Environmental Epigenetics of Diesel Particulate Matter Toxicogenomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. ATAC-Seq Analysis
2.3. Database Analysis
2.4. ATAC-Seq Differential Accessibility Analysis and Visualization
2.5. Genomic Variant Extraction
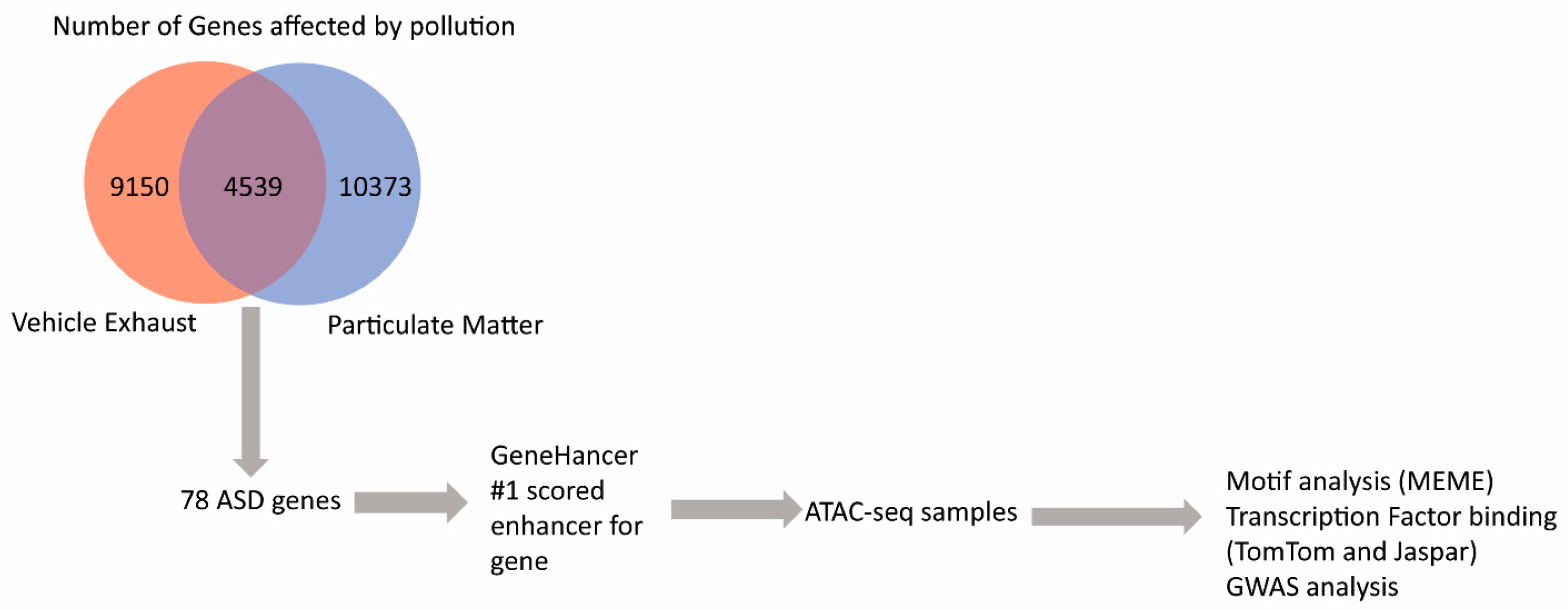
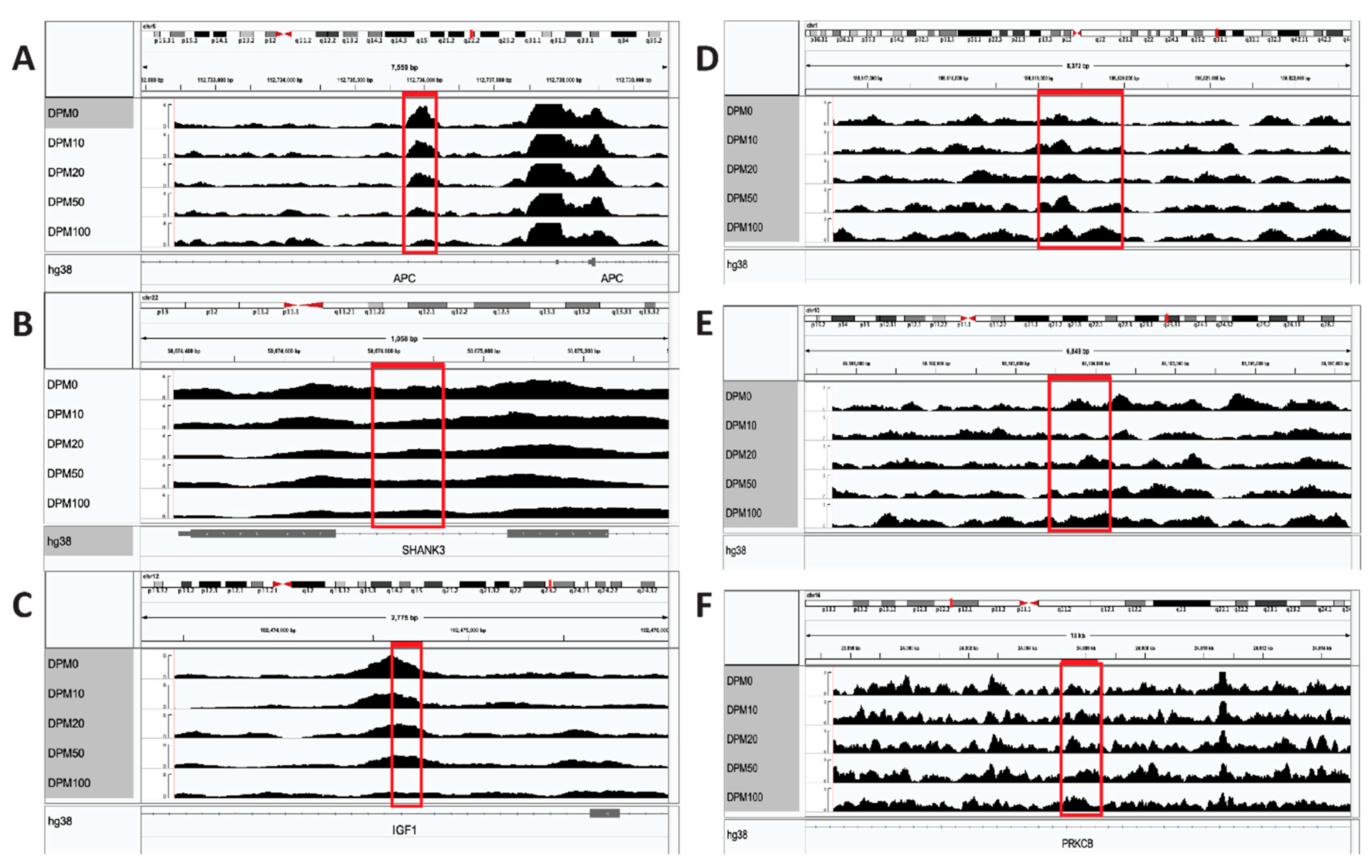
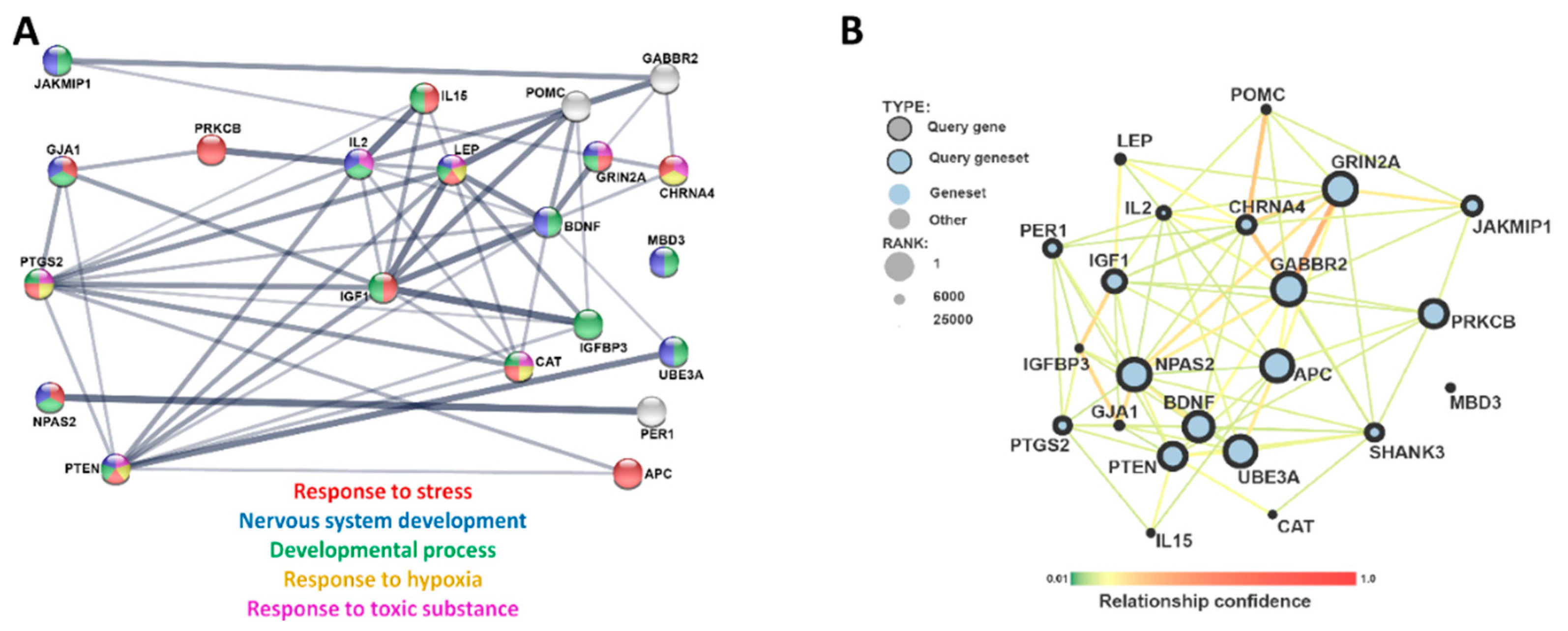
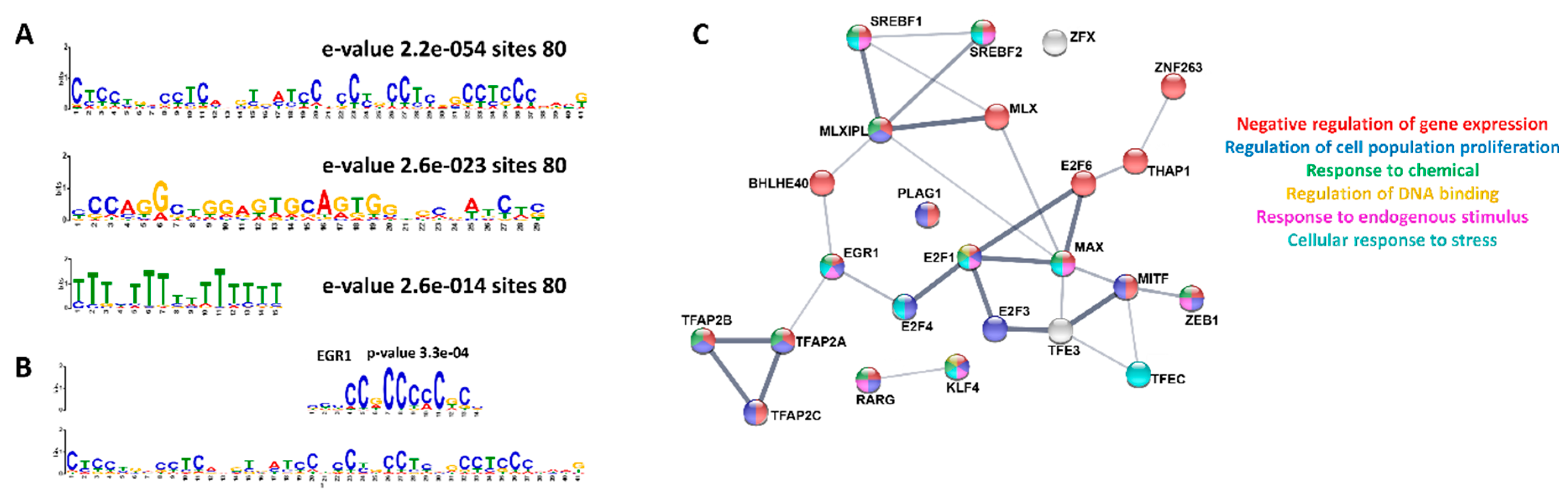
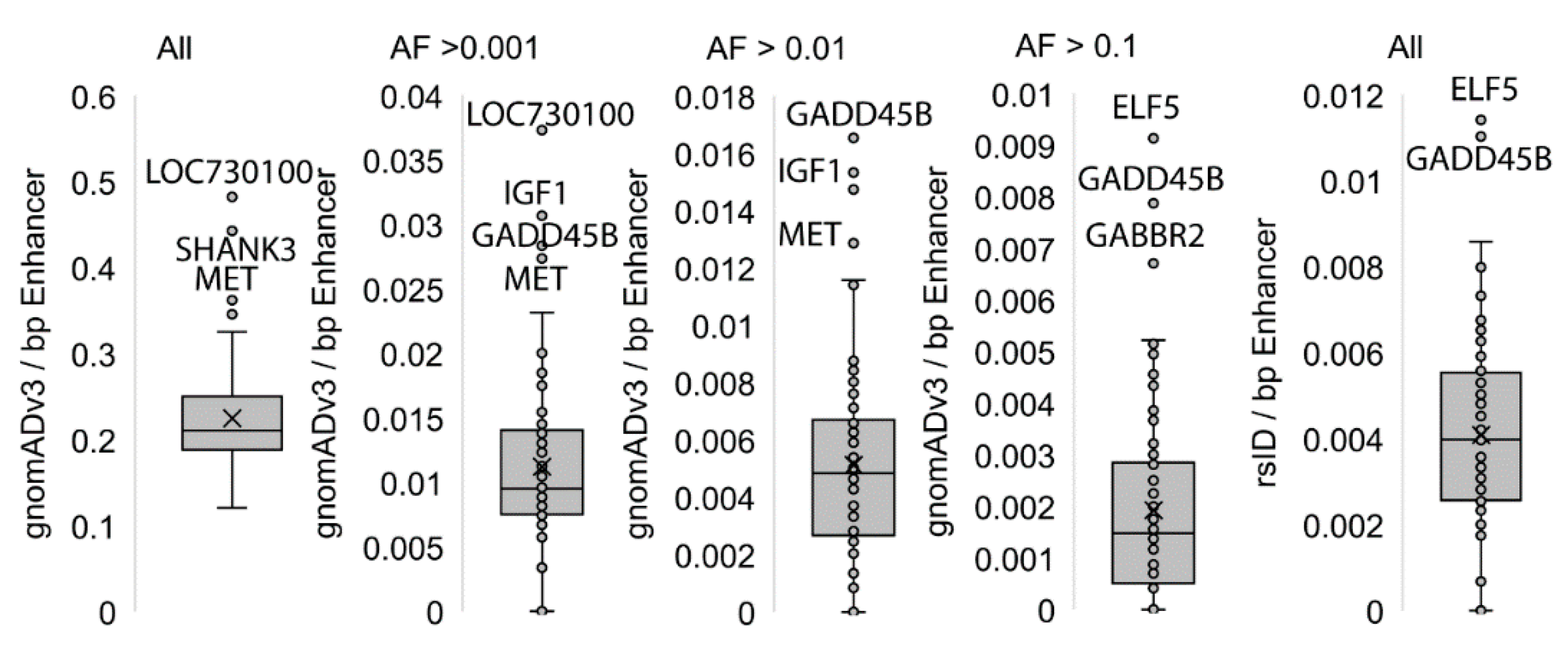
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moorthy, B.; Chu, C.; Carlin, D.J. Polycyclic Aromatic Hydrocarbons: From Metabolism to Lung Cancer. Toxicol. Sci. 2015, 145, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Sciaraffa, R.; Borghini, A.; Montuschi, P.; Gerosa, G.A.; Ricciardi, W.; Moscato, U. Impact of air pollution on respiratory diseases in urban areas: A systematic review Daniele Ignazio La Milia. Eur. J. Public Health 2017, 27. [Google Scholar] [CrossRef]
- Rock, K.D.; Patisaul, H.B. Environmental Mechanisms of Neurodevelopmental Toxicity. Curr. Environ. Health Rep. 2018, 5, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Gluckman, P.D.; Hanson, M.A.; Cooper, C.; Thornburg, K.L. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 2008, 359, 61–73. [Google Scholar] [CrossRef]
- Lee, B.-J.; Kim, B.; Lee, K. Air Pollution Exposure and Cardiovascular Disease. Toxicol. Res. 2014, 30, 71–75. [Google Scholar] [CrossRef]
- Bell, M.L.; Belanger, K.; Ebisu, K.; Gent, J.F.; Lee, H.J.; Koutrakis, P.; Leaderer, B.P. Prenatal Exposure to Fine Particulate Matter and Birth Weight. Epidemiol. Camb. Mass 2010, 21, 884–891. [Google Scholar] [CrossRef]
- Volk, H.E.; Lurmann, F.; Penfold, B.; Hertz-Picciotto, I.; McConnell, R. Traffic-Related Air Pollution, Particulate Matter, and Autism. JAMA Psychiatry 2013, 70, 71–77. [Google Scholar] [CrossRef]
- MohanKumar, S.M.J.; Campbell, A.; Block, M.; Veronesi, B. Particulate matter, oxidative stress and neurotoxicity. Neuro Toxicol. 2008, 29, 479–488. [Google Scholar] [CrossRef]
- Motta, V.; Angelici, L.; Nordio, F.; Bollati, V.; Fossati, S.; Frascati, F.; Tinaglia, V.; Bertazzi, P.A.; Battaglia, C.; Baccarelli, A.A. Integrative Analysis of miRNA and Inflammatory Gene Expression After Acute Particulate Matter Exposure. Toxicol. Sci. 2013, 132, 307–316. [Google Scholar] [CrossRef]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; McMorran, R.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. The Comparative Toxicogenomics Database: Update 2019. Nucleic Acids Res. 2019, 47, D948–D954. [Google Scholar] [CrossRef]
- Buenrostro, J.; Wu, B.; Chang, H.; Greenleaf, W. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. Ed. Frederick M Ausubel Al 2015, 109, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Fullard, J.F.; Hauberg, M.E.; Bendl, J.; Egervari, G.; Cirnaru, M.-D.; Reach, S.M.; Motl, J.; Ehrlich, M.E.; Hurd, Y.L.; Roussos, P. An atlas of chromatin accessibility in the adult human brain. Genome Res. 2018, 28, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Rendeiro, A.F.; Schmidl, C.; Strefford, J.C.; Walewska, R.; Davis, Z.; Farlik, M.; Oscier, D.; Bock, C. Chromatin accessibility maps of chronic lymphocytic leukaemia identify subtype-specific epigenome signatures and transcription regulatory networks. Nat. Commun. 2016, 7, 11938. [Google Scholar] [CrossRef] [PubMed]
- Kagohara, L.T.; Zamuner, F.; Davis-Marcisak, E.F.; Sharma, G.; Considine, M.; Allen, J.; Yegnasubramanian, S.; Gaykalova, D.A.; Fertig, E.J. Integrated single-cell and bulk gene expression and ATAC-seq reveals heterogeneity and early changes in pathways associated with resistance to cetuximab in HNSCC-sensitive cell lines. Br. J. Cancer 2020, 123, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Schep, A.N.; Buenrostro, J.D.; Denny, S.K.; Schwartz, K.; Sherlock, G.; Greenleaf, W.J. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 2015, 25, 1757–1770. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, A.C.; Yeo, R.W.; Buenrostro, J.D.; Greenleaf, W.J.; Kundaje, A.; Brunet, A. Chromatin accessibility dynamics reveal novel functional enhancers in C. elegans. Genome Res. 2017, 27, 2096–2107. [Google Scholar] [CrossRef]
- Li, Z.; Schulz, M.H.; Look, T.; Begemann, M.; Zenke, M.; Costa, I.G. Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 2019, 20, 45. [Google Scholar] [CrossRef]
- Nair, S.; Kim, D.S.; Perricone, J.; Kundaje, A. Integrating regulatory DNA sequence and gene expression to predict genome-wide chromatin accessibility across cellular contexts. Bioinformatics 2019, 35, i108–i116. [Google Scholar] [CrossRef]
- Wilson, M.R.; Reske, J.J.; Holladay, J.; Wilber, G.E.; Rhodes, M.; Koeman, J.; Adams, M.; Johnson, B.; Su, R.-W.; Joshi, N.R.; et al. ARID1A and PI3-kinase pathway mutations in the endometrium drive epithelial transdifferentiation and collective invasion. Nat. Commun. 2019, 10, 3554. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Iny Stein, T.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Lun, A.T.L.; Smyth, G.K. csaw: A Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 2016, 44, e45. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W.S. Quantifying similarity between motifs. Genome Biol. 2007, 8, R24. [Google Scholar] [CrossRef]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef]
- Krishnan, A.; Zhang, R.; Yao, V.; Theesfeld, C.L.; Wong, A.K.; Tadych, A.; Volfovsky, N.; Packer, A.; Lash, A.; Troyanskaya, O.G. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat. Neurosci. 2016, 19, 1454–1462. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Arnold, M.; Raffler, J.; Pfeufer, A.; Suhre, K.; Kastenmüller, G. SNiPA: An interactive, genetic variant-centered annotation browser. Bioinformatics 2015, 31, 1334–1336. [Google Scholar] [CrossRef] [PubMed]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed]
- Curated [Vehicle Exhaust-Autism Spectrum Disorders] Data Were Retrieved from the Comparative Toxicogenomics Database (CTD), MDI Biological Laboratory, Salisbury Cove, Maine, and NC State University, Raleigh, North Carolina. World Wide Web. Available online: http://ctdbase.org/ (accessed on 1 July 2020).
- Curated [Particulate Matter-Autism Spectrum Disorders] Data Were Retrieved from the Comparative Toxicogenomics Database (CTD), MDI Biological Laboratory, Salisbury Cove, Maine, and NC State University, Raleigh, North Carolina. World Wide Web. Available online: http://ctdbase.org/ (accessed on 1 July 2020).
- Abrahams, B.S.; Arking, D.E.; Campbell, D.B.; Mefford, H.C.; Morrow, E.M.; Weiss, L.A.; Menashe, I.; Wadkins, T.; Banerjee-Basu, S.; Packer, A. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 2013, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.-Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180, 568–584.e23. [Google Scholar] [CrossRef]
- Stuart, J.R.; Kawai, H.; Tsai, K.K.C.; Chuang, E.Y.; Yuan, Z.-M. c-Abl regulates Early Growth Response Protein (EGR1) in response to oxidative stress. Oncogene 2005, 24, 8085–8092. [Google Scholar] [CrossRef]
- Curated [EGR1-Chemical] Data Were Retrieved from the Comparative Toxicogenomics Database (CTD), MDI Biological Laboratory, Salisbury Cove, Maine, and NC State University, Raleigh, North Carolina. Available online: http://ctdbase.org/ (accessed on 10 October 2020).
- Ehsanifar, M.; Tameh, A.A.; Farzadkia, M.; Kalantari, R.R.; Zavareh, M.S.; Nikzaad, H.; Jafari, A.J. Exposure to nanoscale diesel exhaust particles: Oxidative stress, neuroinflammation, anxiety and depression on adult male mice. Ecotoxicol. Environ. Saf. 2019, 168, 338–347. [Google Scholar] [CrossRef]
- Kim, Y.-D.; Lantz-McPeak, S.M.; Ali, S.F.; Kleinman, M.T.; Choi, Y.-S.; Kim, H. Effects of ultrafine diesel exhaust particles on oxidative stress generation and dopamine metabolism in PC-12 cells. Environ. Toxicol. Pharmacol. 2014, 37, 954–959. [Google Scholar] [CrossRef]
- Li, K.; Li, L.; Cui, B.; Gai, Z.; Li, Q.; Wang, S.; Yan, J.; Lin, B.; Tian, L.; Liu, H.; et al. Early Postnatal Exposure to Airborne Fine Particulate Matter Induces Autism-like Phenotypes in Male Rats. Toxicol. Sci. 2018, 162, 189–199. [Google Scholar] [CrossRef]
- Hou, L.; Zhang, X.; Tarantini, L.; Nordio, F.; Bonzini, M.; Angelici, L.; Marinelli, B.; Rizzo, G.; Cantone, L.; Apostoli, P.; et al. Ambient PM exposure and DNA methylation in tumor suppressor genes: A cross-sectional study. Part. Fibre Toxicol. 2011, 8, 25. [Google Scholar] [CrossRef]
- Bos, I.; Boever, P.D.; Emmerechts, J.; Buekers, J.; Vanoirbeek, J.; Meeusen, R.; Poppel, M.V.; Nemery, B.; Nawrot, T.; Panis, L.I. Changed gene expression in brains of mice exposed to traffic in a highway tunnel. Inhal. Toxicol. 2012, 24, 676–686. [Google Scholar] [CrossRef]
- Tsamou, M.; Vrijens, K.; Madhloum, N.; Lefebvre, W.; Vanpoucke, C.; Nawrot, T.S. Air pollution-induced placental epigenetic alterations in early life: A candidate miRNA approach. Epigenetics 2018, 13, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Shi, L.; Li, D.; Li, X.; Su, X.; Chen, L.; Jiang, Q.; Jiang, M.; Luo, J.; Ji, A.; et al. Real-Ambient Particulate Matter Exposure-Induced Cardiotoxicity in C57/B6 Mice. Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Baron, V.; Adamson, E.D.; Calogero, A.; Ragona, G.; Mercola, D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGF β 1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006, 13, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Tian, M.; Wang, X.; Zhang, J.; Huang, Q.; Liu, L.; Shen, H. Cortex and hippocampus DNA epigenetic response to a long-term arsenic exposure via drinking water. Environ. Pollut. 2018, 234, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.S.; Talsania, K.; Mettil, W.; Anderson, D.W. Genetic Diversity Influences the Response of the Brain to Developmental Lead Exposure. Toxicol. Sci. 2014, 141, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Rynning, I.; Neca, J.; Vrbova, K.; Libalova, H.; Rossner, P.; Holme, J.A.; Gützkow, K.B.; Afanou, A.K.J.; Arnoldussen, Y.J.; Hruba, E.; et al. In Vitro Transformation of Human Bronchial Epithelial Cells by Diesel Exhaust Particles: Gene Expression Profiling and Early Toxic Responses. Toxicol. Sci. 2018, 166, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Cao, J.; Luo, M.; Che, L.; Li, W.; Ying, S.; Chen, Z.; Shen, H. Early growth response gene 1 is essential for urban particulate matter-induced inflammation and mucus hyperproduction in airway epithelium. Toxicol. Lett. 2018, 294, 145–155. [Google Scholar] [CrossRef]
- Martinez, J.M.; Baek, S.J.; Mays, D.M.; Tithof, P.K.; Eling, T.E.; Walker, N.J. EGR1 Is a Novel Target for AhR Agonists in Human Lung Epithelial Cells. Toxicol. Sci. 2004, 82, 429–435. [Google Scholar] [CrossRef]
- Mo, Y.; Wan, R.; Feng, L.; Chien, S.; Tollerud, D.J.; Zhang, Q. Combination effects of cigarette smoke extract and ambient ultrafine particles on endothelial cells. Toxicol. In Vitro 2012, 26, 295–303. [Google Scholar] [CrossRef]
- Heroux, N.A.; Robinson-Drummer, P.A.; Kawan, M.; Rosen, J.B.; Stanton, M.E. Neonatal ethanol exposure impairs long-term context memory formation and prefrontal immediate early gene expression in rats. Behav. Brain Res. 2019, 359, 386–395. [Google Scholar] [CrossRef]
- Robinson-Drummer, P.A.; Chakraborty, T.; Heroux, N.A.; Rosen, J.B.; Stanton, M.E. Age and experience dependent changes in Egr-1 expression during the ontogeny of the context preexposure facilitation effect (CPFE). Neurobiol. Learn. Mem. 2018, 150, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.L.; Rosen, J.B. Immediate-early gene expression in the central nucleus of the amygdala is not specific for anxiolytic or anxiogenic drugs. Neuropharmacology 2006, 50, 57–68. [Google Scholar] [CrossRef]
- Li, L.; Carter, J.; Gao, X.; Whitehead, J.; Tourtellotte, W.G. The Neuroplasticity-Associated Arc Gene Is a Direct Transcriptional Target of Early Growth Response (Egr) Transcription Factors. Mol. Cell. Biol. 2005, 25, 10286–10300. [Google Scholar] [CrossRef] [PubMed]
- Veyrac, A.; Gros, A.; Bruel-Jungerman, E.; Rochefort, C.; Borgmann, F.B.K.; Jessberger, S.; Laroche, S. Zif268/egr1 gene controls the selection, maturation and functional integration of adult hippocampal newborn neurons by learning. Proc. Natl. Acad. Sci. USA 2013, 110, 7062–7067. [Google Scholar] [CrossRef] [PubMed]
- Duclot, F.; Kabbaj, M. The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders. Front. Behav. Neurosci. 2017, 11. [Google Scholar] [CrossRef]
- Nakayama Wong, L.S.; Aung, H.H.; Lamé, M.W.; Wegesser, T.C.; Wilson, D.W. Fine particulate matter from urban ambient and wildfire sources from California’s San Joaquin Valley initiate differential inflammatory, oxidative stress, and xenobiotic responses in human bronchial epithelial cells. Toxicol. Vitro 2011, 25, 1895–1905. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, X.; Zhong, M.; Hotchkiss, I.P.; Lewandowski, R.P.; Wagner, J.G.; Bramble, L.A.; Yang, Y.; Wang, A.; Harkema, J.R.; et al. Ambient particulate air pollution induces oxidative stress and alterations of mitochondria and gene expression in brown and white adipose tissues. Part. Fibre Toxicol. 2011, 8, 20. [Google Scholar] [CrossRef]
- Volk, H.E.; Kerin, T.; Lurmann, F.; Hertz-Picciotto, I.; McConnell, R.; Campbell, D.B. Autism Spectrum Disorder: Interaction of Air Pollution with the MET Receptor Tyrosine Kinase Gene. Epidemiology 2014, 25, 44–47. [Google Scholar] [CrossRef]
- Lintas, C.; Sacco, R.; Garbett, K.; Mirnics, K.; Militerni, R.; Bravaccio, C.; Curatolo, P.; Manzi, B.; Schneider, C.; Melmed, R.; et al. Involvement of the PRKCB1 gene in autistic disorder: Significant genetic association and reduced neocortical gene expression. Mol. Psychiatry 2009, 14, 705–718. [Google Scholar] [CrossRef]
- Li, X.; Lv, Y.; Hao, J.; Sun, H.; Gao, N.; Zhang, C.; Lu, R.; Wang, S.; Yin, L.; Pu, Y.; et al. Role of microRNA-4516 involved autophagy associated with exposure to fine particulate matter. Oncotarget 2016, 7, 45385–45397. [Google Scholar] [CrossRef]
- Kong, A.; Frigge, M.L.; Masson, G.; Besenbacher, S.; Sulem, P.; Magnusson, G.; Gudjonsson, S.A.; Sigurdsson, A.; Jonasdottir, A.; Jonasdottir, A.; et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 2012, 488, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Janecka, M.; Mill, J.; Basson, M.A.; Goriely, A.; Spiers, H.; Reichenberg, A.; Schalkwyk, L.; Fernandes, C. Advanced paternal age effects in neurodevelopmental disorders—Review of potential underlying mechanisms. Transl. Psychiatry 2017, 7, e1019. [Google Scholar] [CrossRef] [PubMed]
- Grabrucker, A.M. Environmental Factors in Autism. Front. Psychiatry 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Chaste, P.; Leboyer, M. Autism risk factors: Genes, environment, and gene-environment interactions. Dialogues Clin. Neurosci. 2012, 14, 281–292. [Google Scholar]
- Guan, J.; Cai, J.J.; Ji, G.; Sham, P.C. Commonality in dysregulated expression of gene sets in cortical brains of individuals with autism, schizophrenia, and bipolar disorder. Transl. Psychiatry 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Schwartz, C.E. Intellectual disability and autism spectrum disorders: Causal genes and molecular mechanisms. Neurosci. Biobehav. Rev. 2014, 46, 161–174. [Google Scholar] [CrossRef]
- Srivastava, S.; Sahin, M. Autism spectrum disorder and epileptic encephalopathy: Common causes, many questions. J. Neurodev. Disord. 2017, 9. [Google Scholar] [CrossRef]
- Karnuta, J.M.; Scacheri, P.C. Enhancers: Bridging the gap between gene control and human disease. Hum. Mol. Genet. 2018, 27, R219–R227. [Google Scholar] [CrossRef]
- Gangwar, R.S.; Bevan, G.H.; Palanivel, R.; Das, L.; Rajagopalan, S. Oxidative stress pathways of air pollution mediated toxicity: Recent insights. Redox Biol. 2020, 34, 101545. [Google Scholar] [CrossRef]
- Haghani, A.; Johnson, R.; Safi, N.; Zhang, H.; Thorwald, M.; Mousavi, A.; Woodward, N.C.; Shirmohammadi, F.; Coussa, V.; Wise, J.P.; et al. Toxicity of urban air pollution particulate matter in developing and adult mouse brain: Comparison of total and filter-eluted nanoparticles. Environ. Int. 2020, 136, 105510. [Google Scholar] [CrossRef]
- Campbell, A.; Oldham, M.; Becaria, A.; Bondy, S.C.; Meacher, D.; Sioutas, C.; Misra, C.; Mendez, L.B.; Kleinman, M. Particulate Matter in Polluted Air May Increase Biomarkers of Inflammation in Mouse Brain. Neuro Toxicol. 2005, 26, 133–140. [Google Scholar] [CrossRef] [PubMed]
| Linked Regulatory Region | PubMed_ID | MAPPED_TRAIT | RISK ALLELE | CHR_ID | CHR_POS | p-Valume |
|---|---|---|---|---|---|---|
| chr2:51031736-51032889 | 30038396 | self-reported educational attainment | rs12620796-A | 2 | 51060711 | 9.00 × 10−11 |
| chr5:148730602-148971200 | 30287806 | nicotine dependence symptom count, depressive symptom measurement | rs57108954-T | 5 | 148857822 | 3.00 × 10−6 |
| chr5:148730602-148971200 | 26242244 | exploratory eye movement measurement | rs17108911-? | 5 | 148903759 | 6.00 × 10−6 |
| chr7:22600600-22602886 | 29071344 | unipolar depression, alcohol dependence | rs2905347-G | 7 | 22580700 | 6.00 × 10−6 |
| chr7:116685939-116778343 | 19010793 | multiple sclerosis | rs10243024-? | 7 | 116706549 | 6.00 × 10−6 |
| chr8:117846474-117850990 | 30038396 | mathematical ability | rs17430287-A | 8 | 117844068 | 4.00 × 10−8 |
| chr9:4506589-4512250 | 29503163 | schizophrenia, response to risperidone | rs16921385-A | 9 | 4507513 | 4.00 × 10−8 |
| chr10:88060002-88390829 | 31604244 | multiple sclerosis | rs1819577-A | 10 | 88067364 | 1.00 × 10−7 |
| chr10:88060002-88390829 | 30038396 | self-reported educational attainment | rs1426619-T | 10 | 88331783 | 1.00 × 10−11 |
| chr10:88060002-88390829 | 30038396 | self-reported educational attainment | rs1426619-T | 10 | 88331783 | 1.00 × 10−10 |
| chr11:27720334-28567694 | 30643258 | risk-taking behavior | rs16918024-T | 11 | 28566879 | 8.00 × 10−9 |
| chr12:102474603-102536836 | 31676860 | brain volume measurement | rs703545-? | 12 | 102549222 | 1.00 × 10−8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bilinovich, S.M.; Lewis, K.; Thompson, B.L.; Prokop, J.W.; Campbell, D.B. Environmental Epigenetics of Diesel Particulate Matter Toxicogenomics. Int. J. Environ. Res. Public Health 2020, 17, 7386. https://doi.org/10.3390/ijerph17207386
Bilinovich SM, Lewis K, Thompson BL, Prokop JW, Campbell DB. Environmental Epigenetics of Diesel Particulate Matter Toxicogenomics. International Journal of Environmental Research and Public Health. 2020; 17(20):7386. https://doi.org/10.3390/ijerph17207386
Chicago/Turabian StyleBilinovich, Stephanie M., Kristy Lewis, Barbara L. Thompson, Jeremy W. Prokop, and Daniel B. Campbell. 2020. "Environmental Epigenetics of Diesel Particulate Matter Toxicogenomics" International Journal of Environmental Research and Public Health 17, no. 20: 7386. https://doi.org/10.3390/ijerph17207386
APA StyleBilinovich, S. M., Lewis, K., Thompson, B. L., Prokop, J. W., & Campbell, D. B. (2020). Environmental Epigenetics of Diesel Particulate Matter Toxicogenomics. International Journal of Environmental Research and Public Health, 17(20), 7386. https://doi.org/10.3390/ijerph17207386





