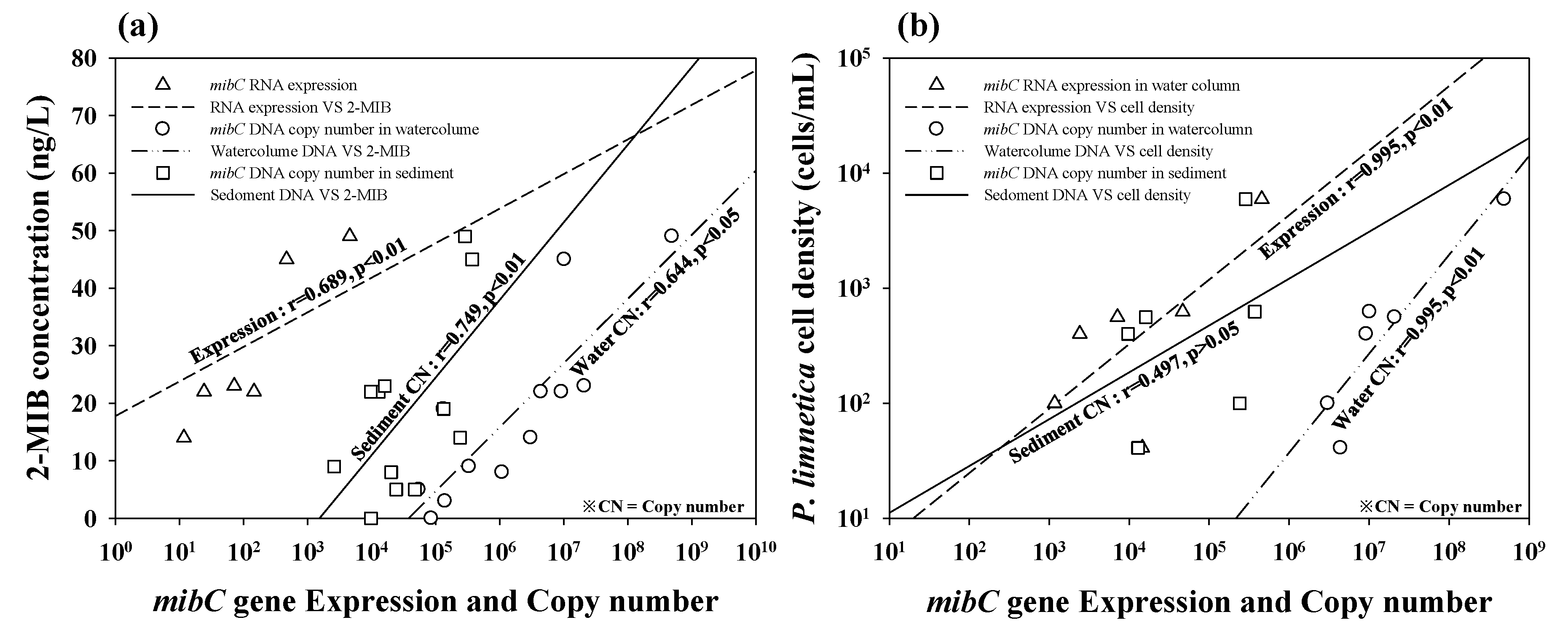
3.3. Quantitative Analysis of In Vivo and In Vitro Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jüttner, F.; Watson, S.B. Biochemical and ecological control of geosmin and 2-methylisoborneol in source waters. Appl. Environ. Microbiol. 2007, 73, 4395–4406. [Google Scholar] [CrossRef]
- Suffet, I.M.; Khiari, D.; Bruchet, A. The drinking water taste and odor wheel for the millennium: Beyond geosmin and 2-methylisoborneol. Water Sci. Technol. 1999, 40, 1–13. [Google Scholar] [CrossRef]
- Srinivasan, R.; Sorial, G.A. Treatment of taste and odor causing compounds 2-methyl isoborneol and geosmin in drinking water: A critical review. J. Environ. Sci. 2011, 23, 1–13. [Google Scholar] [CrossRef]
- Worley, J.L.; Dietrich, A.M.; Hoehn, R.C. Dechlorination techniques to improve sensory odor testing of geosmin and 2-MIB. Am. Water Work. Assoc. J. 2003, 95, 109. [Google Scholar] [CrossRef]
- Oikawa, T.; Tsunoda, T.; Nakahigashi, H.; Shimoriku, M.; Kanami, T.; Kimura, S. Musty odor producing benthic cyanobacteria in the Tama River (Japan) and identification of species by genetic analysis. J. Water Supply Res. Technol. -Aqua 2015, 64, 839–846. [Google Scholar] [CrossRef]
- Su, M.; Yu, J.; Zhang, J.; Chen, H.; An, W.; Vogt, R.D.; Andersen, T.; Jia, D.; Wang, J.; Yang, M. MIB-producing cyanobacteria (Planktothrix sp.) in a drinking water reservoir: Distribution and odor producing potential. Water Res. 2015, 68, 444–453. [Google Scholar] [CrossRef]
- Wang, R.; Li, D.; Jin, C.X.; Yang, B.W. Seasonal occurrence and species specificity of fishy and musty odor in Huajiang Reservoir in winter, China. Water Resour. Ind. 2015, 11, 13–26. [Google Scholar] [CrossRef]
- HRWEMD. Investigation of Causes of Off-Flavor Material Production by Harmful Algae and Management Strategy; Final Report; HEWEMD: Hanam, Korea, 2016. [Google Scholar]
- Choi, B.-G. Effects of Meteoro-Hydrological Fluctuation on Water Quality Change and Occurrence of Cyanobacterial Off-Flavors in Euiam Reservoir; Konkuk University: Seoul, Korea, 2017. [Google Scholar]
- Backer, L.; McGillicuddy, D. Harmful algal blooms. Oceanography 2006, 19, 94–106. [Google Scholar] [CrossRef]
- Watson, S.B. Cyanobacterial and eukaryotic algal odour compounds: Signals or by-products? A review of their biological activity. Phycologia 2003, 42, 332–350. [Google Scholar] [CrossRef]
- Watson, S.B.; Monis, P.; Baker, P.; Giglio, S. Biochemistry and genetics of taste-and odor-producing cyanobacteria. Harmful Algae 2016, 54, 112–127. [Google Scholar] [CrossRef]
- Otten, T.G.; Graham, J.L.; Harris, T.D.; Dreher, T.W. Elucidation of Taste-and Odor-Producing Bacteria and Toxigenic Cyanobacteria in a Midwestern Drinking Water Supply Reservoir by Shotgun Metagenomic Analysis. Appl. Environ. Microbiol. 2016, 82, 5410–5420. [Google Scholar] [CrossRef]
- Wang, Z.; Song, G.; Shao, J.; Tan, W.; Li, Y.; Li, R. Establishment and field applications of real-time PCR methods for the quantification of potential MIB-producing cyanobacteria in aquatic systems. J. Appl. Phycol. 2016, 28, 325–333. [Google Scholar] [CrossRef]
- Jiang, J.; Saint, C.P.; Cane, D.E.; Monis, P.T. Isolation and characterization of the gene associated with geosmin production in cyanobacteria. Environ. Sci. Technol. 2008, 42, 8027–8032. [Google Scholar] [CrossRef]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 100, 1541–1546. [Google Scholar] [CrossRef]
- Zuo, Y.; Li, L.; Wu, Z.; Song, L. Isolation, identification and odour-producing abilities of geosmin/2-MIB in actinomycetes from sediments in Lake Lotus, China. J. Water Supply Res. Technol. -Aqua 2009, 58, 552–561. [Google Scholar] [CrossRef]
- Chiu, Y.-T.; Yen, H.-K.; Lin, T.-F. An alternative method to quantify 2-MIB producing cyanobacteria in drinking water reservoirs: Method development and field applications. Environ. Res. 2016, 151, 618–627. [Google Scholar] [CrossRef]
- Jørgensen, N.O.; Podduturi, R.; Burford, M.A. Relations between abundance of potential geosmin-and 2-MIB-producing organisms and concentrations of these compounds in water from three Australian reservoirs. J. Water Supply Res. Technol. -Aqua 2016, 65, 504–513. [Google Scholar]
- Kim, K.; Lim, B.-J.; You, K.-A.; Park, M.-H.; Park, J.H.; Kim, B.-H.; Hwang, S.-J. Identification and Analysis of Geosmin Production Potential of Anabaena stain Isolated from North Han River using Genetic Methods. Korean J. Ecol. Environ. 2014, 47, 342–349. [Google Scholar] [CrossRef]
- HRWEMD. Investigation of the Outbreak Causes and Management Measures of the Taste and Odor Compound (2-Methylisoborneol) in the North Han River Water System; 1st Year Final Report; HEWEMD: Hanam, Korea, 2019. [Google Scholar]
- Allen, M.M. Simple conditions for growth of unicellular blue-green algae on plates1, 2. J. Phycol. 1968, 4, 1–4. [Google Scholar] [CrossRef]
- Lazarevic, V.; Gaïa, N.; Girard, M.; Francois, P.; Schrenzel, J. Comparison of DNA extraction methods in analysis of salivary bacterial communities. PLoS ONE 2013, 8, e67699. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- IDT. IDT’s PrimerQuest Tool, 2.2.3; Integrated DNA Technology: Coralville, IA, USA, 2012. [Google Scholar]
- Tuji, A.; Niiyama, Y. The identity and phylogeny of Pseudanabaena strain, NIES-512, producing 2-methylisoborneol (2-MIB). Bull. Natl. Mus. Nat. Sci. 2016, 42, 83–89. [Google Scholar]
- Iwase, S.; Abe, T. Identification and change in concentration of musty-odor compounds during growth in blue-green algae. Grad. Sch. Oceanogr. Tokai Univ. Sea-Nat. Cult. 2010, 8, 27–33. [Google Scholar]
- Sakai, M.; Hosoda, A.; Kanazawa, S. Isolation and characterization of two cyanobacteria-lytic bacterial strains from the submerged paddy soil. Soil Microorg. 2003, 57, 21–28. [Google Scholar]
- Matsushita, T.; Kobayashi, Y.; Nakamura, K.; Matsui, Y.; Ohno, K. Effect of transmembrane pressure on geosmin release from cyanobacterial cell during microfiltration. Water Pract. Technol. 2010, 5, wpt2010049. [Google Scholar] [CrossRef]
- Choi, G.G.; Yoon, S.K.; Kim, H.S.; Ahn, C.Y.; Oh, H.M. Morphological and Molecular Analyses of Anabaena variabilis and Trichormus variabilis (Cyanobacteria) from Korea. Korean J. Environ. Biol. 2012, 30, 54–63. [Google Scholar]
- Kyung Hee, O. Screening of Conserved DNA Sequences in Microcystin-Producing Cyanobacteria and Application of the Sequences to Monitor Microcystins in Water Reservoirs; Seoul National University of Graduate School: Seoul, Korea, 2009. [Google Scholar]
- Vaitomaa, J.; Rantala, A.; Halinen, K.; Rouhiainen, L.; Tallberg, P.; Mokelke, L.; Sivonen, K. Quantitative real-time PCR for determination of microcystin synthetase E copy numbers for Microcystis and Anabaena in lakes. Appl. Environ. Microbiol. 2003, 69, 7289–7297. [Google Scholar] [CrossRef]
- Ministry-of-Environment. Standard Methods for the Examination of Drinking Water Surveillance; Ministry-of-Environment: Sejong, Korea, 2016.
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Suda, S.; Watanabe, M.M.; Otsuka, S.; Mahakahant, A.; Yongmanitchai, W.; Nopartnaraporn, N.; Liu, Y.; Day, J.G. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int. J. Syst. Evol. Microbiol. 2002, 52, 1577–1595. [Google Scholar]
- PrimierBiosoft. PCR Primer Design Guidelines. Available online: http://www.premierbiosoft.com/ (accessed on 29 December 2019).
- Rychlik, W.; Spencer, W.; Rhoads, R. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990, 18, 6409–6412. [Google Scholar] [CrossRef]
- John, M.W.; Rapley, R. Molecular Biomethods Handbook, 2nd ed.; Humana Press: Totowa, NJ, USA, 2008. [Google Scholar]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.; Smith, J.A.; Struhl, K. Short Protocols in Molecular Biology; Jordan University of Science and Technology: Amman, Jordan, 1992. [Google Scholar]
- Zimba, P.V.; Dionigi, C.P.; Millie, D.F. Evaluating the relationship between photopigment synthesis and 2-methylisoborneol accumulation in cyanobacteria. J. Phycol. 1999, 35, 1422–1429. [Google Scholar] [CrossRef]
- Biosystems, A. Real-Time PCR Handbook; Life Thecnologies: Carlsbad, CA, USA, 2015. [Google Scholar]
- Debode, F.; Marien, A.; Janssen, É.; Bragard, C.; Berben, G. The influence of amplicon length on real-time PCR results. Base 2017, 21. [Google Scholar] [CrossRef]
- Komatsu, M.; Tsuda, M.; Ōmura, S.; Oikawa, H.; Ikeda, H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc. Natl. Acad. Sci. USA 2008, 105, 7422–7427. [Google Scholar] [CrossRef]
- Giglio, S.; Chou, W.; Ikeda, H.; Cane, D.; Monis, P. Biosynthesis of 2-methylisoborneol in cyanobacteria. Environ. Sci. Technol. 2010, 45, 992–998. [Google Scholar] [CrossRef]
- Byun, J.-H.; Hwang, S.-J.; Kim, B.-H.; Park, J.-R.; Lee, J.-K.; Lim, B.-J. Relationship between a Dense Population of Cyanobacteria and Odorous Compounds in the North Han River System in 2014 and 2015. Korean J. Ecol. Environ. 2015, 48, 263–271. [Google Scholar] [CrossRef]
| No. | Name | Direction | Length (mer) | Sequence (5′ → 3′) | GC Contents (%) | Purpose | Amplicon Size (bp) | Melting Temp. |
|---|---|---|---|---|---|---|---|---|
| 1 | mibC117 | Forward | 19 | CCG AGC GAT TCA ACG AGC C | 63.2 | Real-time PCR | 117 | 60 °C |
| Reverse | 22 | CCT CGT CCT CAT CAA ACC CGC A | 59.1 | |||||
| 2 | mibC127 | Forward | 20 | CCC CTG TTA CGC CAC CTT CT | 60.0 | Real-time PCR | 127 | 60 °C |
| Reverse | 24 | TCA TGG AGG TGT AGA AGC TGT CGT | 50.0 | |||||
| 3 | mibC132 | Forward | 22 | CGY ACC TGT TAC GCC ACC TTC T | 56.8 | Real-time PCR | 132 | 60 °C |
| Reverse | 24 | TCA TGG AGG TGT AGA AGC TGT CGT | 50.0 | |||||
| 4 | mibC196 | Forward | 24 | ACG ACA GCT TCT ACA CCT CCA TGA | 50.0 | Real-time PCR Screening | 196 | 64 °C |
| Reverse | 26 | AAT CTG TAG CAC CAT GTT GAC WGG TG | 46.2 | |||||
| 5 | mibC200 | Forward | 24 | ACA TGG TGC TAC AGA TTG CGG CGG | 58.3 | Real-time PCR Screening | 200 | 64 °C |
| Reverse | 24 | GCT GTT ATG CCA TTC AAA TGC GCC | 50.0 | |||||
| 6 | mibC300 | Forward | 23 | TGT TAC GCC ACC TTC TCT ATG TT | 43.5 | Screening | 300 | 64 °C |
| Reverse | 21 | CAA TCT GTA GCA CCA TGT TGA | 42.9 | |||||
| 7 | mibC564 | Forward | 22 | TCG GCT GTT GAT YGG KGC CAA G | 59.1 | Screening | 564 | 64 °C |
| Reverse | 25 | AAT CTG TAG CAC CAT GTT GAC WGG T | 44.0 | |||||
| 8 | mibC719 | Forward | 19 | CCG AGC GAT TCA ACG AGC C | 63.2 | Screening | 719 | 60 °C |
| Reverse | 25 | AAT CTG TAG CAC CAT GTT GAC WGG T | 44.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Yoon, Y.; Cho, H.; Hwang, S.-J. Molecular Probes to Evaluate the Synthesis and Production Potential of an Odorous Compound (2-methylisoborneol) in Cyanobacteria. Int. J. Environ. Res. Public Health 2020, 17, 1933. https://doi.org/10.3390/ijerph17061933
Kim K, Yoon Y, Cho H, Hwang S-J. Molecular Probes to Evaluate the Synthesis and Production Potential of an Odorous Compound (2-methylisoborneol) in Cyanobacteria. International Journal of Environmental Research and Public Health. 2020; 17(6):1933. https://doi.org/10.3390/ijerph17061933
Chicago/Turabian StyleKim, Keonhee, Youngdae Yoon, Hyukjin Cho, and Soon-Jin Hwang. 2020. "Molecular Probes to Evaluate the Synthesis and Production Potential of an Odorous Compound (2-methylisoborneol) in Cyanobacteria" International Journal of Environmental Research and Public Health 17, no. 6: 1933. https://doi.org/10.3390/ijerph17061933
APA StyleKim, K., Yoon, Y., Cho, H., & Hwang, S.-J. (2020). Molecular Probes to Evaluate the Synthesis and Production Potential of an Odorous Compound (2-methylisoborneol) in Cyanobacteria. International Journal of Environmental Research and Public Health, 17(6), 1933. https://doi.org/10.3390/ijerph17061933






