1. Introduction
The recent pandemic has affected more than seventeen million people across 215 countries and territories and caused deaths of more than 751.399 people as per reported data from John Hopkins University Coronavirus Resource Center on August 13, 2020. World Health Organization declared this pandemic a Public Health Emergency of International Concern (PHEIC) on 31 January 2020 [
1]. The novel coronavirus responsible for this pandemic was named “2019-nCoV” and was later changed to “SARS-CoV-2” owing to its high similarity to severe acute respiratory syndrome coronavirus (SARS-CoV) [
2]. Finally, it became popular as Coronavirus Disease 2019 (COVID-19) [
3]. Scientists and governments around the globe are putting forward their best efforts and resources for the effective treatment of human coronavirus (HCoV) infections. However, neither vaccines nor antiviral drugs are approved for the treatment of HCoV infections such as SARS, MERS (Middle Eastern respiratory syndrome), and the COVID-19 [
4,
5,
6,
7]. The word “corona” is derived from Latin, meaning crown, as it possesses crown-like structure, hence called coronavirus. Coronaviruses possess a small size of approximately 65–125 nm in diameter and hold the nucleic material as single-stranded RNA. The similarity of the SARS-CoV-2 genome with SARS-CoV is about 79% [
8]. SARS-CoV-2 is a type of beta coronavirus [
9], belonging to the Coronavirinae subfamily and Coronaviridae family in order Nidovirales [
10]. Alpha-, beta-, gamma-, and delta-coronaviruses are the subgroups of the coronavirus family. The alpha- and beta-coronaviruses originated from mammals, while gamma- and delta-coronaviruses originated from birds and pigs [
11].
SARS-CoV and MERS-CoV originated from China in 2003 [
12] and Saudi Arabia [
13] in 2012, respectively, while the outbreak of COVID-19 began in December 2019 in Wuhan, China. Coronaviruses (CoVs) have shown significant pathogenicity against humans and other vertebrates [
14]. These coronaviruses are highly contagious [
15] and can cause severe respiratory infection [
16] and acute respiratory distress syndrome (ARDS), followed by acute lung injury (ALI), leading to pulmonary failure and, ultimately, death [
17]. The current strain of COVID-19 is less deadly than the previous MERS-CoV infection, with a mortality rate of 40%, and SARS-CoV, with a mortality rate of 10% [
18,
19]. Epidemiological investigations conducted by the World Health Organization (WHO) confirmed that the COVID-19 triggered by the Huanan South China seafood marketplace in Wuhan. However, no substantial evidence to exhibit that animal connection has been identified [
20,
21].
Further studies concluded that the genome sequence of COVID-19 is highly related to CoV infecting bats [
22]. Recently, it has been shown that bat CoVs can infect humans without an intermediate carrier [
23,
24]. It is suggested that SARS-CoV-2 originated from the Chinese chrysanthemum bat as their genomic sequence is very much similar to SARS-CoV-2. Furthermore, there may be an intermediate host between humans and bats. The pangolin, an animal used by the Chinese for its meat and having specific medicinal value, is thought to be the source of this virus. Epidemiologists speculate that someone who purchased a pangolin from one of the wet markets in Wuhan got infected after consumption, setting forward the chain of transmission [
25].
A plethora of research and review papers have been published since the onset of this pandemic [
7]. Moreover, the mass media is bombarding the public with conflicting information that is often misleading and confusing. There is an urgent need for accurate and reliable information related to COVID-19. In this review, we have tried our best to compile up to date and accurate information, citing reliable and reputed journals, that will be helpful for the scientific community as well as the researchers working in this field. First, we thoroughly explained the origins and evolution of SARS-CoV-2, followed by a detailed account of its epidemiology and pathogenesis. There are very few reports that address the comprehensive clinical features of the disease. Furthermore, information about treatment options for COVID-19 is not adequate.
In this report, we have tried to gather the most updated and accurate data concerning this disease. In addition, information about all ongoing clinical trials of vaccines and other potential therapeutics has been addressed comprehensively. This review covers multiples aspects of COVID-19, including the structure of the virus, evolution process of the disease, epidemiologic outcomes, pathogenetic data, clinical features, diagnosis tools, treatment strategies, and prevention and control pathways.
4. Epidemiology
As described earlier, SARS-CoV-2 originated from Wuhan, China, and has affected more than 20 million people across 212 countries and territories [
14]. The transmission rate in some countries is exceptionally high compared with others, and the number of infected patients is increasing gradually [
36]. Chronologically, the first virus was obtained from a patient on 7 January 2020. In response, the sequence of the viral genome was published by Chinese scientists on 10 January 2020, on Global Initiative on Sharing All Influenza Data (GISAID) [
37]. In China, five patients were admitted to the hospital between 18 and 29 December 2019, out of which one patient expired [
38].
Additionally, 41 more patients were admitted to the hospitals by 2 January 2020. The diagnosis of these patients confirmed COVID-19. Thailand was the first country that was infected after China, and the first case was reported in Thailand on 13 January 2020, in a patient who traveled to Thailand. Five hundred and seventy-one new cases of COVID-19 were reported by 22 January 2020, in 25 provinces of China [
39]. National Health Commission of China (NHC) confirmed the first 17 deaths due to COVID-19 by 22 January 2020. On 25 January 2020, there were a total of 1975 infected patients in China, out of which 56 expired [
40]. According to another report from 30 January 2020, a total of 7734 cases were reported in China, and 90 cases were confirmed from other countries including Vietnam, Nepal, Sri Lanka, Japan, Thailand, Republic of Korea, United States, United Arab Emirates, Malaysia, The Philippines, India, Australia, Canada, Cambodia, Finland, Singapore, France, Germany, and Taiwan [
41].
The Chinese Center for Disease Control and Prevention reported 44,672 confirmed cases by 11 February 2020, in which 81% of deaths were seen in patients age over 60 years. Mortality rates in patients aged 70–79 and over 80 years were 8.0% and 14.8%, respectively [
42,
43]. On 28 February 2020, WHO reported 82,000 confirmed cases worldwide, and the outbreak reached 45 countries other than China. Following the rapid transmission course, 90,000 infected cases and 60 countries were dealing with the COVID-19 outbreak by 2 March 2020. Previously, it was thought that children were less susceptible to infection owing to a lack of evidence, but on 5 March 2020, Chinese studies concluded that children are as susceptible as adults. By 13 March 2020, Europe was declared as the epicenter of the outbreak by WHO. There were a greater number of reported cases in Europe as compared with the rest of the world. In Europe, Italy was the largest outbreak territory. The United States President declared the national emergency in the United States on 13 March 2020. By 27 March 2020, the total number of infected patients reached half a million, and pandemic spanned about 175 countries. The one million mark globally was reached on 2 April 2020. The next million cases were reported in less than two weeks, and there were 2 million cases worldwide on 15 April 2020, with the United States being on the top of the list, followed by Italy and Spain [
44]. Currently, there are almost 21 million cases with 752,225 deaths around the globe, accessed on August 13, 2020 (
https://coronavirus.jhu.edu/map.html).
Rapid transmission of COVID-19 reflects the high transmissibility and reproductive number (R
o). Chinese reports show a R
o value of 2.2–2.7, meaning the infected cases double every 6–7 days [
45]. The rate of transmission was inconceivably high, so several medical facilities were established, and guidelines were reported by the CDC and the WHO to manage the escalating disease [
46].
Another parameter for understanding the severity and prognosis of infection is the case fatality ratio (CFR), which is calculated by dividing the number of expired patients by the total number of diagnosed patients, multiplied by 100. According to the WHO situation report-185 of COVID-19, the overall CFR was 5.12 as of 23 July 2020. A remarkable difference in CFR was noted in some countries presented in
Table 1. The exact cause for this variability is still a topic of investigation in the scientific community. CFR is useful in estimating the risk of death within a population owing to a specific disease. Data suggest increased mortality with advanced age has a well-known impact on the prognosis of the disease. The median age of any population reflects the variation in fatality rates [
47]. One of the Chinese reports revealed that the mortality rate could be 3% high in geriatrics, especially over 80 [
48].
5. Pathogenesis
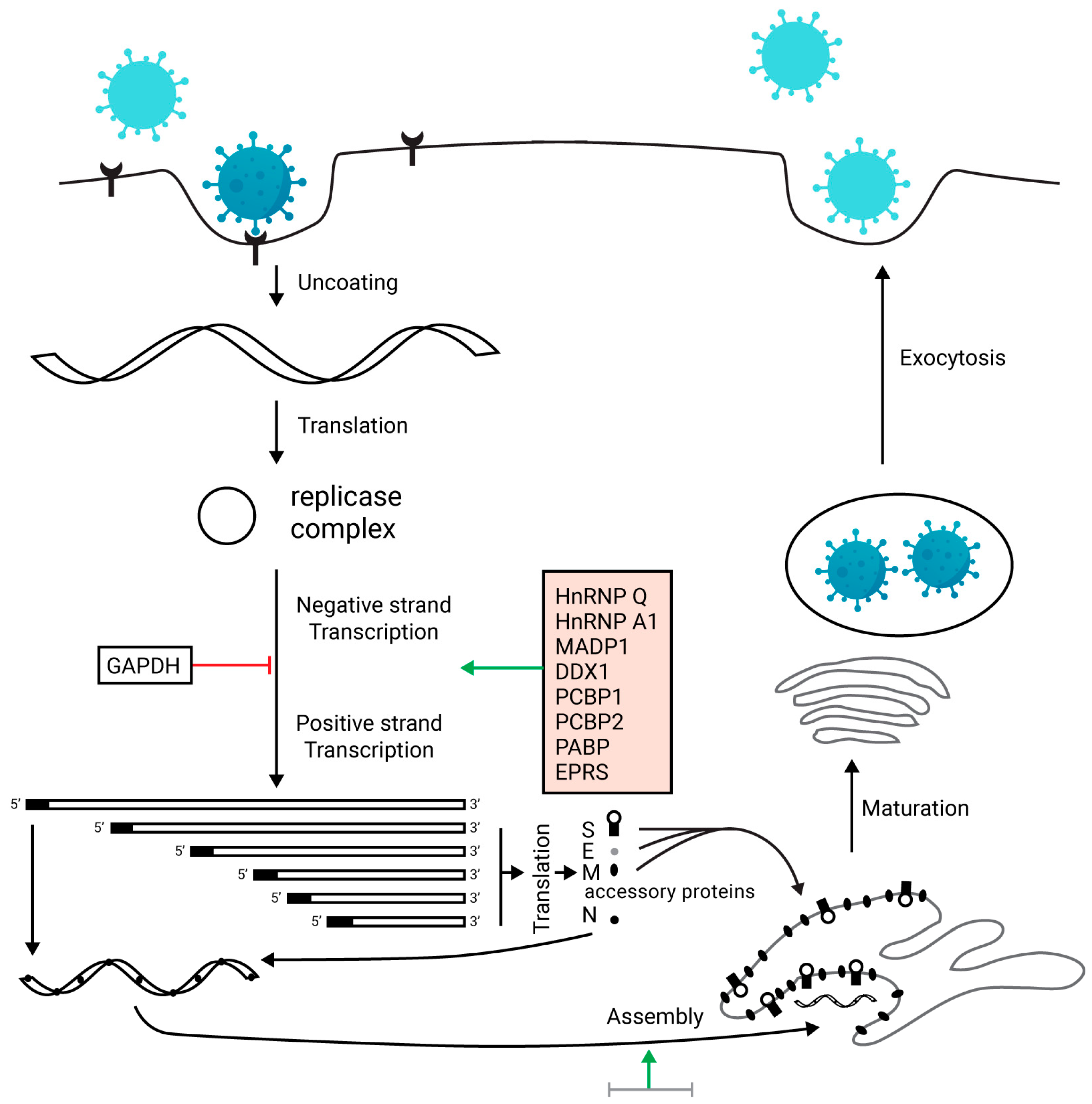
SARS-CoV-2 targets the lower respiratory tract and causes mild to severe symptoms. COVID-19 patients show significant clinical symptoms, including non-productive cough, myalgia, reduced leukocyte, dyspnea, fever, fatigue, and pneumonia. These symptoms are also shown by the patients infected by SARS-CoV and MERS-CoV [
49]. Although the pathogenesis of novel coronavirus disease is not fully understood, the similarity of novel coronavirus with SARS-CoV can provide significant information about the pathogenesis of COVID-19 infection. The body shows aggressive inflammatory responses after viral entry, and it may cause severe damage to airways. The pathogenicity is increased by a large number of viral copies replicated in the cell. The life cycle of SARS-CoV-2 is divided into four stages: (i) entry of the virus, (ii) protein expression, (iii) transcription, and (iv) release of the virus from the cell [
50].
Coronavirus enters the cell by its sole determinant viral S-protein and undergoes several steps to accomplish the replication. While S-protein is divided into two domains, S1 and S2 domain, the S1 domain is responsible for binding with the receptor, and the S2 domain accounts for the fusion of the viral membrane with the cell membrane [
51]. The viral spike glycoproteins attach themselves to the ACE2 receptor of the cell, for both viruses SARS-CoV [
52] and SARS-CoV-2 [
53]. Initially, the virus binds to the cell receptor and enters the cell through the process of membrane fusion of the virus and plasma membrane [
54]. Belouzard et al. [
55] showed that the proteolytic processes occur in the S2 domain, which accounts for fusion and infection. After the viral entry, the RNA genome of the virus is released in the cytoplasm and then undergoes translation, followed by transcription, through which the virus continues to replicate [
56]. After the formation of viral proteins by translation, these new proteins are inserted into the endoplasmic reticulum or Golgi apparatus. When viral RNA is combined with the proteins, the nucleocapsid is formed. Finally, the newly formed viruses enclosed in the vesicles are released by exocytosis [
56]. The viral release marks the time when infected patients start displaying significant symptoms and laboratory values. Higher leukocyte count, respiratory problems, and exaggerated pro-inflammatory cytokines are exhibited by COVID-19 patients. In a case report, a patient presented with fever for the past two days displayed abnormal breathing sounds and a temperature of 39.0 °C. The real time-polymerase chain reaction (RT-PCR) conducted on the sputum culture showed a positive result for COVID-19 and confirmed the disease [
57].
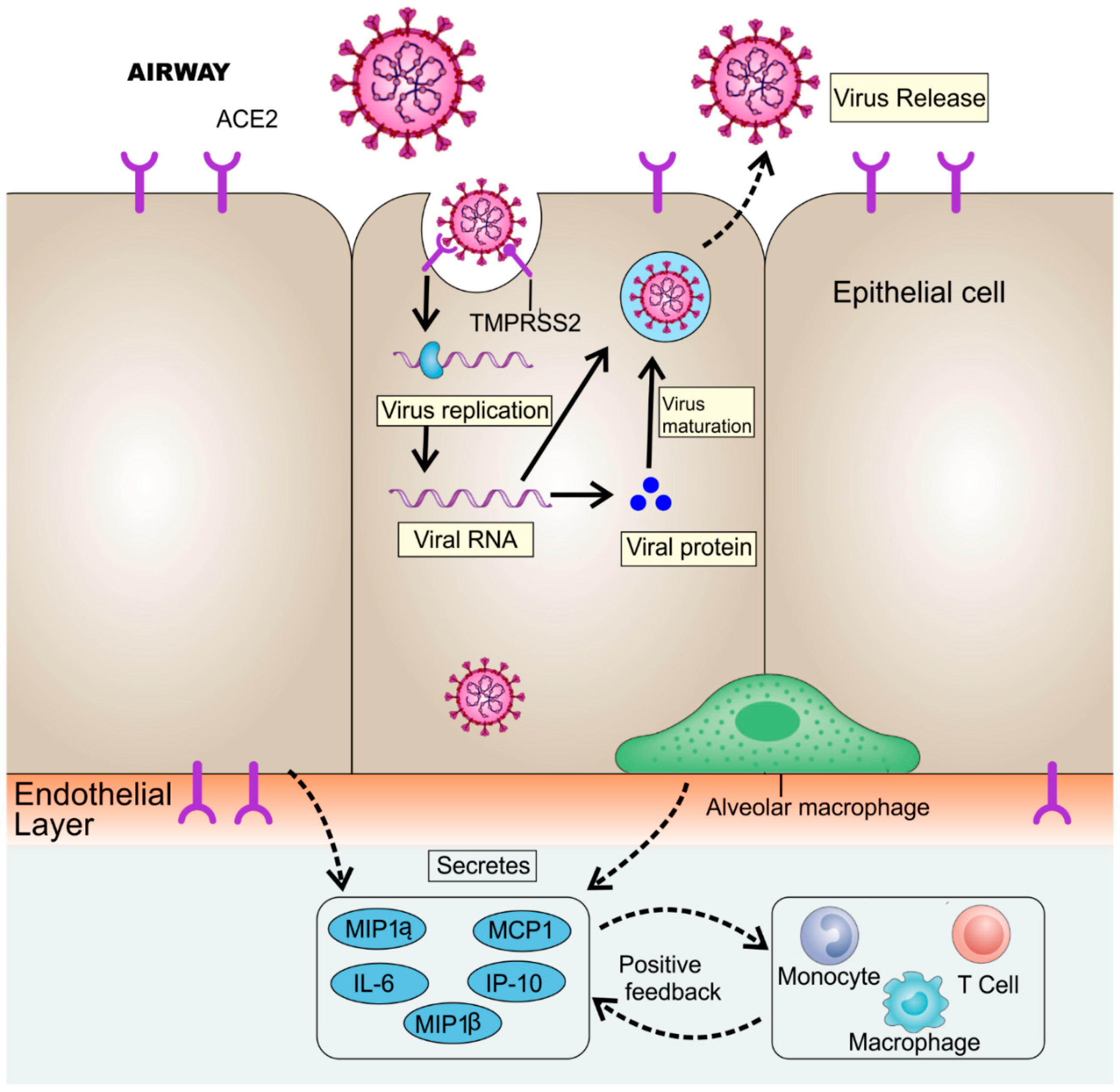
The laboratory analysis showed a leukocyte count of 2.91 × 10
9 cells/L, of which neutrophils were 70%. Further, the C-reactive protein was 16.16 mg/L (normal range = 0–10 mg/L). The erythrocyte sedimentation rate (ESR) was also high with D-dimer. The primary pathogenesis of the nCoV is associated with respiratory infections, pneumonia, ground-glass opacities (GGOs), acute cardiac injury, ARDS, and RNAaemia [
38]. Additionally, high blood values of chemokines and cytokines were also observed in patients, including IL1-β, IL7, IL8, IL9, IL10, IL1RA basic FGF2, GMCSF, IFNγ, IP10, GCSF MCP1, MIP1α, MIP1β, PDGFB, and TNFα, as shown in
Figure 2. Critical patients showed a high value of pro-inflammatory cytokines as a cytokine storm, including IL2, IL7, IL10, GCSF, IP10, MCP1, MIP1α, GCSF, and TNFα, which exaggerated the severity of the infection [
58]. This exaggerated immune response destroys the lung’s infrastructure [
59].
Current models give information about the three stages of the immune response. The first stage involves the early activation and persuasive interferon response for clearing the virus. The second stage exhibits the delayed response of interferon that may lead to tissue damage. The third stage leads to hyper-inflammation, followed by exaggerated macrophage activation, resulting in fibrosis dysregulation of tissue repair processes [
59]. For each step, potential therapeutic options can be developed. Some are under clinical trials to evaluate their efficacy along with safety. Initially, the virus attaches to the ACE2 receptor. TMPRSS2 is responsible for the viral protein cleavage. Protease inhibitors can be designed and developed to prevent spike protein cleavage. Blocking the viral fusion through either TMPRSS2 or receptor ACE2 can prevent the infection. For removing pro-inflammatory cytokines, several novel models have been proposed in which the blood of a COVID-19 patient is passed through customized columns that recognize and trap the pro-inflammatory cytokines, and then the purified blood is administered back to the patient [
60]. ARDS seen in several COVID-19 patients may lead to secondary infections, and respiratory failure leads to death in 79% of critical cases. Furthermore, the cytokine storm may lead to sepsis and cause death in 28% of critical cases [
61]. Scientists are trying to design therapeutics to prevent this exaggerated immune response responsible for the enhanced severity of the infection [
60,
62].
As discussed earlier, ACE2 is the receptor for SARS-CoV-2. It has been reported that the involvement of ACE2 receptors is also related to the renin–angiotensin system (RAS) in 2019-nCoV pathogenesis [
63]. Very recently, a meta-analysis was conducted to get insight into the relationship between the prevalence of COVID-19 disease and the genetic differences in the genes involved in the RAS system [
63]. The results of the study suggest that the increase of the I/D allele frequency ratio significantly increases the recovery rate (point estimate: 0.48, confidence interval (CI) 95%: 0.05–0.91,
p = 0.027), but the I/D allele frequency ratio has no significant difference in the case of death rate (point estimate: 1.74, CI 95%: 4.5–1.04,
p = 0.22). The I/D allele ratio of ACE receptors varies significantly across world regions, accounting for the different recovery rates across regions, but it also raises concerns that ethnic and genetic differences can impact the effectiveness of currently investigated RAS-associated medications in different regions [
63].
6. Clinical Features
SARS-CoV-2 represents an incubation period of 6.4 days, fluctuating between 2.1 and 11.1 days with possible asymptomatic transmission [
64]. Infection is manifested after the incubation period. The period from the commencement of disease leading to death fluctuates between 6 and 41 days with an average of 14 days. The age of patient and condition of the immune system are the sole determinants of this period. Persons with a weaker immune system are more susceptible to infection. This period is shorter in geriatrics >60 years, as older patients have a weaker immune system [
65]. The most common symptoms of COVID-19 are dry cough, myalgia, diarrhea, headache, and high-grade fever [
66]; fever is reported in more than 90% of cases, cough in 75%, and dyspnea in 50% of the patients [
8]. Fewer patients may also develop kidney damage, ARDS, septic shock, and acute cardiac injury requiring hospitalization [
67]. The clinical symptoms are presented in
Table 2.
Mutual laboratory findings include lymphopenia and leucopenia. These findings were common and observed in a large number of cases [
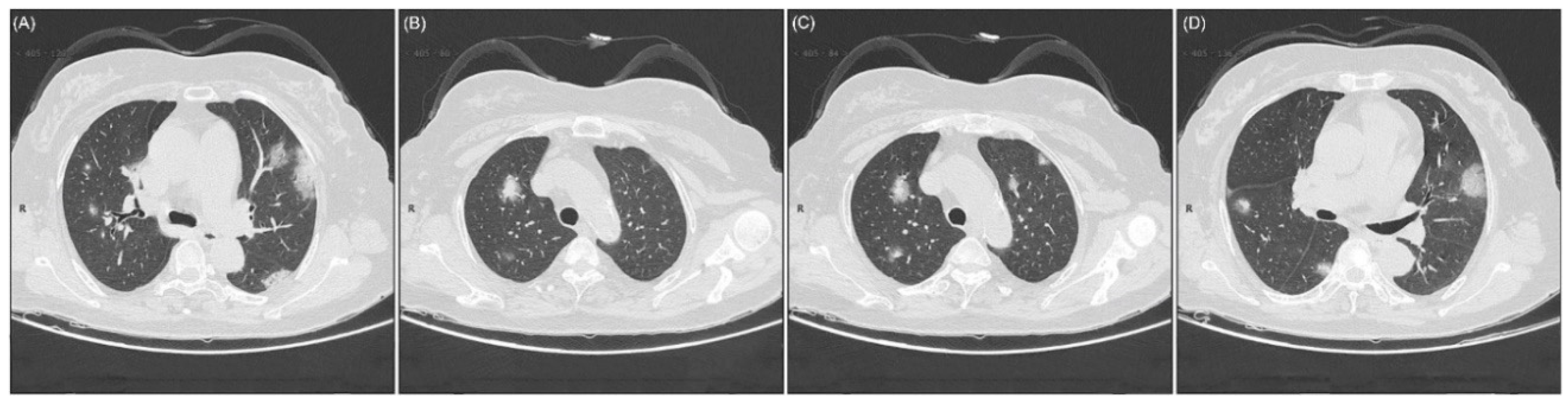
68]. Chest computed tomography (CT) scan showed clinical features including pneumonia and other abnormal features, for instance, RNAaemia, acute cardiac injury, and occurrence of GGO, that set the path towards death. In several cases, multiple marginal GGOs were observed in both lungs, which expectedly triggers the systemic and localized immune reaction, causing severe inflammation [
65]. Atypical CT scan showed single, multiple solid, and consolidated nodules in the middle of the lobe of right or left or both lungs, fenced by GGOs in the number of COVID-19 patients (
Figure 3) [
69].
Although there are some similarities, the symptoms manifested by earlier coronaviruses are not precisely similar to those of COVID-19. For instance, the distinctive features of COVID-19 are fever, fatigue, and dry cough. In contrast, upper respiratory tract symptoms like rhinorrhea, sore throat, and sneezing are uncommon, which were commonly observed in previous coronavirus infections. Moreover, COVID-19 patients present with intestinal symptoms like diarrhea, which were less prevalent in MERS-CoV and SARS-CoV infections [
70].
In addition, patients may also report anosmia and dysgeusia as early symptoms of COVID-19 [
71]. Anosmia (loss of sense of smell) and dysgeusia (alteration of the sense of taste) are also associated with COVID-19 patients. The American Academy of Otolaryngology-Head and Neck Surgery developed the COVID-19 Anosmia Reporting Tool for clinicians to conduct a pilot study. This tool allows clinicians to submit the anosmia and dysgeusia associated COVID-19 patients. They analyzed the first 237 entries and revealed that anosmia was observed in 73% of patients prior to the diagnosis of COVID-19 and was the early symptom in 26.6% of COVID-19 patients. Conclusively, this study suggested that anosmia can be the representative symptom of COVID-19 [
72,
73].
7. Diagnosis
The diagnosis and treatment program for COVID-19 was formulated by NHC of China [
69,
74] and was previously suggested by WHO for SARS and MERS [
75,
76,
77,
78]. The specific diagnostic measures have been implemented for suspected as well as confirmed cases: a patient with a minimum of two clinical symptoms and at least one exposure history is accounted as a suspected case. The suspected case should show at least three clinical symptoms if there is not any clear exposure history.
The serology testing for COVID-19 is helpful and provides the basis for diagnosis. In the case of the unavailability of molecular testing, the serological test provides a means to triage the suspected cases of COVID-19. A fast-serological test with specific performance features is highly important to avoid missing factual cases of COVID-19. A positive test for IgM or IgG antibody strongly indicates SARS-CoV-2 infection. This approach is extremely effective in individuals 5–10 days after the onset of symptoms [
79]. IgG or IgM positive patients are quarantined, and those who need critical care are referred to the hospital. Those with a negative serological antibody test have a swab collected for molecular testing. The experience in China has shown that the use of a serological antibody test can improve the sensitivity of COVID-19 case detection [
80]. This approach allowed to test a huge number of symptomatic individuals rapidly in the community, minimizing the waiting time for molecular testing and preventing the health-management system from being overwhelmed [
79].
Subsequently, when nucleic acids of SARS-CoV-2 by RT-PCR results come back positive, it is accounted for as a confirmed case [
76]. Meanwhile, a CT scan of the chest provides confirmed diagnostic evidence of COVID-19 disease as it has greater sensitivity and high specificity. Thus, the chest CT has been recognized as a significant indicator for COVID-19 diagnosis in different epidemic areas [
15,
31,
81]. It can be said that the chest CT is an essential procedure for the timely detection and management of this infection. However, it should be noted that a patient with an RT-PCR may have a negative chest CT at the time of admission [
82]. In this context, a study compared three RT-PCR analyses, targeting the RNA-dependent RNA polymerase RdRp/Helicase, spike, and nucleocapsid genes of the virus with testified RdRp-P2 assay, which is extensively used in more than 30 European countries. According to this study, RdRp/Hel is more specific and gives accurate results for SARS-CoV-2. Moreover, the RdRp/helicase assay of COVID-19 does not cross-react with other types of coronaviruses and respiratory microorganisms in the cellular medium. In contrast, the RdRp-P2 assay, reported for previous coronaviruses, reacts with the pathogens of the respiratory tract in the cell culture. Thus, the COVID-19-RdRp/Hel can be considered sensitive and specific, and this information helps fortify the diagnosis of COVID-19 [
83].
Laboratory findings can be helpful in predicting COVID-19 cases with positive RT-PCR. ALT, C-reactive protein, neutrophils, AST, and lymphocytes counts have very good accuracy in predicting cases with positive RT-PCR for COVID-19 patients [
84]. A study suggested that the number and percentage of white blood cells (WBCs), lymphocytes, and neutrophils were significantly different between positive and negative RT-PCR cases for COVID-19 patients [
85]. Another cohort study suggested that lymphopenia (low levels of lymphocytes) occurred in 80% of COVID-19 patients [
86]. An increased level of AST and ALT was also noted in severely affected patients [
87]. High levels of LDH, C-reactive protein, neutrophils, and erythrocyte sedimentation rate (ESR) were also notable in COVID-19 patients [
88]. Thus, these laboratory findings have great importance in terms of predicting COVID-19 cases with positive RT-PCR [
84].
8. Treatment
Currently, no specific and sensitive therapeutic regimen is available for the treatment of COVID-19. However, the primary step is isolation to prevent any type of contact that later may cause transmission of the disease. If the patient experiences any mild symptoms, it should be managed at their residence with proper counseling to patients about the critical signs and symptoms. The primary management protocols include the management of cough and fever [
89]. First-line therapy to manage the fever is acetaminophen, while expectorants such as guaifenesin are used for non-productive cough [
90]. Patients with SARS, hypoxemia, respiratory distress or shock should be treated by the administration of instant oxygen therapy. The frequency of oxygen therapy should be 5 L/min to obtain SpO
2 of ≥92–95% in pregnant women, and ≥90% in adults and children [
91]. Antiviral therapy is also being tried to manage the infection [
92].
Critical patients show an unhealthy immune response that eventually causes hyper-inflammation. This exaggerated immune response should be controlled and checked by different approaches and procedures. As discussed earlier, inflammatory cytokines are released at the site of infection and cause infiltration of lungs. Clinical trials are underway to identify the benefits of blocking cytokine storm targeting IL-1β and IL-6, as shown in
Table 3.
Interfering cytokine storm signaling can prevent hyper-inflammation, thereby reducing infection. Current evidence shows that the overproduction of macrophages linked to monocytes accumulates in tissue that can cause lung infiltration. For instance, it is found that CCR2 activation contributes to the accumulation of monocytes in the tissue, causing severe inflammation. So monocytes’ accumulation can be reduced by blocking the CCR2, resulting in minimizing hyper-inflammation [
96]. Different clinical trials are underway to reduce the hyper-inflammation by JAK inhibitors [
97]. Other relevant clinical trials are ongoing to evaluate the blocking of myeloid-derived cytokines, for example, TNF [
98] and GM–CSF. By targeting this cytokine signaling, the hyper-inflammation can be reduced [
99].
Remdesivir is a broad-spectrum antiviral prodrug, which is a nucleotide analog and is metabolized in the cell to adenosine triphosphate analog, which inhibits the viral RNA polymerases. Remdesivir is the drug of choice against several viral families, including Ebola, SARS-CoV, and MERS-CoV. Remdesivir is being used for prophylactic and therapeutic purposes and has shown remarkable efficacy in the non-clinical model of coronaviruses [
100,
101]. It has also shown promising activity against SARS-CoV-2 [
102,
103]. A large clinical trial of remdesivir is currently underway for patients with mild to moderate symptoms [
104]. The critically ill patients treated with amicable use of remdesivir showed better efficacy, and the clinical condition of the patients was improved [
105]. On 1 May 2020, the Food and Drug Administration (FDA) of the USA authorized the emergency use of remdesivir for the treatment of COVID-19 [
106]. The recent version of treatment guidelines by NHC recommends the use of lopinavir/ritonavir and IFN-α. The first version of treatment guidelines was issued on 15 January 2020, later revised five times, and the 6th edition is the most recent one that was issued on 18 February 2020. Lopinavir/ritonavir, ribavirin, and IFN-α were added in the 5th edition of treatment guidelines for combating COVID-19. Later, arbidol and chloroquine phosphate have also been added in the 6th edition, as these drugs show better clinical efficacy. Potential drugs included in the 6th edition of treatment guidelines by NHC of China, as shown in
Table 4, presented promising therapeutic effects [
107]. Although the recent study was an open-label, individually randomized, controlled trial representing patients admitted in hospitals, treated with lopinavir/ritonavir (400 mg/100 mg), no clinical improvement was observed by this treatment beyond standard care [
108]. On the basis of the guidelines, ritonavir is used as a booster to enhance the efficacy of lopinavir. These medications are prescribed to be taken for a short period of time (<2 weeks), suggesting their acute response.
Hydroxychloroquine, lopinavir/ritonavir, chloroquine, and darunavir/cobicistat are being used for patients with severe conditions. Interferon-beta B1 is extensively used for critical patients [
110]. Another drug, oseltamivir, is being used in China by the medical personnel for suspected patients, but there is a lack of evidence on the specific activity of oseltamivir against COVID-19. A 54-year-old female patient from Wuhan city presented with fever for the past two days. Two tests were conducted, the first test was not positive, but the second test confirmed the disease. Meanwhile, this woman was treated with oseltamivir for three days, and her chest CT indicated reminiscent improvement in the density of GGO [
90].
Patients with vigorous immune reactions can be treated with glucocorticoids such as methylprednisolone, which is used at 1–2 mg/kg/day dose in children and 25–150 mg/day in adults. It is believed to reduce the prognosis of the disease in patients with impaired oxygen index, that is, less than 300 mm Hg, limited to 5 days [
69]. For patients with premature disease stage, without hypoxia, use of corticosteroids should be avoided. However, in critical situations where mechanical ventilation is mandatory, anti-inflammatory therapy is possibly useful and can be employed [
59]. A glucocorticoids-based drug dexamethasone may be employed against COVID-19. Dexamethasone is a synthetic corticosteroid approved by the FDA in 1958 as a broad-spectrum immunosuppressor, which is about 30 times as active as cortisone and possesses a longer duration of action (2–3 days). Dexamethasone may not only limit the production and damaging effect of the cytokines, but may also inhibit the protective function of T cells and reduce the ability of B cells to synthesize antibodies [
111]. Dexamethasone may be a useful agent for short-term therapy in severe intubated COVID-19 patients [
112]. A recent controlled open-label, randomized trial of dexamethasone in hospitalized patients, concluded that the use of dexamethasone with the aid of mechanical ventilation or oxygen alone resulted in lower mortality at the dose of 6 mg once daily for 10 days. However, dexamethasone was not effective in those patients who were not receiving respiratory support [
113]. Dexamethasone is on an essential medicines list recommended by the World Health Organization and is readily accessible worldwide at a low cost. Guidelines issued by the U.K.’s chief medical officers and by the National Institutes of Health in the United States have already been updated to recommend the use of glucocorticoids such as dexamethasone in hospitalized patients with COVID-19 [
114,
115].
As discussed earlier, a variety of drugs are recommended and tested in non-clinical models; among them, penciclovir, ribavirin, and favipiravir require high doses to decrease the infection and are supposed to be less effective [
90]. Chloroquine and its derivative hydroxychloroquine are considered to be effective against COVID-19 and are currently used in Chinese patients to reduce viral infection. Potential drug therapy for COVID-19 is listed in
Table 4 to gain a better understanding of treatment [
116]. An open-label, non-randomized clinical trial showed that hydroxychloroquine (600 mg/day) treatment is magnificently related to a decreased rate of viral replication in patients, and its effectiveness is reinforced by azithromycin (500 mg/day) [
116]. Hydroxychloroquine showed superior sensitivity over chloroquine against COVID-19 [
117]. Contrary to the previous results, at the time of publication, an open-label, randomized controlled clinical trial showed that the use of hydroxychloroquine alone or with azithromycin did not improve the clinical status of hospitalized patients with mild to moderate COVID-19 as compared with standard care [
118].
According to many reports, the COVID-19 patients are at high risk for venous thromboembolism, a term that associates deep vein thrombosis (DVT) [
119]. In this context, patients are treated prophylactically by giving low molecular weight heparin (LMWH), when multiple risk factors exist. Hence, hospitalized COVID-19 patients are generally treated with higher doses of LMWH than recommended for thromboprophylaxis [
120]. A recent document by the Italian Drug Agency (AIFA) suggested the use of 80 to 100 mg enoxaparin daily, instead of the usual 40 mg, while in some hospitals, even higher, up to full anticoagulant doses of LMWH or unfractionated heparin are used [
121].
SARS-CoV-2 targets the ACE2 receptor via spike-like protein, so it is an efficient target for the drugs, including chloroquine, promazine, and emodine, to prevent the viral attachment with the surface of the host cell [
122]. S protein priming is the next precarious phase of viral entry into the host cell, where it is required to be cleaved by intracellular proteases including TMPRSS2, cathepsins, and furin for the fusion of viral membrane with the cell membrane of the host. S-protein priming by TMPRSS2 is the most crucial phase for the prognosis of infection into the host, so the viral infection can be controlled with the use of protease inhibitors, including camostat and nafamostat, which target TMPRSS2, and thereby inhibit the viral fusion [
123]. The immune system represents the defensive mechanism in attacking the virus; major immune cells include T-lymphocytes and natural killer cells [
122]. Apart from antiviral therapy, the use of antibiotics can be used for the associated co-infections, community-acquired, or hospital-acquired pneumonia, and it depends on the clinical results of the patients. These broad-spectrum antibiotics, including azithromycin and fluoroquinolones, can be given as empirical therapy to cover all attainable pathogens, continuing the therapy until all pathogenic bacteria are cleared from the body. The combination therapy of azithromycin with hydroxychloroquine showed significant activity against COVID-19 infection [
124].
Table 5 delineates the potential therapeutics against COVID-19.
Some researchers recommend the use of vitamin C supplements that may help to treat the clinical symptoms of the patient by potentially activating the immune system. These supplements help to reduce the severity of pneumonia; however, this requires more clinical evaluation [
131]. Supplements of vitamin D
3 can also be useful for combating the infection. While vitamin D can activate the innate immune system, its decreased value indicates enhanced autoimmunity and susceptibility towards infection. Grant et al. indicated the use of vitamin D in decreasing the probability of respiratory infections caused by COVID-19. As vitamin D induces the antimicrobial peptides such as defensins and cathelicidins that eventually interfere with the viral growth and pro-inflammatory cytokines, more evaluation is needed by randomized controlled trials on a large number of individuals to reach to a conclusion [
132].
Presently, no vaccine exists against COVID-19; however, several clinical trials are underway, as discussed in
Table 6. Scientific research is ongoing by various medical institutes and companies to produce effective vaccines all over the world that can combat COVID-19.
A most recent study published in Lancet claims the production of a vaccine named PittCoVacc developed by the University of Pittsburgh School of Medicine. PittCoVacc uses S-protein, which provides protection by the induction of specific antibodies in the body. Researchers are conducting different analyses on the vaccine, and they are planning a series of tests in humans in the coming months [
129]. Another vaccine named mRNA-1273 is being developed by the National Institute of Allergy and Infectious Disease researchers combined with biotechnology institution Moderna. In this case, mRNA is enclosed in lipid nanoparticles that encode the S-protein of the virus, and then it takes the antigen into the body. In response to antigen, specific antibodies are produced that provide protection against SARS-CoV-2. The clinical trial of the mRNA vaccine is underway, while scientists are working together to evaluate the pathogenesis and clinical features for combating the disease by approaching a series of therapeutic options and development of other vaccines [
133].
University of Oxford, United Kingdom, conducted a randomized clinical trial of the COVID-19 vaccine named ChAdOx1 nCoV-19 vaccine. The estimated participants in this study were about 10,260, and this vaccine was administered intramuscularly. Fortunately, this vaccine appeared to be most effective in participants showing an effective response and has already entered into second and third clinical trials (clinicaltrials.gov).
Finally, there is an emerging concern that patients recovering from COVID-19 may be at risk of reinfection. To address this concern, Bao et al. investigated acquired immunity to SARS-CoV-2 in rhesus macaques. This study published recently suggests that immunity acquired following primary infection with SARS-CoV-2 may protect upon subsequent exposure to the virus [
134]. However, more studies are needed to reach a definitive conclusion.
10. Conclusions
This review provides insight into various aspects of COVID-19, including disease pathophysiology, its clinical representation, mechanism of drugs actions, and impacted population. However, the major concentration of this report was to emphasize potential therapeutics and provide all essential information related to it in one document. On the basis of the class of drugs, multiple classes of drugs have been shown to work primarily to stabilize the patient and, secondly, target the virus for treatment.
The first class of drugs is anti-inflammatory ones that deal with cytokines signaling through targeting various receptors such as IL-6. The major outcome of the treatment is to reduce the inflammation during the therapy. The second group of medications is classic orally administered drugs that have been used for antiviral or anti-parasite ones such as chloroquine phosphate and lopinavir/ritonavir. Among them, remdesivir has been approved by FDA for the emergency room treatment of COVID-19, suggesting its promising potency. In addition to all the mentioned medications, corticosteroids are used to assist with reducing the pro-inflammatory effect. However, owing to their unfavorable side effect profile, their use should be limited to a short period of time in critical patients.
It needs to be emphasized that, among all tested drugs, antiviral compounds such as remdesivir inhibit RNA transcription, leading to reduced viral load. This class of drugs is potentially the most promising tool with which to treat COVID-19 patients. Thus, although there is no solid evidence available to prove their long-term clinical outcomes as of this day, drugs targeting viral RNA are the most promising tools in this battle. Further investigations are required to test a library of synthesized antiviral agents to obtain the most potent candidate to be used in clinics. The ongoing trials of the vaccine will be a hope to provide a vaccine displaying efficacy without side effects to overcome the COVID-19 pandemic to humankind.