Prenatal Multivitamin Use and MTHFR Genotype Are Associated with Newborn Cord Blood DNA Methylation
Abstract
:1. Introduction
2. Methods
2.1. Study Sample
2.2. Exposure Assessment
2.3. Genetic Measures
2.4. DNA Methylation Measures
2.5. DNA Methylation Data Processing
2.6. Study Sample Descriptive Statistics
2.7. Single Site DNA Methylation Analysis
2.8. Global Methylation Analysis
2.9. Tissue Specificity
3. Results
3.1. Study Sample Characteristics
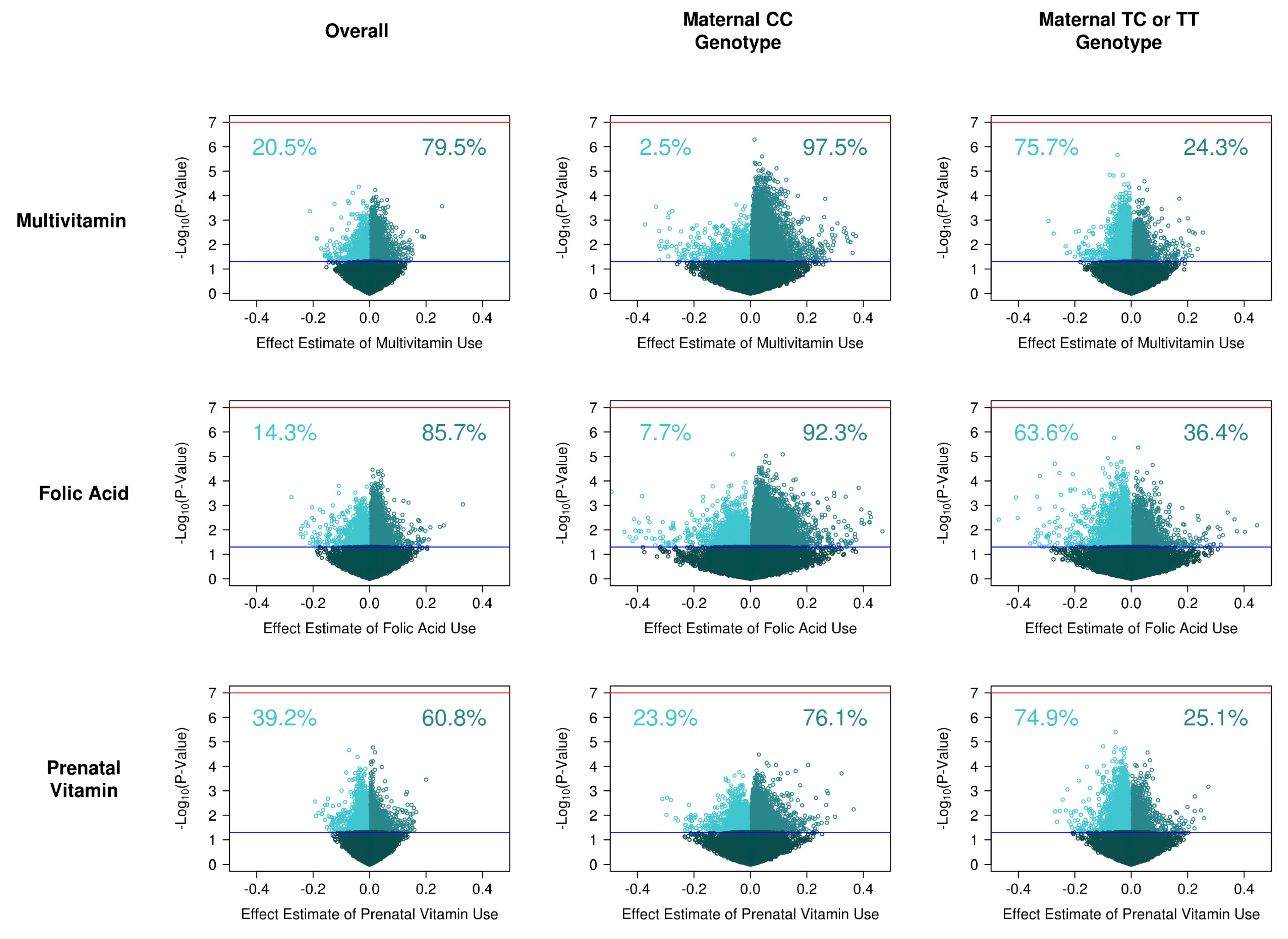
3.2. Supplement Intake on Single Site DNA Methylation
3.3. MTHFR and Single Site DNA Methylation
3.4. Supplement Intake on Single Site DNA Methylation, Stratified by Genotype
3.5. Pathway Analysis
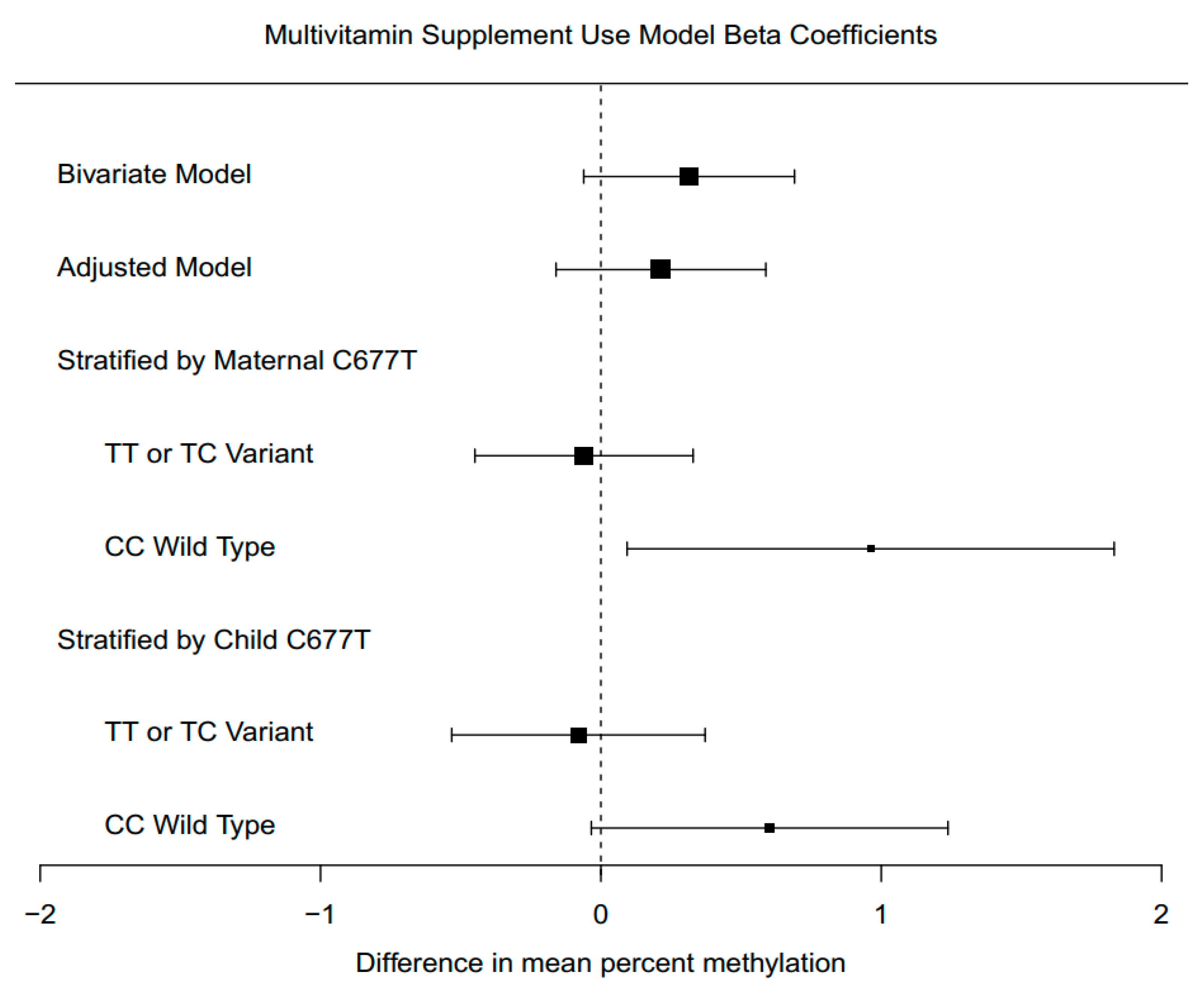
3.6. Supplement Intake Associations with Global DNA Methylation
3.7. Supplement Intake Associations with Global DNA Methylation, Stratified by Genotype
3.8. Tissue Specificity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Acronyms
References
- Institute of Medicine Committee on Nutritional Status During Pregnancy and Lactation. Nutrition During Pregnancy: Part I Weight Gain; Part II Nutrient Supplements; National Academies Press: Washington, DC, USA, 1990. [Google Scholar]
- Aronsson, C.A.; Vehik, K.; Yang, J.; Uusitalo, U.; Hay, K.; Joslowski, G.; Riikonen, A.; Ballard, L.; Virtanen, S.M.; Norris, J.M.; et al. Use of dietary supplements in pregnant women in relation to sociodemographic factors—A report from The Environmental Determinants of Diabetes in the Young (TEDDY) study. Public Health Nutr. 2013, 16, 1390–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, L.; Lee, A.H.; Yau, K.K.W.; Hui, Y.V.; Binns, C.W. Consumption of dietary supplements by Chinese women during pregnancy and postpartum: A prospective cohort study. Matern. Child Nutr. 2017, 13. [Google Scholar] [CrossRef] [Green Version]
- Ray, J.G.; Singh, G.; Burrows, R.F. Evidence for suboptimal use of periconceptional folic acid supplements globally. BJOG 2004, 111, 399–408. [Google Scholar] [CrossRef] [PubMed]
- MRC Vitamin Study Research Group. Prevention of neural tube defects: Results of the Medical Research Council Vitamin Study. Lancet 1991, 338, 131–137. [Google Scholar] [CrossRef]
- Roza, S.J.; van Batenburg-Eddes, T.; Steegers, E.A.; Jaddoe, V.W.; Mackenbach, J.P.; Hofman, A.; Verhulst, F.C.; Tiemeier, H. Maternal folic acid supplement use in early pregnancy and child behavioural problems: The Generation R Study. Br. J. Nutr. 2010, 103, 445–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julvez, J.; Fortuny, J.; Mendez, M.; Torrent, M.; Ribas-Fitó, N.; Sunyer, J. Maternal use of folic acid supplements during pregnancy and four-year-old neurodevelopment in a population-based birth cohort. Paediatr. Perinat. Epidemiol. 2009, 23, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Schlotz, W.; Jones, A.; Phillips, D.I.; Gale, C.R.; Robinson, S.M.; Godfrey, K.M. Lower maternal folate status in early pregnancy is associated with childhood hyperactivity and peer problems in offspring. J Child. Psychol. Psychiatry 2010, 51, 594–602. [Google Scholar] [CrossRef] [Green Version]
- Roth, C.; Magnus, P.; Schjølberg, S.; Stoltenberg, C.; Surén, P.; McKeague, I.W.; Davey Smith, G.; Reichborn-Kjennerud, T.; Susser, E. Folic acid supplements in pregnancy and severe language delay in children. JAMA 2011, 306, 1566–1573. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, R.J.; Iosif, A.M.; Guerrero Angel, E.; Ozonoff, S. Association of Maternal Prenatal Vitamin Use With Risk for Autism Spectrum Disorder Recurrence in Young Siblings. JAMA Psychiatry 2019. [Google Scholar] [CrossRef]
- Ulrich, C.M.; Kampman, E.; Bigler, J.; Schwartz, S.M.; Chen, C.; Bostick, R.; Fosdick, L.; Beresford, S.A.; Yasui, Y.; Potter, J.D. Lack of association between the C677T MTHFR polymorphism and colorectal hyperplastic polyps. Cancer Epidemiol. Biomark. Prev. 2000, 9, 427–433. [Google Scholar]
- Hiraoka, M.; Kagawa, Y. Genetic polymorphisms and folate status. Congenit. Anom. (Kyoto) 2017, 57, 142–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Yan, J.; West, A.A.; Perry, C.A.; Malysheva, O.V.; Devapatla, S.; Pressman, E.; Vermeylen, F.; Caudill, M.A. Maternal choline intake alters the epigenetic state of fetal cortisol-regulating genes in humans. FASEB J. 2012, 26, 3563–3574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauwels, S.; Ghosh, M.; Duca, R.C.; Bekaert, B.; Freson, K.; Huybrechts, I.; Langie, S.A.S.; Koppen, G.; Devlieger, R.; Godderis, L. Dietary and supplemental maternal methyl-group donor intake and cord blood DNA methylation. Epigenetics 2017, 12, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joubert, B.R.; den Dekker, H.T.; Felix, J.F.; Bohlin, J.; Ligthart, S.; Beckett, E.; Tiemeier, H.; van Meurs, J.B.; Uitterlinden, A.G.; Hofman, A.; et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat. Commun. 2016, 7, 10577. [Google Scholar] [CrossRef] [Green Version]
- Newschaffer, C.J.; Croen, L.A.; Fallin, M.D.; Hertz-Picciotto, I.; Nguyen, D.V.; Lee, N.L.; Berry, C.A.; Farzadegan, H.; Hess, H.N.; Landa, R.J.; et al. Infant siblings and the investigation of autism risk factors. J. Neurodev. Disord. 2012, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Abecasis, G.R.; Auton, A.; Brooks, L.D.; DePristo, M.A.; Durbin, R.M.; Handsaker, R.E.; Kang, H.M.; Marth, G.T.; McVean, G.A.; Consortium, G.P. An integrated map of genetic variation from 1092 human genomes. Nature 2012, 491, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Delaneau, O.; Zagury, J.F.; Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 2013, 10, 5–6. [Google Scholar] [CrossRef]
- Howie, B.; Fuchsberger, C.; Stephens, M.; Marchini, J.; Abecasis, G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012, 44, 955–959. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [Green Version]
- Triche, T.J., Jr.; Weisenberger, D.J.; Van Den Berg, D.; Laird, P.W.; Siegmund, K.D. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013, 41, e90. [Google Scholar] [CrossRef] [Green Version]
- Bakulski, K.M.; Feinberg, J.I.; Andrews, S.V.; Yang, J.; Brown, S.; McKenney, S.L.; Witter, F.; Walston, J.; Feinberg, A.P.; Fallin, M.D. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics 2016, 11, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.A.; Lemire, M.; Choufani, S.; Butcher, D.T.; Grafodatskaya, D.; Zanke, B.W.; Gallinger, S.; Hudson, T.J.; Weksberg, R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013, 8, 203–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Phipson, B.; Maksimovic, J.; Oshlack, A. missMethyl: An R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics 2016, 32, 286–288. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Zhao, N.; Bell, D.A.; Maity, A.; Staicu, A.M.; Joubert, B.R.; London, S.J.; Wu, M.C. Global analysis of methylation profiles from high resolution CpG data. Genet. Epidemiol. 2015, 39, 53–64. [Google Scholar] [CrossRef] [Green Version]
- James, P.; Sajjadi, S.; Tomar, A.S.; Saffari, A.; Fall, C.H.D.; Prentice, A.M.; Shrestha, S.; Issarapu, P.; Yadav, D.K.; Kaur, L.; et al. Candidate genes linking maternal nutrient exposure to offspring health via DNA methylation: A review of existing evidence in humans with specific focus on one-carbon metabolism. Int. J. Epidemiol. 2018, 47, 1910–1937. [Google Scholar] [CrossRef]
- Richmond, R.C.; Joubert, B.R. Contrasting the effects of intra-uterine smoking and one-carbon micronutrient exposures on offspring DNA methylation. Epigenomics 2017, 9, 351–367. [Google Scholar] [CrossRef] [Green Version]
- Araya, C.; Carrasco, D. Mechanisms of vertebrate neural plate internalization. Int. J. Dev. Biol. 2020. [Google Scholar] [CrossRef]
- Rolo, A.; Savery, D.; Escuin, S.; de Castro, S.C.; Armer, H.E.; Munro, P.M.; Molè, M.A.; Greene, N.D.; Copp, A.J. Regulation of cell protrusions by small GTPases during fusion of the neural folds. Elife 2016, 5, e13273. [Google Scholar] [CrossRef]
- Boeke, C.E.; Baccarelli, A.; Kleinman, K.P.; Burris, H.H.; Litonjua, A.A.; Rifas-Shiman, S.L.; Tarantini, L.; Gillman, M. Gestational intake of methyl donors and global LINE-1 DNA methylation in maternal and cord blood: Prospective results from a folate-replete population. Epigenetics 2012, 7, 253–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andraos, S.; de Seymour, J.V.; O’Sullivan, J.M.; Kussmann, M. The Impact of Nutritional Interventions in Pregnant Women on DNA Methylation Patterns of the Offspring: A Systematic Review. Mol. Nutr. Food Res. 2018, 62, e1800034. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, S.; Duca, R.C.; Devlieger, R.; Freson, K.; Straetmans, D.; Van Herck, E.; Huybrechts, I.; Koppen, G.; Godderis, L. Maternal Methyl-Group Donor Intake and Global DNA (Hydroxy)Methylation before and during Pregnancy. Nutrients 2016, 8, 474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ly, A.; Ishiguro, L.; Kim, D.; Im, D.; Kim, S.E.; Sohn, K.J.; Croxford, R.; Kim, Y.I. Maternal folic acid supplementation modulates DNA methylation and gene expression in the rat offspring in a gestation period-dependent and organ-specific manner. J. Nutr. Biochem. 2016, 33, 103–110. [Google Scholar] [CrossRef]
- Friso, S.; Choi, S.W.; Girelli, D.; Mason, J.B.; Dolnikowski, G.G.; Bagley, P.J.; Olivieri, O.; Jacques, P.F.; Rosenberg, I.H.; Corrocher, R.; et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl. Acad. Sci. USA 2002, 99, 5606–5611. [Google Scholar] [CrossRef] [Green Version]
- Axume, J.; Smith, S.S.; Pogribny, I.P.; Moriarty, D.J.; Caudill, M.A. The MTHFR 677TT genotype and folate intake interact to lower global leukocyte DNA methylation in young Mexican American women. Nutr. Res. 2007, 27, 1365-1317. [Google Scholar] [CrossRef] [Green Version]
- Crider, K.S.; Quinlivan, E.P.; Berry, R.J.; Hao, L.; Li, Z.; Maneval, D.; Yang, T.P.; Rasmussen, S.A.; Yang, Q.; Zhu, J.H.; et al. Genomic DNA methylation changes in response to folic acid supplementation in a population-based intervention study among women of reproductive age. PLoS ONE 2011, 6, e28144. [Google Scholar] [CrossRef] [Green Version]
- Aarabi, M.; San Gabriel, M.C.; Chan, D.; Behan, N.A.; Caron, M.; Pastinen, T.; Bourque, G.; MacFarlane, A.J.; Zini, A.; Trasler, J. High-dose folic acid supplementation alters the human sperm methylome and is influenced by the MTHFR C677T polymorphism. Hum. Mol. Genet. 2015, 24, 6301–6313. [Google Scholar] [CrossRef] [Green Version]
- Sae-Lee, C.; Corsi, S.; Barrow, T.M.; Kuhnle, G.G.C.; Bollati, V.; Mathers, J.C.; Byun, H.M. Dietary Intervention Modifies DNA Methylation Age Assessed by the Epigenetic Clock. Mol. Nutr. Food Res. 2018, 62, e1800092. [Google Scholar] [CrossRef] [Green Version]
- Vryer, R.; Saffery, R. What’s in a name? Context-dependent significance of ’global’ methylation measures in human health and disease. Clin. Epigenetics 2017, 9, 2. [Google Scholar] [CrossRef] [Green Version]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, D.I.; Blair, J.D.; Lott, P.; Yu, H.O.; Hong, D.; Crary, F.; Ashwood, P.; Walker, C.; Korf, I.; Robinson, W.P.; et al. The human placenta methylome. Proc. Natl. Acad. Sci. USA 2013, 110, 6037–6042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardenas, A.; Rifas-Shiman, S.L.; Godderis, L.; Duca, R.C.; Navas-Acien, A.; Litonjua, A.A.; DeMeo, D.L.; Brennan, K.J.; Amarasiriwardena, C.J.; Hivert, M.F.; et al. Prenatal Exposure to Mercury: Associations with Global DNA Methylation and Hydroxymethylation in Cord Blood and in Childhood. Environ. Health Perspect 2017, 125, 087022. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Gong, Y.J.; Cao, W.C.; Wang, R.X.; Wang, Y.X.; Liu, C.; Chen, Y.J.; Huang, L.L.; Ai, S.H.; Lu, W.Q.; et al. Prenatal urinary polycyclic aromatic hydrocarbon metabolites, global DNA methylation in cord blood, and birth outcomes: A cohort study in China. Environ. Pollut 2018, 234, 396–405. [Google Scholar] [CrossRef]
- Plusquin, M.; Guida, F.; Polidoro, S.; Vermeulen, R.; Raaschou-Nielsen, O.; Campanella, G.; Hoek, G.; Kyrtopoulos, S.A.; Georgiadis, P.; Naccarati, A.; et al. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ. Int. 2017, 108, 127–136. [Google Scholar] [CrossRef]
- Levine, S.Z.; Kodesh, A.; Viktorin, A.; Smith, L.; Uher, R.; Reichenberg, A.; Sandin, S. Association of Maternal Use of Folic Acid and Multivitamin Supplements in the Periods Before and During Pregnancy With the Risk of Autism Spectrum Disorder in Offspring. JAMA Psychiatry 2018, 75, 176–184. [Google Scholar] [CrossRef] [Green Version]
| Study Sample (n = 130) | Multivitamin Use Preconception | p-Value | ||
|---|---|---|---|---|
| No (n = 86) | Yes (n = 44) | |||
| Child/Cord Blood Characteristics | ||||
| DNA Methylation (mean, sd) | ||||
| Overall | 50.80 (0.94) | 50.7 (1.0) | 51.0 (0.9) | 0.05 |
| Open Sea | 73.61 (1.30) | 73.4 (1.4) | 74.0 (1.1) | 0.02 |
| Shelf | 78.06 (1.40) | 77.9 (1.5) | 78.4 (1.2) | 0.018 |
| Shore | 46.76 (0.87) | 46.7 (0.8) | 47.0 (0.9) | 0.061 |
| Island | 19.40 (0.58) | 19.4 (0.6) | 19.4 (0.6) | 0.59 |
| Cell Type Percent (mean, sd) | ||||
| Nucleated red blood cells | 55 (42.3%) | 10.1 (5.3) | 9.14 (5.0) | 0.30 |
| Granulocytes | 75 (57.7%) | 43.5 (11.6) | 43.3 (13.8) | 0.96 |
| Methylation Measurement Round | 0.58 | |||
| 1 | 63 (48.5%) | 59 (68.6%) | 33 (75.0%) | |
| 2 | 67 (51.5%) | 27 (31.4%) | 11 (25.0%) | |
| Sex | 0.16 | |||
| Female | 64 (49.2%) | 38 (44.2%) | 26 (59.1%) | |
| Male | 66 (50.8%) | 48 (55.8%) | 18 (40.9%) | |
| Child MTHFR C677T Genotype | 0.76 | |||
| CC | 63 (48.5%) | 43 (50.0%) | 20 (45.5%) | |
| TT or TC | 67 (51.5%) | 43 (50.0%) | 24 (54.5%) | |
| Maternal Characteristics | ||||
| Age (years) | 33.62 (4.63) | 33.4 (4.4) | 34.0 (5.1) | 0.48 |
| Education | 0.66 | |||
| High School or Less | 26 (20.0%) | 19 (22.1%) | 7 (15.9%) | |
| Less Than 4 Years College | 18 (13.8%) | 10 (11.6%) | 8 (18.2%) | |
| 4 Years of College | 57 (43.8%) | 37 (43.0%) | 20 (45.5%) | |
| Post Graduate | 29 (22.3%) | 20 (23.3%) | 9 (20.5%) | |
| Race/Ethnicity | 0.99 | |||
| Non-Hispanic White | 66 (50.8%) | 43 (50.0%) | 23 (52.3%) | |
| Non-Hispanic Black | 12 (9.2%) | 8 (9.3%) | 4 (9.09%) | |
| Hispanic/Latino | 14 (10.8%) | 10 (11.6%) | 4 (9.09%) | |
| Other | 38 (29.2%) | 25 (29.1%) | 13 (29.5%) | |
| Folic Acid Use Preconception | 0.81 | |||
| No | 115 (88.5%) | 77 (89.5%) | 38 (86.4%) | |
| Yes | 15 (11.5%) | 9 (10.5%) | 6 (13.6%) | |
| Prenatal Vitamin Use Preconception | 0.001 | |||
| No | 79 (60.8%) | 43 (50.0%) | 36 (81.8%) | |
| Yes | 51 (39.2%) | 43 (50.0%) | 8 (18.2%) | |
| Maternal MTHFR C677T Genotype | 0.24 | |||
| CC | 55 (42.3%) | 40 (46.5%) | 15 (34.1%) | |
| TT or TC | 75 (57.7%) | 46 (53.5%) | 29 (65.9%) | |
| MTHFR C677T Allele Present in Child | MTHFR C677T Allele Present in Mother | |||||
|---|---|---|---|---|---|---|
| No (n = 63) | Yes (n = 67) | p-Value | No (n = 55) | Yes (n = 75) | p-Value | |
| Child/Cord Blood Characteristics | ||||||
| DNA Methylation (mean, sd) | ||||||
| All | 50.6 (1.04) | 50.9 (0.81) | 0.07 | 50.7 (1.14) | 50.9 (0.75) | 0.24 |
| Open Sea | 73.4 (1.52) | 73.8 (1.02) | 0.045 | 73.4 (1.67) | 73.8 (0.93) | 0.16 |
| Shelf | 77.8 (1.62) | 78.3 (1.12) | 0.038 | 77.9 (1.78) | 78.2 (1.03) | 0.17 |
| Shore | 46.6 (0.92) | 46.9 (0.80) | 0.124 | 46.7 (1.00) | 46.8 (0.75) | 0.46 |
| Island | 19.3 (0.54) | 19.4 (0.61) | 0.342 | 19.4 (0.58) | 19.4 (0.58) | 0.58 |
| Cell Type Percent (mean, sd) | ||||||
| nRBC | 9.84 (5.0) | 9.74 (5.4) | 0.91 | 10.1 (5.9) | 9.54 (4.6) | 0.54 |
| Granulocytes | 43.7 (12.8) | 43.2 (11.9) | 0.82 | 44.1 (12.5) | 42.9 (12.2) | 0.58 |
| DNA Methylation Round | 0.97 | 0.58 | ||||
| 1 | 44 (69.8%) | 48 (71.6%) | 37 (67.3%) | 55 (73.3%) | ||
| 2 | 19 (30.2%) | 19 (28.4%) | 18 (32.7%) | 20 (26.7%) | ||
| Child Sex | 0.87 | 0.39 | ||||
| Female | 32 (50.8%) | 32 (47.8%) | 30 (54.5%) | 34 (45.3%) | ||
| Male | 31 (49.2%) | 35 (52.2%) | 25 (45.5%) | 41 (54.7%) | ||
| Child MTHFR C677T | - | - | - | <0.001 | ||
| CC Genotype | 41 (74.5%) | 22 (29.3%) | ||||
| TT or TC Genotype | 14 (25.5%) | 53 (70.7%) | ||||
| Maternal Characteristics | ||||||
| Age (years) | 33.9 (4.50) | 33.4 (4.76) | 0.58 | 33.9 (4.3) | 33.4 (4.9) | 0.57 |
| Multivitamins | 0.63 | 0.19 | ||||
| None Pre- Conception | 43 (68.3%) | 42 (62.7%) | 40 (72.7%) | 45 (60.0%) | ||
| Used Pre- Conception | 20 (31.7%) | 25 (37.3%) | 15 (27.3%) | 30 (40.0%) | ||
| Folic Acid | 0.56 | 0.61 | ||||
| None Preconception | 55 (87.3%) | 55 (82.1%) | 45 (81.8%) | 65 (86.7%) | ||
| Used Preconception | 8 (12.7%) | 12 (17.9%) | 10 (18.2%) | 10 (13.3%) | ||
| Prenatal Vitamins | 0.41 | 0.37 | ||||
| None Preconception | 35 (55.6%) | 43 (64.2%) | 30 (54.5%) | 48 (64.0%) | ||
| Used Pre-conception | 28 (44.4%) | 24 (35.8%) | 25 (45.5%) | 27 (36.0%) | ||
| Education | 0.63 | 0.53 | ||||
| High school or less | 12 (19.0%) | 14 (20.9%) | 9 (16.4%) | 17 (22.7%) | ||
| <4 Yrs College | 9 (14.3%) | 9 (13.4%) | 10 (18.2%) | 8 (10.7%) | ||
| 4 Yrs of College | 25 (39.7%) | 32 (47.8%) | 25 (45.5%) | 32 (42.7%) | ||
| Post Graduate | 17 (27.0%) | 12 (17.9%) | 11 (20.0%) | 18 (24.0%) | ||
| Race/Ethnicity | 0.002 | 0.006 | ||||
| Non-Hispanic White | 26 (41.3%) | 40 (59.7%) | 22 (40.0%) | 44 (58.7%) | ||
| Non-Hispanic Black | 11 (17.5%) | 1 (1.49%) | 10 (18.2%) | 2 (2.67%) | ||
| Hispanic/Latino | 4 (6.35%) | 10 (14.9%) | 4 (7.27%) | 10 (13.3%) | ||
| Other | 22 (34.9%) | 16 (23.9%) | 19 (34.5%) | 19 (25.3%) | ||
| Maternal MTHFR C677T | <0.001 | - | - | - | ||
| CC Genotype | 41 (65.1%) | 14 (20.9%) | ||||
| TT or TC Genotype | 22 (34.9%) | 53 (79.1%) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakulski, K.M.; Dou, J.F.; Feinberg, J.I.; Brieger, K.K.; Croen, L.A.; Hertz-Picciotto, I.; Newschaffer, C.J.; Schmidt, R.J.; Fallin, M.D. Prenatal Multivitamin Use and MTHFR Genotype Are Associated with Newborn Cord Blood DNA Methylation. Int. J. Environ. Res. Public Health 2020, 17, 9190. https://doi.org/10.3390/ijerph17249190
Bakulski KM, Dou JF, Feinberg JI, Brieger KK, Croen LA, Hertz-Picciotto I, Newschaffer CJ, Schmidt RJ, Fallin MD. Prenatal Multivitamin Use and MTHFR Genotype Are Associated with Newborn Cord Blood DNA Methylation. International Journal of Environmental Research and Public Health. 2020; 17(24):9190. https://doi.org/10.3390/ijerph17249190
Chicago/Turabian StyleBakulski, Kelly M., John F. Dou, Jason I. Feinberg, Katharine K. Brieger, Lisa A. Croen, Irva Hertz-Picciotto, Craig J. Newschaffer, Rebecca J. Schmidt, and M. Daniele Fallin. 2020. "Prenatal Multivitamin Use and MTHFR Genotype Are Associated with Newborn Cord Blood DNA Methylation" International Journal of Environmental Research and Public Health 17, no. 24: 9190. https://doi.org/10.3390/ijerph17249190
APA StyleBakulski, K. M., Dou, J. F., Feinberg, J. I., Brieger, K. K., Croen, L. A., Hertz-Picciotto, I., Newschaffer, C. J., Schmidt, R. J., & Fallin, M. D. (2020). Prenatal Multivitamin Use and MTHFR Genotype Are Associated with Newborn Cord Blood DNA Methylation. International Journal of Environmental Research and Public Health, 17(24), 9190. https://doi.org/10.3390/ijerph17249190





