Bisphenol a Exposure, DNA Methylation, and Asthma in Children
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Determination of Cases
2.3. Laboratory Method
2.3.1. Exposure Monitoring
2.3.2. Selection of Candidate Genes by Methylation-Dependent Fragment Separation (MDFS)
2.3.3. Analysis of the Methylation Status by Quantitative PCR (qPCR) and Pyrosequencing in a Cohort of 228 Children
2.3.4. Analysis of Plasma Level of MAPK1 Protein
2.4. Statistical Analysis
3. Results
3.1. Selection of the Most Relevant Candidate Genes
3.2. Relationship of MAPK1 5′CGI Methylation Status with BPA Exposure and Asthma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Masoli, M.; Fabian, D.; Holt, S.; Beasley, R. Global Initiative for Asthma (GINA) Program, 2004. The global burden of asthma: Executive summary of the GINA Dissemination Committee Report. Allergy 2004, 59, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.Y.; Chen, Y.J.; Lin, M.W.; Chen, T.J.; Chu, S.Y.; Chen, C.C.; Lee, D.D.; Chang, Y.T.; Wang, W.J.; Liu, H.N. Prevalence of atopic dermatitis, allergic rhinitis and asthma in Taiwan: A national study 2000 to 2007. Acta Derm. Venereol. 2010, 90, 589–594. [Google Scholar] [PubMed]
- Asher, M.I.; Keil, U.; Anderson, H.R.; Beasley, R.; Crane, J.; Martinez, F.; Mitchell, E.A.; Pearce, N.; Sibbald, B.; Stewart, A.W.; et al. International study of asthma and allergies in childhood (ISAAC): Rationale and methods. Eur. Respir. J. 1995, 8, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Brozek, J.L.; Bousquet, J.; Baena-Cagnani, C.E.; Bonini, S.; Canonica, G.W.; Casale, T.B.; van Wijk, R.G.; Ohta, K.; Zuberbier, T.; Schunemann, H.J. Global Allergy and Asthma European Network; Grading of Recommendations Assessment, Development and Evaluation Working Group. Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines: 2010 Revision. J. Allergy Clin. Immunol. 2010, 126, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.F.; Yang, C.C.; Wang, I.J. Association between allergic diseases, allergic sensitization and attention-deficit/hyperactivity disorder in children: A large-scale, population-based study. J. Chin. Med. Assoc. 2017, 81, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Li, S.S.L. Bisphenol A and phthalates exhibit similar toxicogenomics and health effects. Gene 2012, 494, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Donohue, K.M.; Miller, R.L.; Perzanowski, M.S.; Just, A.C.; Hoepner, L.A.; Arunajadai, S.; Canfield, S.; Resnick, D.; Calafat, A.M.; Perera, F.P.; et al. Prenatal and postnatal bisphenol A exposure and asthma development among inner-city children. J. Allergy Clin. Immunol. 2013, 131, 736–742. [Google Scholar] [CrossRef]
- Kwak, E.S.; Just, A.; Whyatt, R.; Miller, R.L. Phthalates, Pesticides, and Bisphenol-A Exposure and the Development of Nonoccupational Asthma and Allergies: How Valid Are the Links? Open Allergy J. 2009, 2, 45–50. [Google Scholar] [CrossRef]
- Wang, I.J.; Chen, C.Y.; Bornehag, C.G. Bisphenol A exposure may increase the risk of development of atopic disorders in children. Int. J. Hyg. Environ. Health 2016, 219, 311–316. [Google Scholar] [CrossRef]
- Newbold, R.R.; Jefferson, W.N.; Padilla-Banks, E. Prenatal Exposure to Bisphenol A at Environmentally Relevant Doses Adversely Affects the Murine Female Reproductive Tract Later in Life. Environ. Health Perspect. 2009, 117, 879–885. [Google Scholar] [CrossRef]
- Onuzulu, C.D.; Rotimi, O.A.; Rotimi, S.O. Epigenetic modifications associated with in utero exposure to endocrine disrupting chemicals BPA, DDT and Pb. Rev. Environ. Health 2019, 34, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Alavian-Ghavanini, A.; Ruegg, J. Understanding epigenetic effects of endocrine disrupting chemicals: From mechanisms to novel test methods. Basic Clin. Pharmacol. Toxicol. 2018, 122, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Chianese, R.; Troisi, J.; Richards, S.; Scafuro, M.; Fasano, S.; Guida, M.; Pierantoni, R.; Meccariello, R. Bisphenol A in reproduction: Epigenetic effects. Curr. Med. Chem. 2018, 25, 748–770. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.J.; Karmaus, W.J.J.; Chen, S.L.; Holloway, J.W.; Ewart, S. Effects of phthalate exposure on asthma may be mediated through alterations in DNA methylation. Clin. Epigenet. 2015, 7, 27. [Google Scholar] [CrossRef]
- Wilson, N.K.; Chuang, J.C.; Morgan, M.K.; Lordo, R.A.; Sheldon, L.S. An observational study of the potential exposures of preschool children to pentachlorophenol, bisphenol-A, and nonylphenol at home and daycare. Environ. Res. 2007, 103, 9–20. [Google Scholar] [CrossRef]
- Stahlhut, R.W.; Welshons, W.V.; Swan, S.H. Bisphenol A data in NHANES suggest longer than expected half-life, substantial nonfood exposure, or both. Environ. Health Perspect. 2009, 117, 784–789. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Hunt, P.A.; Myers, J.P.; VomSaal, F.S. Human exposures to bisphenol A: Mismatches between data and assumptions. Rev. Environ. Health 2013, 28, 37–58. [Google Scholar] [CrossRef]
- Chen, W.L.; Gwo, J.C.; Wang, G.S.; Chen, C.Y. Distribution of feminizing compounds in the aquatic environment and bioaccumulation in wild tilapia tissues. Environ. Sci. Pollut. Res. Int. 2014, 21, 11349–11360. [Google Scholar] [CrossRef]
- Baccarelli, A.; Bollati, V. Epigenetics and environmental chemicals. Curr. Opin. Pediatrics 2009, 21, 243–251. [Google Scholar] [CrossRef]
- Peluso, M.; Bollati, V.; Munnia, A.; Srivatanakul, P.; Jedpiyawongse, A.; Sangrajrang, S.; Piro, S.; Ceppi, M.; Bertazzi, P.A.; Boffetta, P.; et al. DNA methylation differences in exposed workers and nearby residents of the Ma Ta Phut industrial estate, Rayong, Thailand. Int. J. Epidemiol. 2012, 41, 1753–1760. [Google Scholar] [CrossRef]
- Hou, L.; Zhang, X.; Wang, D.; Baccarelli, A. Environmental chemical exposures and human epigenetics. Int. J. Epidemiol. 2012, 41, 79–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.J.; Lin, T.J. FLG P478S polymorphisms and environmental risk factors for the atopic march in Taiwanese children: A prospective cohort study. Ann. Allergy Asthma Immunol. 2015, 114, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Soto-Martinez, M.E.; Avila, L.; Soto-Quiros, M.E. New criteria for the diagnosis and management of asthma in children under 5 years old: GINA Guidelines 2009 [In Spanish]. An. Pediatr. 2009, 71, 91–94. [Google Scholar]
- Niggemann, B.; Jacobsen, L.; Dreborg, S.; Ferdousi, H.A.; Halken, S.; Host, A.; Koivikko, A.; Koller, D.; Norberg, L.A.; Urbanek, R.; et al. PAT Investigator Group Five-year follow-up on the PAT study: Specific immunotherapy and long-term prevention of asthma in children. Allergy 2006, 61, 855–859. [Google Scholar] [CrossRef]
- Gerona, R.R.; Pan, J.; Zota, A.R.; Schwartz, J.M.; Friesen, M.; Taylor, J.A.; Hunt, P.A.; Woodruff, T.J. Direct measurement of Bisphenol A (BPA), BPA glucuronide and BPA sulfate in a diverse and low-income population of pregnant women reveals high exposure, with potential implications for previous exposure estimates: A cross-sectional study. Environ. Health 2016, 15, 50. [Google Scholar] [CrossRef]
- Cayman Chemical Company, 2012. Creatinine (Urinary) Assay Kit. Available online: https://www.caymanchem.com/pdfs/500701.pdf (accessed on 22 April 2015).
- Singh, S.; Li, S.S. Epigenetic effects of environmental chemicals bisphenol A and phthalates. Int. J. Mol. Sci. 2012, 13, 10143–10153. [Google Scholar] [CrossRef]
- Lin, T.J.; Karmaus, W.J.; Chen, M.L.; Hsu, J.C.; Wang, I.J. Interactions between bisphenol A exposure and GSTP1 polymorphisms in childhood asthma. Allergy Asthma Immunol. Res. 2018, 10, 172–179. [Google Scholar]
- Candiloro, I.L.M.; Mikeska, T.; Dobrovic, A. Assessing combined methylation-sensitive high resolution melting and pyrosequencing for the analysis of heterogeneous DNA methylation. Epigenetics 2011, 6, 500–507. [Google Scholar] [CrossRef]
- Ditlevsen, S.; Christensen, U.; Lynch, J.; Damsgaard, M.T.; Keiding, N. The mediation proportion: A structural equation approach for estimating the proportion of exposure effect on outcome explained by an intermediate variable. Epidemiology 2005, 16, 114–120. [Google Scholar] [CrossRef]
- Sager, H.B.; Hulsmans, M.; Lavine, K.J.; Moreira, M.B.; Heidt, T.; Courties, G.; Sun, Y.; Iwamoto, Y.; Tricot, B.; Khan, O.F.; et al. Proliferation and recruitment contribute to myocardial macrophage expansion in chronic heart failure. Circ. Res. 2016, 119, 853–864. [Google Scholar] [CrossRef]
- O’Brien, E.; Dolinoy, D.C.; Mancuso, P. Perinatal bisphenol a exposures increase production of pro-inflammatory mediators in bone marrow-derived mast cells of adult mice. J. Immunotoxicol. 2014, 11, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Rezg, R.; El-Fazaa, S.; Gharbi, N.; Mornagui, B. Bisphenol A and human chronic diseases: Current evidences, possible mechanisms, and future perspectives. Environ. Int. 2014, 64, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Takamoto, M.; Sugane, K. Exposure to bisphenol A prenatally or in adulthood promotes TH2 cytokine production associated with reduction of CD4+ CD25+ regulatory T cells. Environ. Health Perspect. 2008, 116, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Sawai, C.; Anderson, K.; Walser-Kuntz, D. Effect of bisphenol A on murine immune function: Modulation of interferon-γ, IgG2a, and disease symptoms in NZB × NZW F1 mice. Environ. Health Perspect. 2003, 111, 1883–1887. [Google Scholar] [CrossRef] [PubMed]
- Midoro-Horiuti, T.; Tiwari, R.; Watson, C.S.; Goldblum, R.M. Maternal bisphenol a exposure promotes the development of experimental asthma in mouse pups. Environ. Health Perspect. 2010, 118, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Berger, K.; Eskenazi, B.; Balmes, J.; Kogut, K.; Holland, N.; Calafat, A.M.; Harley, K.G. Prenatal high molecular weight phthalates and bisphenol A, and childhood respiratory and allergic outcomes. Pediatric Allergy Immunol. 2019, 30, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Spanier, A.J.; Kahn, R.S.; Kunselman, A.R.; Hornung, R.; Xu, Y.; Calafat, A.M.; Lanphear, B.P. Prenatal exposure to bisphenol A and child wheeze from birth to 3 years of age. Environ. Health Perspect. 2012, 120, 916–920. [Google Scholar] [CrossRef]
- Gascon, M.; Casas, M.; Morales, E.; Valvi, D.; Ballesteros-Gomez, A.; Luque, N.; Rubio, S.; Monfort, N.; Ventura, R.; Martinez, D.; et al. Prenatal exposure to bisphenol A and phthalates and childhood respiratory tract infections and allergy. J. Allergy Clin. Immunol. 2015, 135, 370–378. [Google Scholar] [CrossRef]
- Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Skinner, M.K. Plastics Derived Endocrine Disruptors (BPA, DEHP and DBP) Induce Epigenetic Transgenerational Inheritance of Obesity, Reproductive Disease and Sperm Epimutations. PLoS ONE 2013, 8, e55387. [Google Scholar] [CrossRef]
- Hung, C.H.; Yang, S.N.; Kuo, P.L.; Chu, Y.T.; Chang, H.W.; Wei, W.J.; Huang, S.K.; Jong, Y.J. Modulation of cytokine expression in human myeloid dendritic cells by environmental endocrine-disrupting chemicals involves epigenetic regulation. Environ. Health Perspect. 2010, 118, 67–72. [Google Scholar] [CrossRef]
- Drewes, G.; Ebneth, A.; Preuss, U.; Mandelkow, E.M.; Mandelkow, E. MARK, a novel family of protein kinases that phosphorylate microtubule- associated proteins and trigger microtubule disruption. Cell 1997, 89, 297–308. [Google Scholar] [CrossRef]
- Qi, S.; Fu, W.; Wang, C.; Liu, C.; Quan, C.; Kourouma, A.; Yan, M.; Yu, T.; Duan, P.; Yang, K. BPA-induced apoptosis of rat Sertoli cells through Fas/FasL and JNKs/p38 MAPK pathways. Reprod. Toxicol. 2014, 50, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.W.; Bloom, M.S.; Robinson, W.P.; Kim, D.; Parsons, P.; vom Saal, F.S.; Taylor, J.A.; Steuerwald, A.J.; Fujimoto, V.Y. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum. Reprod. 2012, 27, 1401–1410. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, S.; Mousavi, S.N.; Aghapour, F.; Rezaee, B.; Sadeghi, F.; Moghadamnia, A.A. Induction Effect of Bisphenol A on Gene Expression Involving Hepatic Oxidative Stress in Rat. Oxid. Med. Cell. Longev. 2016, 2016, 6298515. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Oh, P.S.; Lim, K.T. Allergy-related cytokines (IL-4 and TNF-??) are induced by Di(2-ethylhexyl) phthalate and attenuated by plant-originated glycoprotein (75 kDa) in HMC-1 cells. Environ. Toxicol. 2011, 26, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Cruz, I.; Alegria-Torres, J.A.; Montes-Castro, N.; Jimenez-Garza, O.; Quintanilla-Vega, B. Environmental Epigenetic Changes, as Risk Factors for the Development of Diseases in Children: A Systematic Review. Ann. Glob. Health 2018, 84, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Susiarjo, M.; Sasson, I.; Mesaros, C.; Bartolomei, M.S. Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse. PLoS Genet. 2013, 9, e1003401. [Google Scholar] [CrossRef]
- Martin, E.M.; Fry, R.C. Environmental Influences on the Epigenome: Exposure Associated DNA Methylation in Human Populations. Annu. Rev. Public Health 2018, 39, 309–333. [Google Scholar] [CrossRef]
- Viera, L.; Chen, K.; Nel, A.; Lloret, M.G. The impact of air pollutants as an adjuvant for allergic sensitization and asthma. Curr. Allergy Asthma Rep. 2009, 9, 327–333. [Google Scholar] [CrossRef]
- Yu, H.S.; Angkasekwinai, P.; Chang, S.H.; Chung, Y.; Dong, C. Protease allergens induce the expression of IL-25 via Erk and p38 MAPK pathway. J. Korean Med. Sci. 2010, 25, 829–834. [Google Scholar] [CrossRef]
- Wolstenholme, J.T.; Rissman, E.F.; Connelly, J.J. The role of Bisphenol A in shaping the brain, epigenome and behavior. Horm. Behav. 2011, 59, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Yamasaki, A.; Yang, J.; Shan, L.; Halayko, A.J.; Gounni, A.S. IL-17A induces eotaxin-1/CC chemokine ligand 11 expression in human airway smooth muscle cells: Role of MAPK (Erk1/2, JNK, and p38) pathways. J. Immunol. 2006, 177, 4064–4071. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Ci, X.; He, J.; Wei, M.; Yang, X.; Cao, Q.; Li, H.; Guan, S.; Deng, Y.; Pang, D.; et al. A novel anti-inflammatory role for ginkgolide B in asthma via inhibition of the ERK/MAPK signaling pathway. Molecules 2011, 16, 7634–7648. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.H.; Chuang, C.H.; Liu, S.L.; Shen, H.D. Genetic polymorphism of transforming growth factor β1 and tumor necrosis factor α is associated with asthma and modulates the severity of asthma. Respir. Care 2013, 58, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Hermsdorff, H.H.; Mansego, M.L.; Campion, J.; Milagro, F.I.; Zulet, M.A.; Martinez, J.A. TNF-alpha promoter methylation in peripheral white blood cells: Relationship with circulating TNFα, truncal fat and n-6 PUFA intake in young women. Cytokine 2013, 64, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Lakind, J.S.; Levesque, J.; Dumas, P.; Bryan, S.; Clarke, J.; Naiman, D.Q. Comparing United States and Canadian population exposures from national biomonitoring surveys: Bisphenol A intake as a case study. J. Expo. Sci. Environ. Epidemiol. 2012, 22, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Adoamnei, E.; Mendiola, J.; Vela-Soria, F.; Fernasndez, M.F.; Olea, N.; Jorgensen, N.; Swan, S.H.; Torres-Cantero, A.M. Urinary bisphenol A concentrations are associated with reproductive parameters in young men. Environ. Res. 2018, 161, 122–128. [Google Scholar] [CrossRef]
- Ye, X.; Wong, L.Y.; Bishop, A.M.; Calafat, A.M. Variability of urinary concentrations of bisphenol A in spot samples, first morning voids, and 24-h collections. Environ. Health Perspect. 2011, 119, 983–988. [Google Scholar] [CrossRef]
- Remes, S.T.; Pekkanen, J.; Remes, K.; Salonen, R.O.; Korppi, M. In search of childhood asthma: Questionnaire, tests of bronchial hyperresponsiveness, and clinical evaluation. Thorax 2002, 57, 120–126. [Google Scholar] [CrossRef]
| Category | Subjects with Urine and Blood Specimens (N = 228) | Initial Cohort with Urine Specimens (N = 453) | p-Value |
|---|---|---|---|
| Gender (male) (%) | 55.9 | 57.7 | 0.667 |
| Prematurity <37week (%) | 7.6 | 9.0 | 0.702 |
| Maternal age ≥34 years (%) | 24.0 | 17.8 | 0.126 |
| Maternal history of atopy (%Yes) | 40.7 | 35.1 | 0.408 |
| Maternal education College (%) | 25.4 | 30.8 | 0.370 |
| Breast feeding (%Yes) | 67.2 | 76.4 | 0.106 |
| ETS exposure (%Yes) | 59.5 | 46.2 | 0.066 |
| Family income per year >1,500,000 (NT dollars) (%) | 9.1 | 8.0 | 0.531 |
| Asthma (% Yes) | 24.6 | 26.9 | 0.507 |
| Gene (ID) | CpG Island Location | Gene Function | Map |
|---|---|---|---|
| TSS Position | Promoter Methylation Percentage (Met%) (mean ± SD) upon Low and High BPA Exposure | ||
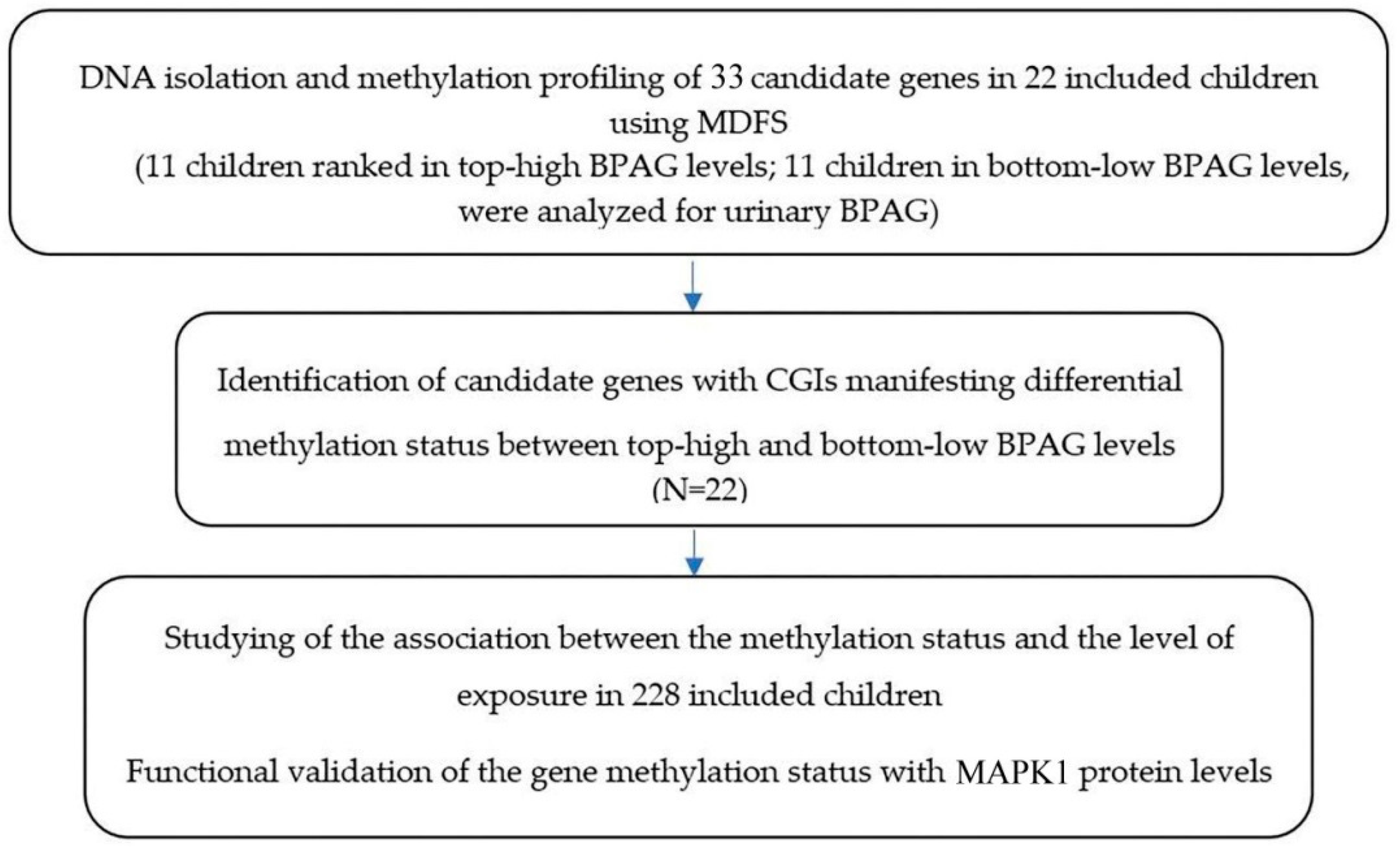
| AR (367) | ChrX: 66763684–66764077 | Development and maintenance of the male sexual phenotype, DNA-binding transcription factor that regulates gene expression | NM_000044 Genome Position: chX 66680589–66860844(+) 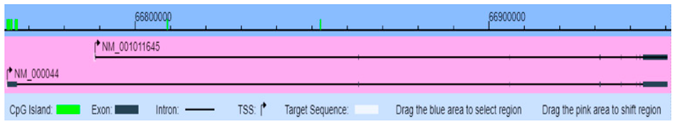 |
| 66763873 | Bottom-low vs. top-high exposure 37.76 ± 23.87 vs. 23.73 ± 18.33 p = 0.138 | ||
| TNFα (7124) | Chr6: 31543344–31544344 | Pro-inflammatory cytokine- stimulates the acute phase reaction and airway inflammation and regulates immune cells | NM_000594 Genome Position: chr6 31651328–31654089(+)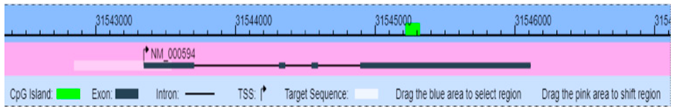 |
| 31543350 | Bottom-low vs. top-high exposure 42.15 ± 36.60 vs. 23.20 ± 22.37 p = 0.16 | ||
| IL-4 (16189) | Chr5: 132035956–132036176 | Activates B-cell and T-cell proliferation induces B-cell class switching to IgE | NM_000589 Genome Position: chr5 132037271–132046267(+)  |
| 132040541 Specific primers for IL-4 gene as below: Forward 5′-GTTGATTGGTTTTAAGTGATTGATAATT-3’ and backward 5′-Biotinylated ATACCCAAATAAATACTCACCTTTCACT-3’. | Bottom-low vs. top-high exposure 89.36 ± 7.65 vs. 85.73 ± 6.99 p = 0.26 | ||
| MAPK1 (5594) | Chr22: 20443948–20551970 | Mediates cell growth, adhesion, survival, and differentiation. Regulates meiosis, mitosis and postmitotic functions | NM_002745 Genome Position: chr22 2044394–20551970(-)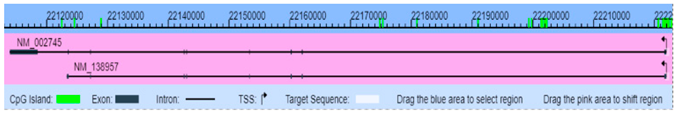 |
| 20447613 | Bottom-low vs. top-high exposure 79.82 ± 5.56 vs. 69.82 ± 5.88 p = 0.001 |
| Sample | BPAG Level (ng/mL) | TNFα | p-Value | AR | p-Value | IL-4 | p-Value | MAPK1 | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| Met% | Met% | Met% a | Met% | ||||||
| Bottom-Low Exposure | |||||||||
| L1 | 0.81 | 11.07 | 0.158 | 50.00 | 0.138 | 87.00 | 0.258 | 82.00 | 0.001 |
| L2 | 0.81 | 85.22 | 33.86 | 88.00 | 87.00 | ||||
| L3 | 0.81 | 73.88 | 6.57 | 71.00 | 81.00 | ||||
| L4 | 0.81 | 14.41 | 50.00 | 88.00 | 75.00 | ||||
| L5 | 6.55 | 67.60 | 8.36 | 100.00 | 86.00 | ||||
| L6 | 6.55 | 14.38 | 79.64 | 89.00 | 79.00 | ||||
| L7 | 6.55 | 11.04 | 27.36 | 91.00 | 75.00 | ||||
| L8 | 6.55 | 94.64 | 52.27 | 89.00 | 76.00 | ||||
| L9 | 6.55 | 76.97 | 65.62 | 88.00 | 84.00 | ||||
| L10 | 6.55 | 6.85 | 28.80 | 92.00 | 84.00 | ||||
| L11 | 6.55 | 7.57 | 12.91 | 100.00 | 69.00 | ||||
| Top-High Exposure | |||||||||
| H1 | 86.55 | 34.31 | 51.32 | 81.00 | 71.00 | ||||
| H2 | 96.26 | 5.59 | 4.57 | 84.00 | 66.00 | ||||
| H3 | 99.03 | 10.16 | 11.95 | 71.00 | 73.00 | ||||
| H4 | 115.60 | 6.14 | 50.43 | 91.00 | 74.00 | ||||
| H5 | 137.20 | 23.49 | 14.76 | 91.00 | 69.00 | ||||
| H6 | 143.20 | 22.76 | 13.19 | 87.00 | 72.00 | ||||
| H7 | 147.40 | 21.59 | 36.34 | 90.00 | 69.00 | ||||
| H8 | 155.10 | 84.09 | 4.78 | 82.00 | 77.00 | ||||
| H9 | 239.50 | 28.02 | 13.66 | 80.00 | 60.00 | ||||
| H10 | 260.50 | 10.39 | 14.32 | 90.00 | 60.00 | ||||
| H11 | 392.00 | 8.63 | 45.65 | 96.00 | 77.00 | ||||
| MAPK1 Promoter Methylation Percentage (Met %) | Ln-BPAG | p-Value |
|---|---|---|
| Adjusted β a | 0.83 | 0.022 * |
| Association Between the MAPK1 5′CGI Methylation Status | Asthma (N = 56) | Non-Asthma (N = 172) | Subjects (N = 228) | OR (95% CIs) | Adjusted OR b (95% CIs) |
|---|---|---|---|---|---|
| Lower methylated MARK1 5′CGI a | 35 (62.5) | 93 (46.5) | 114 (50.0) | 2.17 (1.27–3.68) * | 2.33 (1.01–5.39) * |
| Higher methylated MARK1 5′CGI | 21 (37.5) | 107 (53.5) | 114 (50.0) | 1 | 1 |
| BPA Levels | Ln-BPAG |
|---|---|
| Asthma Adjusted OR (95% CI) 1 | 1.52 (1.12–2.05) * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-F.; Karmaus, W.J.J.; Yang, C.-C.; Chen, M.-L.; Wang, I.-J. Bisphenol a Exposure, DNA Methylation, and Asthma in Children. Int. J. Environ. Res. Public Health 2020, 17, 298. https://doi.org/10.3390/ijerph17010298
Yang C-F, Karmaus WJJ, Yang C-C, Chen M-L, Wang I-J. Bisphenol a Exposure, DNA Methylation, and Asthma in Children. International Journal of Environmental Research and Public Health. 2020; 17(1):298. https://doi.org/10.3390/ijerph17010298
Chicago/Turabian StyleYang, Chia-Feng, Wilfried J. J. Karmaus, Chen-Chang Yang, Mei-Lien Chen, and I-Jen Wang. 2020. "Bisphenol a Exposure, DNA Methylation, and Asthma in Children" International Journal of Environmental Research and Public Health 17, no. 1: 298. https://doi.org/10.3390/ijerph17010298
APA StyleYang, C.-F., Karmaus, W. J. J., Yang, C.-C., Chen, M.-L., & Wang, I.-J. (2020). Bisphenol a Exposure, DNA Methylation, and Asthma in Children. International Journal of Environmental Research and Public Health, 17(1), 298. https://doi.org/10.3390/ijerph17010298






