Effects of Long Noncoding RNA H19 Polymorphisms on Urothelial Cell Carcinoma Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. DNA Extraction and H19 Genotyping
2.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kamat, A.M.; Hahn, N.M.; Efstathiou, J.A.; Lerner, S.P.; Malmstrom, P.U.; Choi, W.; Guo, C.C.; Lotan, Y.; Kassouf, W. Bladder cancer. Lancet 2016, 388, 2796–2810. [Google Scholar] [CrossRef]
- Tung, M.C.; Wen, Y.C.; Wang, S.S.; Lin, Y.W.; Chow, J.M.; Yang, S.F.; Chien, M.H. Impact of long non-coding rna hotair genetic variants on the susceptibility and clinicopathologic characteristics of patients with urothelial cell carcinoma. J. Clin. Med. 2019, 8, 282. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.F.; Yang, C.K.; Ou, Y.C. Urologic cancer in taiwan. Jpn. J. Clin. Oncol. 2016, 46, 605–609. [Google Scholar] [CrossRef]
- Burger, M.; Catto, J.W.; Dalbagni, G.; Grossman, H.B.; Herr, H.; Karakiewicz, P.; Kassouf, W.; Kiemeney, L.A.; La Vecchia, C.; Shariat, S.; et al. Epidemiology and risk factors of urothelial bladder cancer. Eur. Urol. 2013, 63, 234–241. [Google Scholar] [CrossRef]
- Chang, C.H.; Liu, C.S.; Liu, H.J.; Huang, C.P.; Huang, C.Y.; Hsu, H.T.; Liou, S.H.; Chung, C.J. Association between levels of urinary heavy metals and increased risk of urothelial carcinoma. Int. J. Urol. 2016, 23, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Niu, Y. Association between lncrna h19 (rs217727, rs2735971 and rs3024270) polymorphisms and the risk of bladder cancer in chinese population. Minerva Urologica e Nefrologica = Ital. J. Urol. Nephrol. 2018. [Google Scholar] [CrossRef]
- Verhaegh, G.W.; Verkleij, L.; Vermeulen, S.H.; den Heijer, M.; Witjes, J.A.; Kiemeney, L.A. Polymorphisms in the h19 gene and the risk of bladder cancer. Eur. Urol. 2008, 54, 1118–1126. [Google Scholar] [CrossRef]
- Droop, J.; Szarvas, T.; Schulz, W.A.; Niedworok, C.; Niegisch, G.; Scheckenbach, K.; Hoffmann, M.J. Diagnostic and prognostic value of long noncoding rnas as biomarkers in urothelial carcinoma. PLoS ONE 2017, 12, e0176287. [Google Scholar] [CrossRef]
- Wang, J.; Yang, K.; Yuan, W.; Gao, Z. Determination of serum exosomal h19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 9307–9316. [Google Scholar] [CrossRef]
- Yin, Z.; Cui, Z.; Li, H.; Li, J.; Zhou, B. Polymorphisms in the h19 gene and the risk of lung cancer among female never smokers in shenyang, china. BMC Cancer 2018, 18, 893. [Google Scholar] [CrossRef]
- Poirier, F.; Chan, C.T.; Timmons, P.M.; Robertson, E.J.; Evans, M.J.; Rigby, P.W. The murine h19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development 1991, 113, 1105–1114. [Google Scholar]
- Zhang, Y.; Tycko, B. Monoallelic expression of the human h19 gene. Nat. Genet. 1992, 1, 40–44. [Google Scholar] [CrossRef]
- Ariel, I.; Lustig, O.; Schneider, T.; Pizov, G.; Sappir, M.; De-Groot, N.; Hochberg, A. The imprinted h19 gene as a tumor marker in bladder carcinoma. Urology 1995, 45, 335–338. [Google Scholar] [CrossRef]
- Ariel, I.; Ayesh, S.; Perlman, E.J.; Pizov, G.; Tanos, V.; Schneider, T.; Erdmann, V.A.; Podeh, D.; Komitowski, D.; Quasem, A.S.; et al. The product of the imprinted h19 gene is an oncofetal rna. Mol. Pathol. 1997, 50, 34–44. [Google Scholar] [CrossRef]
- Cooper, M.J.; Fischer, M.; Komitowski, D.; Shevelev, A.; Schulze, E.; Ariel, I.; Tykocinski, M.L.; Miron, S.; Ilan, J.; de Groot, N.; et al. Developmentally imprinted genes as markers for bladder tumor progression. J. Urol. 1996, 155, 2120–2127. [Google Scholar] [CrossRef]
- Elkin, M.; Shevelev, A.; Schulze, E.; Tykocinsky, M.; Cooper, M.; Ariel, I.; Pode, D.; Kopf, E.; de Groot, N.; Hochberg, A. The expression of the imprinted h19 and igf-2 genes in human bladder carcinoma. FEBS Lett. 1995, 374, 57–61. [Google Scholar] [CrossRef]
- Zhu, Z.; Xu, L.; Wan, Y.; Zhou, J.; Fu, D.; Chao, H.; Bao, K.; Zeng, T. Inhibition of e-cadherin expression by lnc-rna h19 to facilitate bladder cancer metastasis. Cancer Biomark. Sect. A Dis. Mark. 2018, 22, 275–281. [Google Scholar] [CrossRef]
- Cui, P.; Zhao, Y.; Chu, X.; He, N.; Zheng, H.; Han, J.; Song, F.; Chen, K. Snp rs2071095 in lincrna h19 is associated with breast cancer risk. Breast Cancer Res. Treat. 2018, 171, 161–171. [Google Scholar] [CrossRef]
- Lin, Y.; Fu, F.; Chen, Y.; Qiu, W.; Lin, S.; Yang, P.; Huang, M.; Wang, C. Genetic variants in long noncoding rna h19 contribute to the risk of breast cancer in a southeast china han population. OncoTargets Ther. 2017, 10, 4369–4378. [Google Scholar] [CrossRef]
- Abdollahzadeh, S.; Ghorbian, S. Association of the study between lncrna-h19 gene polymorphisms with the risk of breast cancer. J. Clin. Lab. Anal. 2018, 33, e22826. [Google Scholar] [CrossRef]
- Guo, Q.Y.; Wang, H.; Wang, Y. Lncrna h19 polymorphisms associated with the risk of oscc in chinese population. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3770–3774. [Google Scholar]
- Gong, W.J.; Yin, J.Y.; Li, X.P.; Fang, C.; Xiao, D.; Zhang, W.; Zhou, H.H.; Li, X.; Liu, Z.Q. Association of well-characterized lung cancer lncrna polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016, 37, 8349–8358. [Google Scholar] [CrossRef]
- Li, L.; Guo, G.; Zhang, H.; Zhou, B.; Bai, L.; Chen, H.; Zhao, Y.; Yan, Y. Association between h19 snp rs217727 and lung cancer risk in a chinese population: A case control study. BMC Med. Genet. 2018, 19, 136. [Google Scholar] [CrossRef]
- Yang, M.L.; Huang, Z.; Wang, Q.; Chen, H.H.; Ma, S.N.; Wu, R.; Cai, W.S. The association of polymorphisms in lncrna-h19 with hepatocellular cancer risk and prognosis. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Hua, Q.; Lv, X.; Gu, X.; Chen, Y.; Chu, H.; Du, M.; Gong, W.; Wang, M.; Zhang, Z. Genetic variants in lncrna h19 are associated with the risk of bladder cancer in a chinese population. Mutagenesis 2016, 31, 531–538. [Google Scholar] [CrossRef]
- Cheng, L.; Montironi, R.; Davidson, D.D.; Lopez-Beltran, A. Staging and reporting of urothelial carcinoma of the urinary bladder. Mod. Pathol. 2009, 22 (Suppl. 2), S70–S95. [Google Scholar] [CrossRef]
- Hung, S.C.; Wang, S.S.; Yang, C.K.; Li, J.R.; Cheng, C.L.; Ou, Y.C.; Ho, H.C.; Chiu, K.Y.; Chen, C.S. Comparison of efficacy of adjuvant mec (methotrexate, epirubicin and cisplatin) and gc (gemcitabine and cisplatin) in advanced upper tract urothelial carcinoma. Anticancer Res. 2017, 37, 1875–1883. [Google Scholar]
- Chen, L.M.; Nergard, J.C.; Ni, L.; Rosser, C.J.; Chai, K.X. Long-term exposure to cigarette smoke extract induces hypomethylation at the runx3 and igf2-h19 loci in immortalized human urothelial cells. PLoS ONE 2013, 8, e65513. [Google Scholar] [CrossRef]
- Marsit, C.J.; Karagas, M.R.; Danaee, H.; Liu, M.; Andrew, A.; Schned, A.; Nelson, H.H.; Kelsey, K.T. Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis 2006, 27, 112–116. [Google Scholar] [CrossRef]
- Wolff, E.M.; Liang, G.; Cortez, C.C.; Tsai, Y.C.; Castelao, J.E.; Cortessis, V.K.; Tsao-Wei, D.D.; Groshen, S.; Jones, P.A. Runx3 methylation reveals that bladder tumors are older in patients with a history of smoking. Cancer Res. 2008, 68, 6208–6214. [Google Scholar] [CrossRef]
- Schulz, W.A.; Goering, W. DNA methylation in urothelial carcinoma. Epigenomics 2016, 8, 1415–1428. [Google Scholar] [CrossRef]
- Volanis, D.; Papadopoulos, G.; Doumas, K.; Gkialas, I.; Delakas, D. Molecular mechanisms in urinary bladder carcinogenesis. J. B.U.ON. Off. J. Balkan Union Oncol. 2011, 16, 589–601. [Google Scholar]
- Byun, H.M.; Wong, H.L.; Birnstein, E.A.; Wolff, E.M.; Liang, G.; Yang, A.S. Examination of igf2 and h19 loss of imprinting in bladder cancer. Cancer Res. 2007, 67, 10753–10758. [Google Scholar] [CrossRef]
- Lv, Z.; Xu, Q.; Yuan, Y. A systematic review and meta-analysis of the association between long non-coding rna polymorphisms and cancer risk. Mutat. Res. 2017, 771, 1–14. [Google Scholar] [CrossRef]
- Stewart, S.L.; Cardinez, C.J.; Richardson, L.C.; Norman, L.; Kaufmann, R.; Pechacek, T.F.; Thompson, T.D.; Weir, H.K.; Sabatino, S.A. Surveillance for Cancers Associated with Tobacco Use—United States, 1999–2004; MMWR Surveill Summ; U.S Department of Health and Human Services: Atlanta, GA, USA, 2008; Volume 57, pp. 1–33.
- Casadevall, D.; Kilian, A.Y.; Bellmunt, J. The prognostic role of epigenetic dysregulation in bladder cancer: A systematic review. Cancer Treat. Rev. 2017, 61, 82–93. [Google Scholar] [CrossRef]
- Kim, W.J.; Bae, S.C. Molecular biomarkers in urothelial bladder cancer. Cancer Sci. 2008, 99, 646–652. [Google Scholar] [CrossRef]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 who classification of tumours of the urinary system and male genital organs—Part A: Renal, penile, and testicular tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef]
- Ariel, I.; Sughayer, M.; Fellig, Y.; Pizov, G.; Ayesh, S.; Podeh, D.; Libdeh, B.A.; Levy, C.; Birman, T.; Tykocinski, M.L.; et al. The imprinted h19 gene is a marker of early recurrence in human bladder carcinoma. Mol. Pathol. 2000, 53, 320–323. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted h19 lncrna antagonizes let-7 micrornas. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef]
- Moran, V.A.; Perera, R.J.; Khalil, A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding rnas. Nucleic Acids Res. 2012, 40, 6391–6400. [Google Scholar] [CrossRef]
- Yang, C.; Tang, R.; Ma, X.; Wang, Y.; Luo, D.; Xu, Z.; Zhu, Y.; Yang, L. Tag snps in long non-coding rna h19 contribute to susceptibility to gastric cancer in the chinese han population. Oncotarget 2015, 6, 15311–15320. [Google Scholar] [CrossRef]
- Wu, H.; Zheng, J.; Deng, J.; Hu, M.; You, Y.; Li, N.; Li, W.; Lu, J.; Zhou, Y. A genetic polymorphism in lincrna-uc003opf.1 is associated with susceptibility to esophageal squamous cell carcinoma in chinese populations. Carcinogenesis 2013, 34, 2908–2917. [Google Scholar] [CrossRef]
| Variable | Controls (N = 431) n (%) | Patients (N = 431) n (%) | p Value |
|---|---|---|---|
| Age (yrs) | <0.001 | ||
| ≤65 | 339 (78.7%) | 166 (38.5%) | |
| >65 | 92 (21.3%) | 265 (61.5%) | |
| Mean ± S.D. | 59.4 ± 7.1 | 68.6 ± 11.8 | <0.001 |
| Gender | |||
| Female | 159 (36.9%) | 159 (36.9%) | 1.000 |
| Male | 272 (63.1%) | 272 (63.1%) | |
| Tobacco consumption | 0.071 | ||
| No | 275 (63.8%) | 300 (69.6%) | |
| Yes | 156 (36.2%) | 131 (30.4%) | |
| Stage | |||
| Non muscle invasive tumor (pTa–pT1) | 235 (54.5%) | ||
| Muscle invasive tumor (pT2–pT4) | 196 (45.5%) | ||
| Tumor T status | |||
| Ta | 90 (20.9%) | ||
| T1-T4 | 341 (79.1%) | ||
| Lymph node status | |||
| N0 | 380 (88.2%) | ||
| N1+N2 | 51 (11.8%) | ||
| Metastasis | |||
| M0 | 417 (96.8%) | ||
| M1 | 14 (3.2%) | ||
| Histopathologic grading | |||
| Low grade | 53 (12.3%) | ||
| High grade | 378 (87.7%) |
| Variable | Controls (N = 431) n (%) | Patients (N = 431) n (%) | OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| rs2177727 | ||||
| CC | 191 (44.3%) | 185 (42.9%) | 1.000 (reference) | 1.000 (reference) |
| CT | 188 (43.6%) | 202 (46.9%) | 1.109 (0.836–1.473) | 1.072 (0.784–1.467) |
| TT | 52 (12.1%) | 44 (10.2%) | 0.874 (0.557–1.369) | 0.758 (0.458–1.255) |
| CT+TT | 240 (55.7%) | 246 (57.1%) | 1.058 (0.808–1.385) | 1.002 (0.744–1.350) |
| rs2107425 | ||||
| CC | 171 (39.7%) | 152 (35.3%) | 1.000 (reference) | 1.000 (reference) |
| CT | 190 (44.1%) | 213 (49.4%) | 1.261 (0.941–1.691) | 1.245 (0.901–1.721) |
| TT | 70 (16.2%) | 66 (15.3%) | 1.061 (0.710–1.584) | 0.978 (0.625–1.529) |
| CT+TT | 260 (60.3%) | 279 (64.7%) | 1.207 (0.916–1.591) | 1.172 (0.865–1.589) |
| rs2839698 | ||||
| CC | 192 (44.5%) | 206 (47.8%) | 1.000 (reference) | 1.000 (reference) |
| CT | 184 (42.7%) | 170 (39.4%) | 0.861 (0.647–1.147) | 0.907 (0.660–1.247) |
| TT | 55 (12.8%) | 55 (12.8%) | 0.932 (0.611–1.422) | 0.881 (0.555–1.399) |
| CT+TT | 239 (55.5%) | 225 (52.2%) | 0.877 (0.671–1.147) | 0.901 (0.669–1.212) |
| rs3024270 | ||||
| CC | 120 (27.8%) | 114 (26.5%) | 1.000 (reference) | 1.000 (reference) |
| GC | 208 (48.3%) | 210 (48.7%) | 1.063 (0.772–1.464) | 1.316 (0.920–1.884) |
| GG | 103 (23.9%) | 107 (24.8%) | 1.094 (0.753–1.587) | 1.189 (0.786–1.797) |
| GC+GG | 311 (72.2%) | 317 (73.5%) | 1.073 (0.795–1.449) | 1.271 (0.909–1.778) |
| rs3741219 | ||||
| AA | 185 (42.9%) | 192 (44.5%) | 1.000 (reference) | 1.000 (reference) |
| GA | 190 (44.1%) | 181 (42%) | 0.918 (0.689–1.223) | 0.978 (0.711–1.345) |
| GG | 56 (13%) | 58 (13.5%) | 0.998 (0.656–1.517) | 0.912 (0.576–1.444) |
| GA+GG | 246 (57.1%) | 239 (55.5%) | 0.936 (0.715–1.225) | 0.962 (0.713–1.296) |
| H19 (rs2177727) | ||||
|---|---|---|---|---|
| Variable | CC (%) (n = 185) | CT+TT (%) (n = 246) | OR (95% CI) | p Value |
| Stage | ||||
| Non muscle invasive tumor (pTa–pT1) | 112 (60.5%) | 123 (50%) | 1.000 (reference) | |
| Muscle invasive tumor (pT2–pT4) | 73 (39.5%) | 123 (50%) | 1.534 (1.042–2.258) | 0.030 |
| Tumor T status | ||||
| Ta | 41 (22.2%) | 49 (19.9%) | 1.000 (reference) | |
| T1–T4 | 144 (77.8%) | 197 (80.1%) | 1.145 (0.717–1.826) | 0.571 |
| Lymph node status | ||||
| N0 | 165 (89.2%) | 215 (87.4%) | 1.000 (reference) | |
| N1+N2 | 20 (10.8%) | 31 (12.6%) | 1.190 (0.654–2.162) | 0.569 |
| Metastasis | ||||
| M0 | 179 (96.8%) | 238 (96.7%) | 1.000 (reference) | |
| M1 | 6 (3.2%) | 8 (3.3%) | 1.003 (0.342–2.941) | 0.996 |
| Histopathologic grading | ||||
| Low grade | 22 (11.9%) | 31 (12.6%) | 1.000 (reference) | |
| High grade | 163 (88.1%) | 215 (87.4%) | 0.936 (0.523–1.677) | 0.824 |
| H19 (rs2107425) | ||||
|---|---|---|---|---|
| Variable | CC (%) (n = 152) | CT+TT (%) (n = 279) | OR (95% CI) | p Value |
| Stage | ||||
| Non muscle invasive tumor (pTa–pT1) | 94 (61.8%) | 141 (50.5%) | 1.000 (reference) | |
| Muscle invasive tumor (pT2–pT4) | 58 (38.2%) | 138 (49.5%) | 1.586 (1.06–2.373) | 0.025 |
| Tumor T status | ||||
| Ta | 37 (24.3%) | 53 (19%) | 1.000 (reference) | |
| T1–T4 | 115 (75.7%) | 226 (81%) | 1.372 (0.852–2.209) | 0.193 |
| Lymph node status | ||||
| N0 | 136 (89.5%) | 244 (87.5%) | 1.000 (reference) | |
| N1+N2 | 16 (10.5%) | 35 (12.5%) | 1.219 (0.651–2.284) | 0.536 |
| Metastasis | ||||
| M0 | 150 (98.7%) | 267 (95.7%) | 1.000 (reference) | |
| M1 | 2 (1.3%) | 12 (4.3%) | 3.371 (0.744–15.262) | 0.115 |
| Histopathologic grading | ||||
| Low grade | 16 (10.5%) | 37 (13.3%) | 1.000 (reference) | |
| High grade | 136 (89.5%) | 242 (86.7%) | 0.769 (0.413–1.435) | 0.410 |
| Variable | N | Number of Disease-Specific Mortality | HR (95% CI) | AHR (95% CI) |
|---|---|---|---|---|
| rs2177727 | ||||
| CC | 112 | 17 | 1.000 (reference) | 1.000 (reference) |
| CT+TT | 152 | 30 | 1.308 (0.721–2.373) | 1.404 (0.770–2.561) |
| rs2107425 | ||||
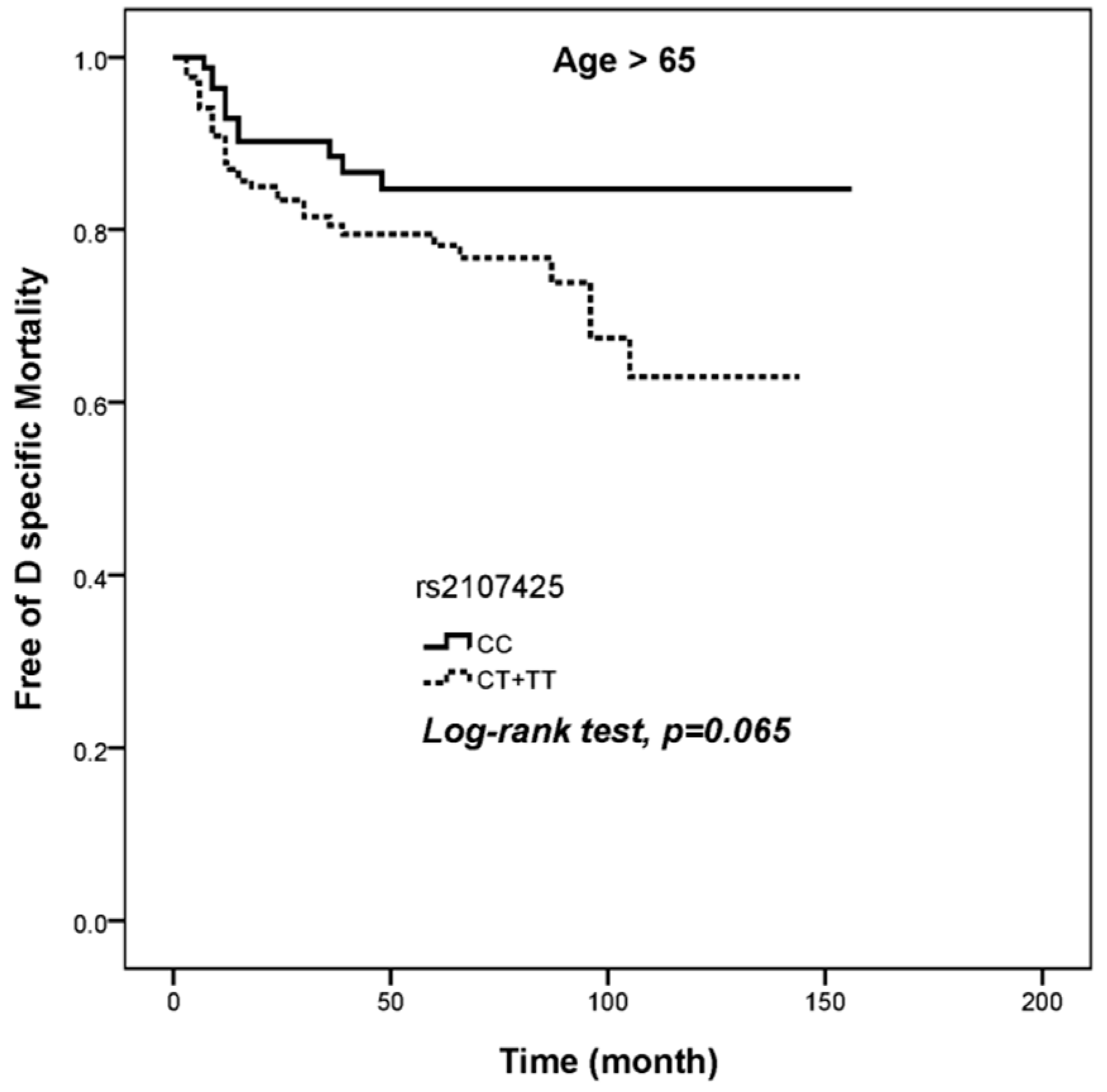
| CC | 93 | 11 | 1.000 (reference) | 1.000 (reference) |
| CT+TT | 171 | 36 | 1.862 (0.948–3.659) | 2.043 (1.029–4.059) |
| rs2839698 | ||||
| CC | 128 | 27 | 1.000 (reference) | 1.000 (reference) |
| CT+TT | 136 | 20 | 0.669 (0.375–1.195) | 0.668 (0.374–1.194) |
| rs3024270 | ||||
| CC | 75 | 14 | 1.000 (reference) | 1.000 (reference) |
| GC+GG | 189 | 33 | 0.934 (0.499–1.749) | 0.892 (0.470–1.691) |
| rs3741219 | ||||
| AA | 120 | 25 | 1.000 (reference) | 1.000 (reference) |
| GA+GG | 144 | 22 | 0.678 (0.382–1.204) | 0.683 (0.383–1.217) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, P.-J.; Hsieh, M.-J.; Hung, T.-W.; Wang, S.-S.; Chen, S.-C.; Lee, M.-C.; Yang, S.-F.; Chou, Y.-E. Effects of Long Noncoding RNA H19 Polymorphisms on Urothelial Cell Carcinoma Development. Int. J. Environ. Res. Public Health 2019, 16, 1322. https://doi.org/10.3390/ijerph16081322
Yang P-J, Hsieh M-J, Hung T-W, Wang S-S, Chen S-C, Lee M-C, Yang S-F, Chou Y-E. Effects of Long Noncoding RNA H19 Polymorphisms on Urothelial Cell Carcinoma Development. International Journal of Environmental Research and Public Health. 2019; 16(8):1322. https://doi.org/10.3390/ijerph16081322
Chicago/Turabian StyleYang, Po-Jen, Ming-Ju Hsieh, Tung-Wei Hung, Shian-Shiang Wang, Shiuan-Chih Chen, Meng-Chih Lee, Shun-Fa Yang, and Ying-Erh Chou. 2019. "Effects of Long Noncoding RNA H19 Polymorphisms on Urothelial Cell Carcinoma Development" International Journal of Environmental Research and Public Health 16, no. 8: 1322. https://doi.org/10.3390/ijerph16081322
APA StyleYang, P.-J., Hsieh, M.-J., Hung, T.-W., Wang, S.-S., Chen, S.-C., Lee, M.-C., Yang, S.-F., & Chou, Y.-E. (2019). Effects of Long Noncoding RNA H19 Polymorphisms on Urothelial Cell Carcinoma Development. International Journal of Environmental Research and Public Health, 16(8), 1322. https://doi.org/10.3390/ijerph16081322








