Density-Based Separation of Microbial Functional Groups in Activated Sludge
Abstract
1. Introduction
2. Materials and Methods
2.1. WRRFs, Sampling and Sample Preservation
2.2. Density-Based Separation of Biomass
2.3. Total DNA Extraction and Quantitative Real-Time PCR
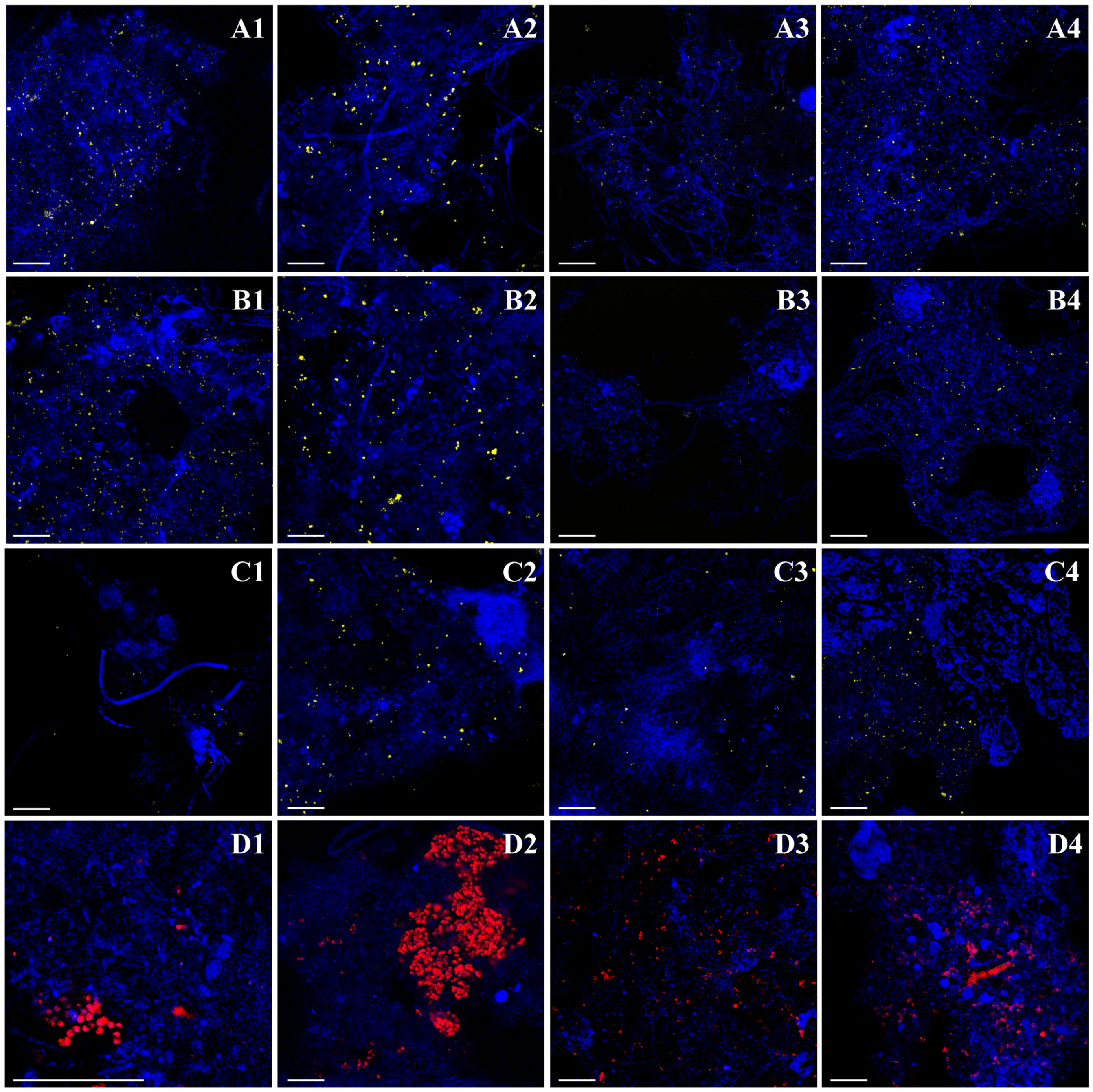
2.4. Fluorescence In Situ Hybridization
2.5. Statistical Analysis
2.6. Nucleotide Sequence Numbers
3. Results
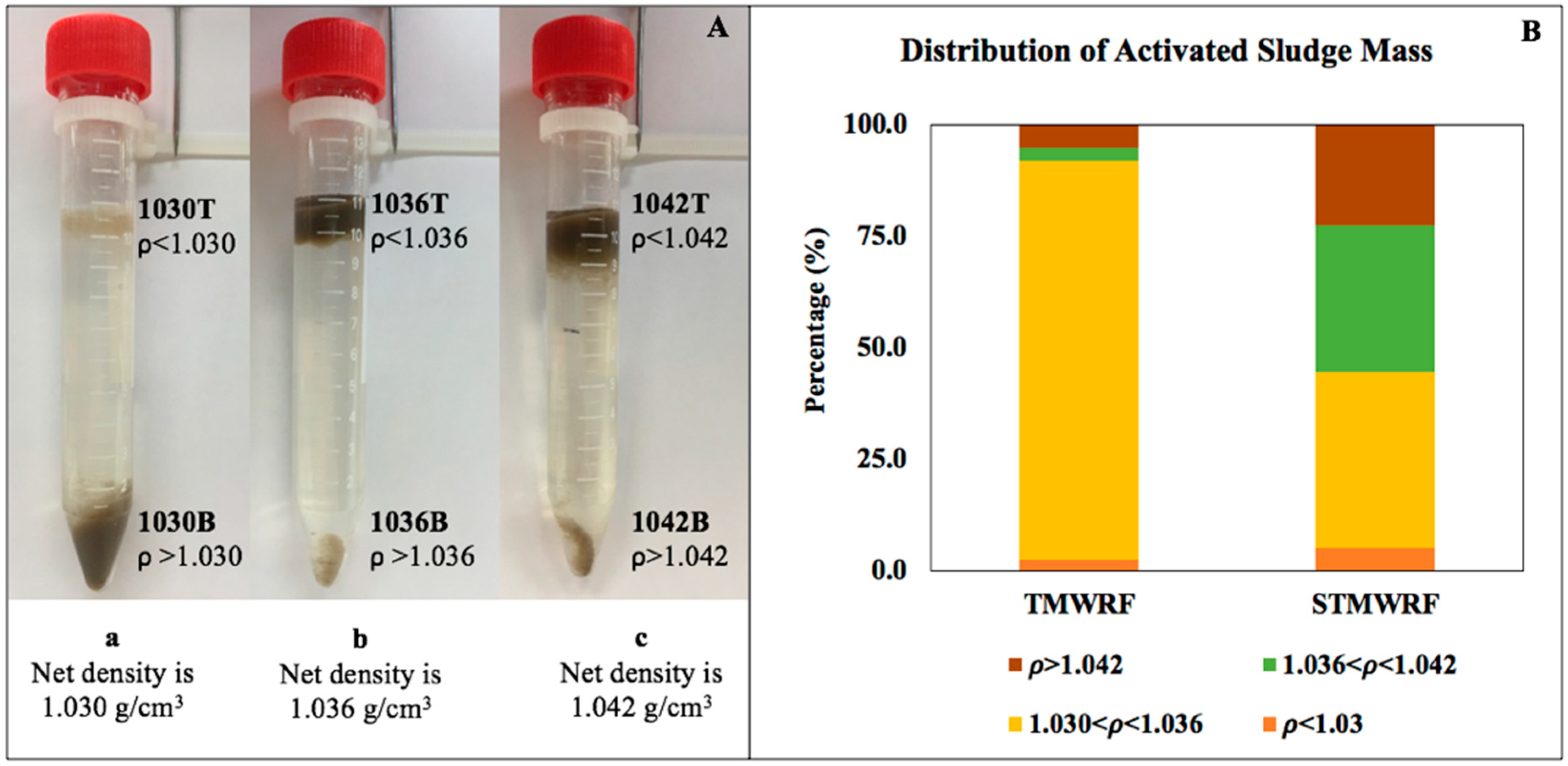
3.1. Density-Based Separation of Biomass
3.2. Microbial Groups in AS of Two Different Domestic WRRFs
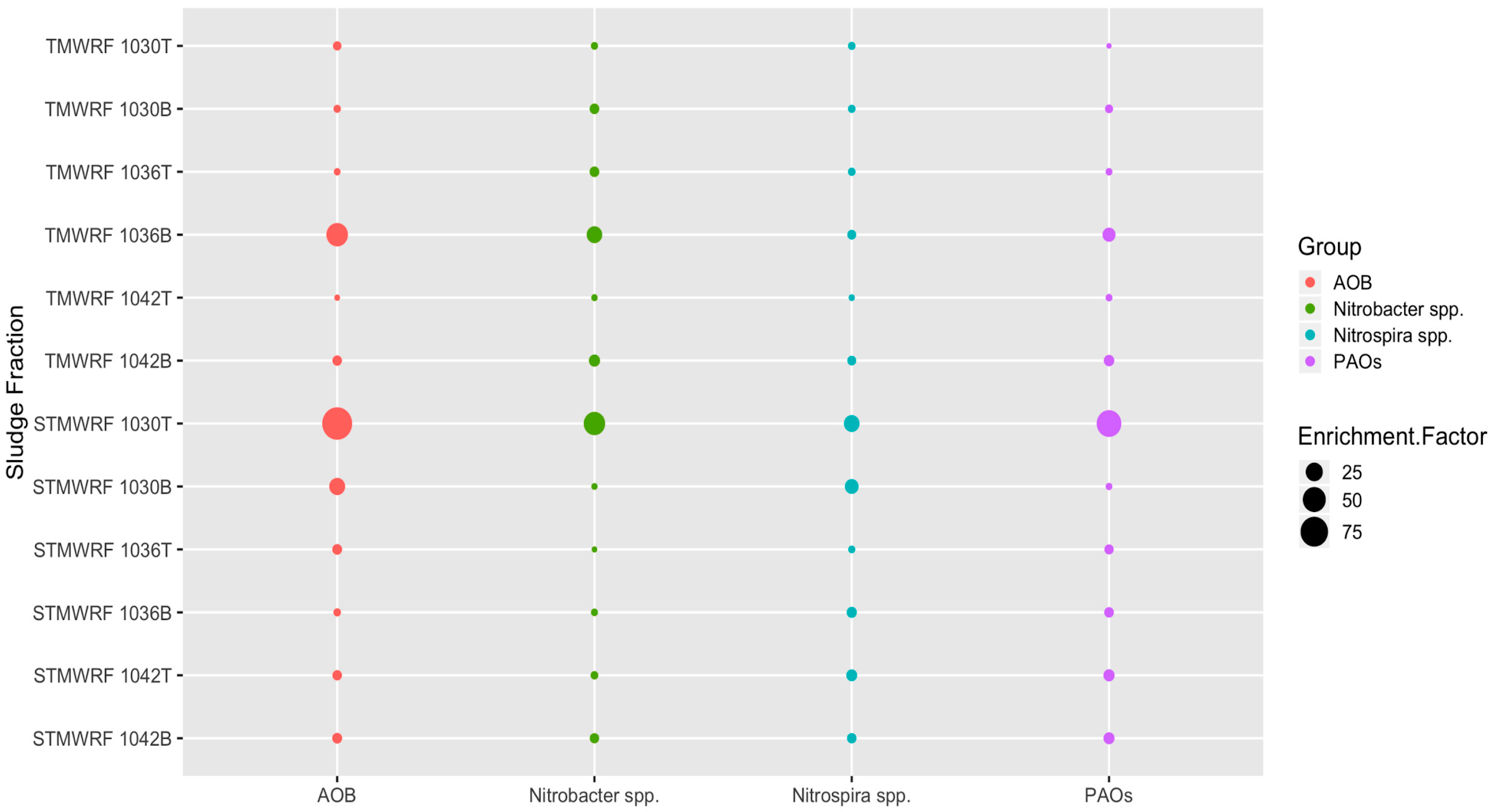
3.3. Abundance of Target Microbial Groups after Density-based Separation
3.3.1. AOB Populations
3.3.2. Nitrobacter spp. NOB Populations
3.3.3. Nitrospira spp. NOB Populations
3.3.4. PAOs Populations
4. Discussion
4.1. AS Density Distribution in Two WRRFs
4.2. Enrichment of Microbial Groups by Density-Based Separation
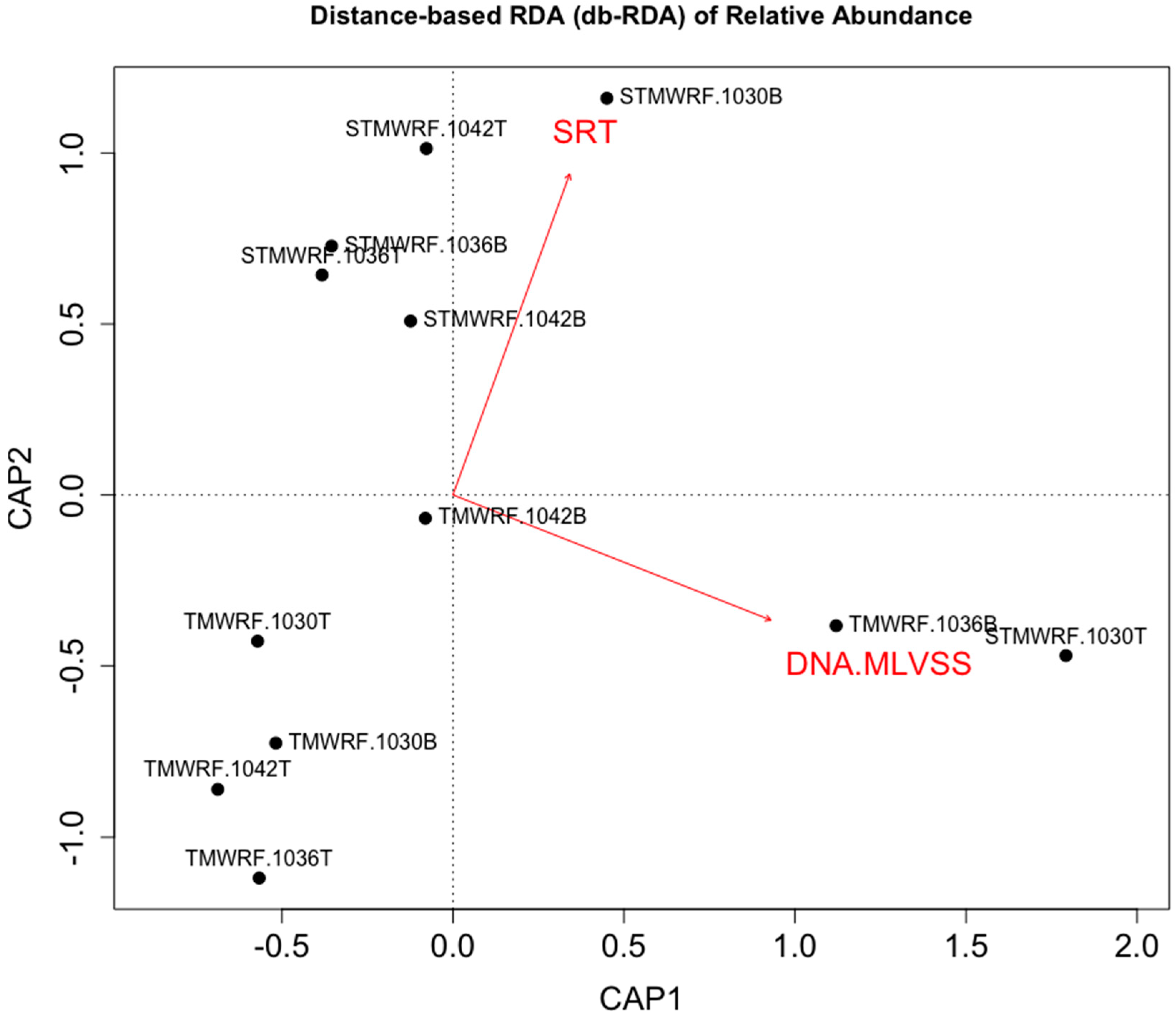
4.3. WRRF Operational Conditions Affecting Density-Based Enrichment of Microbial Functional Groups
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pagilla, K.R.; Jenkins, D.; Kido, W.H. Nocardia control in activated sludge by classifying selectors. Water Environ. Res. 1996, 68, 235–239. [Google Scholar] [CrossRef]
- Li, L.; Pagilla, K.R. Biomass density-function relationships in suspended growth biological processes—A critical review. Water Res. 2017, 111, 274–287. [Google Scholar] [CrossRef] [PubMed]
- Jassby, D.; Xiao, Y.; Schuler, A.J. Biomass density and filament length synergistically affect activated sludge settling: Systematic quantification and modeling. Water Res. 2014, 48, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Schuler, A.J.; Jang, H. Causes of variable biomass density and its effects on settleability in full-scale biological wastewater treatment systems. Environ. Sci. Technol. 2007, 41, 1675–1681. [Google Scholar] [CrossRef]
- Winkler, M.K.H.; Kleerebezem, R.; Strous, M.; Chandran, K.; Van Loosdrecht, M.C.M. Factors influencing the density of aerobic granular sludge. Appl. Microbiol. Biotechnol. 2013, 97, 7459–7468. [Google Scholar] [CrossRef]
- Peng, G.; Ye, F.; Li, Y. Investigation of extracellular polymer substances (EPS) and physicochemical properties of activated sludge from different municipal and industrial wastewater treatment plants. Environ. Technol. 2012, 33, 857–863. [Google Scholar] [CrossRef]
- Badireddy, A.R.; Chellam, S.; Gassman, P.L.; Engelhard, M.H.; Lea, A.S.; Rosso, K.M. Role of extracellular polymeric substances in bioflocculation of activated sludge microorganisms under glucose-controlled conditions. Water Res. 2010, 44, 4505–4516. [Google Scholar] [CrossRef]
- Li, X.Y.; Yang, S.F. Influence of loosely bound extracellular polymeric substances (EPS) on the flocculation, sedimentation and dewaterability of activated sludge. Water Res. 2007, 41, 1022–1030. [Google Scholar] [CrossRef]
- Wilén, B.-M.; Jin, B.; Lant, P. The influence of key chemical constituents in activated sludge on surface and flocculating properties. Water Res. 2003, 37, 2127–2139. [Google Scholar] [CrossRef]
- Fredriksson, N.J.; Hermansson, M.; Wilén, B.-M. Long-term dynamics of the bacterial community in a Swedish full-scale wastewater treatment plant. Environ. Technol. 2017, 40, 912–928. [Google Scholar] [CrossRef]
- Stewart, M.; Palumbo, R.; Rossi, C. Performance testing and long term operations demonstrate successful application of discfilter system for stringent effluent phosphorus limits. Proc. Water Environ. Fed. 2017, 2017, 210–218. [Google Scholar] [CrossRef]
- Schuler, A.J.; Jenkins, D.; Ronen, P. Microbial storage products, biomass density, and settling properties of enhanced biological phosphorus removal activated sludge. Water Sci. Technol. 2001, 43, 173–180. [Google Scholar] [CrossRef]
- Zilles, J.L.; Peccia, J.; Kim, M.W.; Hung, C.H.; Noguera, D.R. Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl. Environ. Microbiol. 2002, 68, 2763–2769. [Google Scholar] [CrossRef]
- Oshiki, M.; Onuki, M.; Satoh, H.; Mino, T. Separation of PHA-accumulating cells in activated sludge based on differences in buoyant density. J. Gen. Appl. Microbiol. 2010, 56, 163–167. [Google Scholar] [CrossRef]
- Schuler, A.J.; Jang, H. Microsphere addition for the study of biomass properties and density effects on settleability in biological wastewater treatment systems. Water Res. 2007, 41, 2163–2170. [Google Scholar] [CrossRef]
- Ju, F.; Zhang, T. Experimental design and bioinformatics analysis for the application of metagenomics in environmental sciences and biotechnology. Environ. Sci. Technol. 2015, 49, 12628–12640. [Google Scholar] [CrossRef]
- Wu, L.; Ning, D.; Zhang, B.; Li, Y.; Zhang, P.; Shan, X.; Zhang, Q.; Brown, M.; Li, Z.; Nostrand, J.D.V.; et al. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol. 2019, 4, 1183–1195. [Google Scholar] [CrossRef]
- Jenkins, D.; Richard, M.G.; Daigger, G.T. Manual on the Causes and Control of Activated Sludge Bulking, Foaming, and Other Solids Separation Problems, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2003; ISBN 978-1-56670-647-6. [Google Scholar]
- Strous, M.; Fuerst, J.A.; Kramer, E.H.M.; Logemann, S.; Van De Pas-Schoonen, K.T.; Webb, R.; Muyzer, G.; Kuenen, J.G. Missing lithotroph identified as new planctomycete. Nature 1999, 400, 446. [Google Scholar] [CrossRef]
- Hathwaik, L.T.; Redelman, D.; Samburova, V.; Zielinska, B.; Shintani, D.K.; Harper, J.F.; Cushman, J.C. Transgressive, reiterative selection by continuous buoyant density gradient centrifugation of Dunaliella salina results in enhanced lipid and starch content. Algal Res. 2015, 9, 194–203. [Google Scholar] [CrossRef]
- Jang, H.; Schuler, A.J. The case for variable density: A new perspective on activated sludge settling. Water Environ. Res. 2007, 79, 2298–2303. [Google Scholar] [CrossRef]
- Zhang, T.; Fang, H.H.P. Applications of real-time polymerase chain reaction for quantification of microorganisms in environmental samples. Appl. Microbiol. Biotechnol. 2006, 70, 281–289. [Google Scholar] [CrossRef]
- You, Y.; Hilpert, M.; Ward, M.J. Detection of a common and persistent tet(L)-carrying plasmid in chicken-waste-impacted farm soil. Appl. Environ. Microbiol. 2012, 78, 3203–3213. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Stubner, S. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen™ detection. J. Microbiol. Methods 2002, 50, 155–164. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Suzuki, M.T.; Taylor, L.T.; DeLong, E.F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 2000, 66, 4605–4614. [Google Scholar] [CrossRef]
- He, S.; Gall, D.L.; McMahon, K.D. “Candidatus Accumulibacter” population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl. Environ. Microbiol. 2007, 73, 5865–5874. [Google Scholar] [CrossRef]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997, 63, 4704–4712. [Google Scholar] [CrossRef]
- Dionisi, H.M.; Layton, A.C.; Harms, G.; Gregory, I.R.; Robinson, K.G.; Sayler, G.S. Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl. Environ. Microbiol. 2002, 68, 245–253. [Google Scholar] [CrossRef]
- Graham, D.W.; Knapp, C.W.; Vleck, E.S.V.; Bloor, K.; Lane, T.B.; Graham, C.E. Experimental demonstration of chaotic instability in biological nitrification. ISME J. 2007, 1, 385. [Google Scholar] [CrossRef]
- Nielsen, P.H.; Daims, H.; Lemmer, H. FISH handbook for biological wastewater treatment: Identification and quantification of microorganisms in activated sludge and biofilms by FISH; IWA Publishing: New York, NY, USA; London, UK, 2009; ISBN 978-1-84339-231-6. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J. Multivariate Analysis of Ecological Communities in R: Vegan Tutorial; University of Oulu: Oulu, Finland, 2007; p. 43. [Google Scholar]
- Mobarry, B.K.; Wagner, M.; Urbain, V.; Rittmann, B.E.; Stahl, D.A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 1996, 62, 2156–2162. [Google Scholar] [CrossRef] [PubMed]
- Juretschko, S.; Timmermann, G.; Schmid, M.; Schleifer, K.-H.; Pommerening-Röser, A.; Koops, H.-P.; Wagner, M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 1998, 64, 3042–3051. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Rath, G.; Koops, H.-P.; Flood, J.; Amann, R. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci. Technol. Lond. 1996, 34, 237–244. [Google Scholar] [CrossRef]
- Crocetti, G.R.; Hugenholtz, P.; Bond, P.L.; Schuler, A.; Keller, J.; Jenkins, D.; Blackall, L.L. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 2000, 66, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Dammel, E.E.; Schroeder, E.D. Density of activated sludge solids. Water Res. 1991, 25, 841–846. [Google Scholar] [CrossRef]
- Sears, K.; Alleman, J.E.; Barnard, J.L.; Oleszkiewicz, J.A. Density and activity characterization of activated sludge flocs. J. Environ. Eng. 2006, 132, 1235–1242. [Google Scholar] [CrossRef]
- Jenkins, D.; Wanner, J. Activated Sludge—100 Years and Counting; IWA Publishing: London, UK, 2014; ISBN 978-1-78040-493-6. [Google Scholar]
- Tandoi, V.; Rossetti, S.; Wanner, J. Activated Sludge Separation Problems: Theory, Control Measures, Practical Experiences; IWA Publishing: London, UK, 2017; ISBN 978-1-78040-863-7. [Google Scholar]
- Dominiak, D.M.; Nielsen, J.L.; Nielsen, P.H. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 2011, 13, 710–721. [Google Scholar] [CrossRef]
- Harms, G.; Layton, A.C.; Dionisi, H.M.; Gregory, I.R.; Garrett, V.M.; Hawkins, S.A.; Robinson, K.G.; Sayler, G.S. Real-Time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 2003, 37, 343–351. [Google Scholar] [CrossRef]
- Rådström, P.; Knutsson, R.; Wolffs, P.; Lövenklev, M.; Löfström, C. Pre-PCR processing. Mol. Biotechnol. 2004, 26, 133–146. [Google Scholar] [CrossRef]
- Saunders, A.M.; Albertsen, M.; Vollertsen, J.; Nielsen, P.H. The activated sludge ecosystem contains a core community of abundant organisms. ISME J. 2016, 10, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Graham, D.W.; Tamaki, H.; Zhang, T. Dominant and novel clades of Candidatus Accumulibacter phosphatis in 18 globally distributed full-scale wastewater treatment plants. Sci. Rep. 2015, 5, 11857. [Google Scholar] [CrossRef] [PubMed]
- Stokholm-Bjerregaard, M.; McIlroy, S.J.; Nierychlo, M.; Karst, S.M.; Albertsen, M.; Nielsen, P.H. A critical assessment of the microorganisms proposed to be important to enhanced biological phosphorus removal in full-scale wastewater treatment systems. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Albertsen, M.; Hansen, L.B.S.; Saunders, A.M.; Nielsen, P.H.; Nielsen, K.L. A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J. 2012, 6, 1094–1106. [Google Scholar] [CrossRef]
- Daims, H.; Lücker, S.; Wagner, M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016, 24, 699–712. [Google Scholar] [CrossRef]
- Zhang, T.; Shao, M.-F.; Ye, L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012, 6, 1137–1147. [Google Scholar] [CrossRef]
- Kong, Y.; Nielsen, J.L.; Nielsen, P.H. Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 2004, 70, 5383–5390. [Google Scholar] [CrossRef]
- Sponza, D.T. Application of toxicity tests into discharges of the pulp-paper industry in Turkey. Ecotoxicol. Environ. Saf. 2003, 54, 74–86. [Google Scholar] [CrossRef]
- Wang, B.-B.; Liu, X.-T.; Chen, J.-M.; Peng, D.-C.; He, F. Composition and functional group characterization of extracellular polymeric substances (EPS) in activated sludge: The impacts of polymerization degree of proteinaceous substrates. Water Res. 2018, 129, 133–142. [Google Scholar] [CrossRef]
| Parameters | TMWRF 1 | STMWRF 2 |
|---|---|---|
| Source of Wastewater | 50% Domestic, 50% industrial | Mainly domestic |
| Biological Process 2 | EBPR | C, N |
| Solids Retention Time (day) | ~2.5 | 12 to 15 |
| Floc size 3 (μm) | 40 | 60 |
| Filament Abundance 4 | Some | Common |
| MLVSS (mg/L) 5 | 31.0 | 32.5 |
| Influent Flow Rate (×1000 m3/day) | 141 | 15 |
| Influent BOD 6 (mg/L) | 250 | 330 |
| Effluent BOD (mg/L) | ~5 | 7 |
| Effluent Total N (mg/L) | 0.2 | 8.4 |
| Effluent Total P (mg/L) | 0.4 | 2.1 |
| Effluent TSS 7 (mg/L) | 2.6 | <5 |
| Suspension Label | Net Density (g/cm3) | Supernatant of AS After Centrifuge (cm3) | Percoll (cm3) | Total Volume (cm3) |
|---|---|---|---|---|
| a | 1.030 | 79.4 | 20.6 | 100 |
| b | 1.036 | 74.6 | 25.4 | 100 |
| c | 1.042 | 69.8 | 30.2 | 100 |
| Target Gene | Primer | Sequence (5′-3′) | Annealing Temp (°C) | Length (bp) | qPCR Performance | Reference | |
|---|---|---|---|---|---|---|---|
| R2 | Efficiency | ||||||
| Eubacterial16S rRNA gene 1 | 27F | AGAGTTTGATCMTGGCTCAG | 60 | ~1500 | -- | -- | [26] |
| 1492R | GGWTACCTTGTTACGACTT | ||||||
| Eubacterial 16S rRNA gene | 1369F | CGGTGAATACGTTCYCGG | 60 | 124 | 0.9966 | 91.09% | [27] |
| 1492R | GGWTACCTTGTTACGACTT | ||||||
| PAOs 16S rRNA gene | 518F | CCAGCAGCCGCGGTAAT | 65 | 351 | 0.9985 | 95.56% | [28] |
| 846R | GTTAGCTACGGCACTAAAAGG | ||||||
| Nitrosomonas spp. amoA gene | amoA-1F | GGGGTTTCTACTGGTGGT | 60 | 491 | 0.9990 | 91.62% | [29] |
| amoA-2R | CCCCTCKGSAAAGCCTTCTTC | ||||||
| Nitrospira spp. 16S rRNA gene | NSR1113F | CCTGCTTTCAGTTGCTACCG | 60 | 150 | 0.9994 | 93.82% | [30] |
| NSR1264R | GTTTGCAGCGCTTTGTACCG | ||||||
| Nitrobacter spp. 16S rRNA gene | Nitro119F | ACCCCTAGCAAATCTCAAAAAACCG | 60 | 227 | 0.9994 | 92.40% | [31] |
| Nitro1423R | CTTCACCCCAGTCGCTGACC | ||||||
| Target Prokaryote | Probe | Sequence (5′-3′) | Formamide (%) | Fluorochrome | Reference |
|---|---|---|---|---|---|
| Eubacteria | EUB 338 | GCTGCCTCCCGTAGGAGT | 35 | ALEX488 | [32] |
| β-Proteobacterial AOB 1 | Nso1225 | CGCCATTGTATTACGTGTGA | 35 | CY3 | [35] |
| Genus Nitrospira (sublineage 1 and 2) 2 | Ntspa1026 | AGCACGCTGGTATTGCTA | 20 | CY3 | [36] |
| Genus Nitrobacter | NIT3 | CCTGTGCTCCATGCTCCG | 40 | CY3 | [37] |
| Candidatus Accumulibacter | PAOmix | PAO462, PAO651 and PAO846 | 35 | CY5 | [38] |
| PAO462 | CCGTCATCTACWCAGGGTTTAAC | 35 | CY5 | ||
| PAO651 | CCCTCTGCCAAACTCCAG | 35 | CY5 | ||
| PAO846 | GTTAGCTACGGCACTAAAAGG | 35 | CY5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; You, Y.; Pagilla, K. Density-Based Separation of Microbial Functional Groups in Activated Sludge. Int. J. Environ. Res. Public Health 2020, 17, 376. https://doi.org/10.3390/ijerph17010376
Li L, You Y, Pagilla K. Density-Based Separation of Microbial Functional Groups in Activated Sludge. International Journal of Environmental Research and Public Health. 2020; 17(1):376. https://doi.org/10.3390/ijerph17010376
Chicago/Turabian StyleLi, Lin, Yaqi You, and Krishna Pagilla. 2020. "Density-Based Separation of Microbial Functional Groups in Activated Sludge" International Journal of Environmental Research and Public Health 17, no. 1: 376. https://doi.org/10.3390/ijerph17010376
APA StyleLi, L., You, Y., & Pagilla, K. (2020). Density-Based Separation of Microbial Functional Groups in Activated Sludge. International Journal of Environmental Research and Public Health, 17(1), 376. https://doi.org/10.3390/ijerph17010376




