Identification of Pathogenic Bacteria from Public Libraries via Proteomics Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Bacterial Cultures, Collection of Bacteria
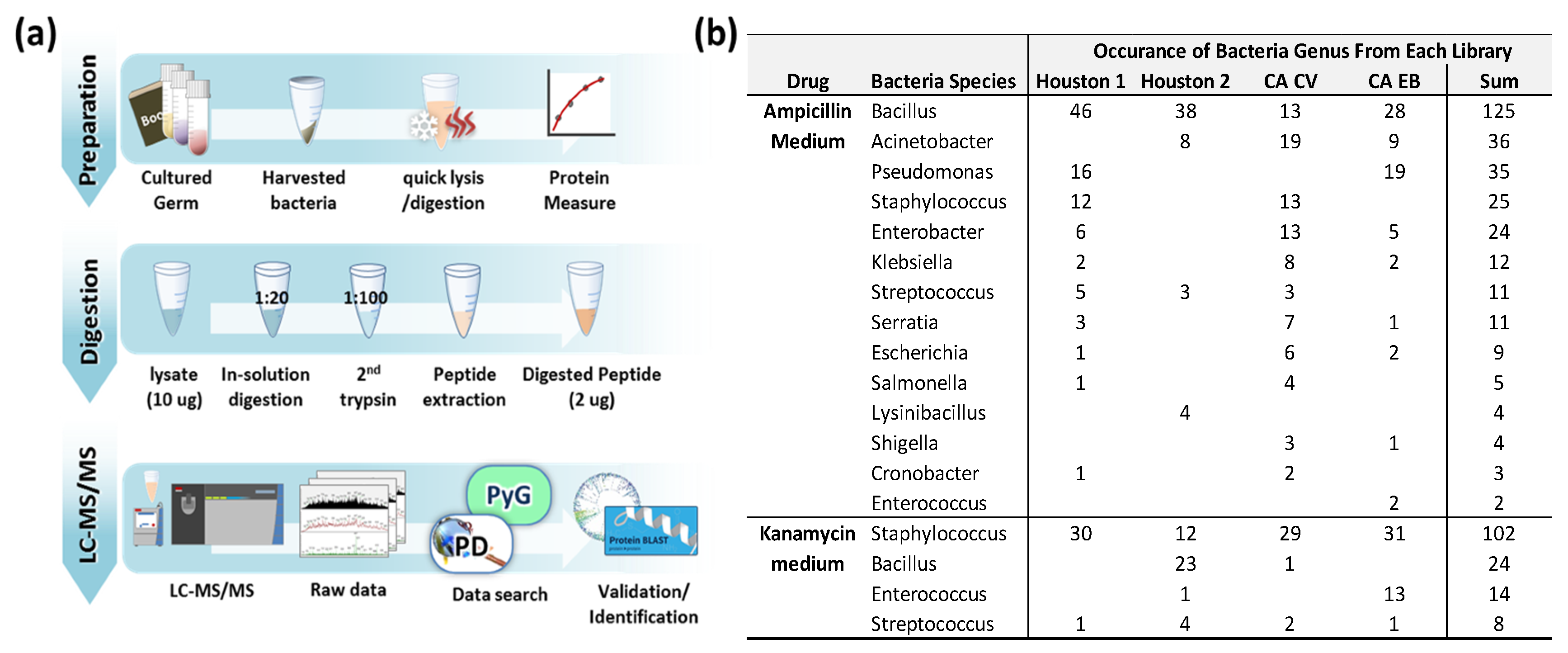
2.3. Proteomics Analysis of Bacteria
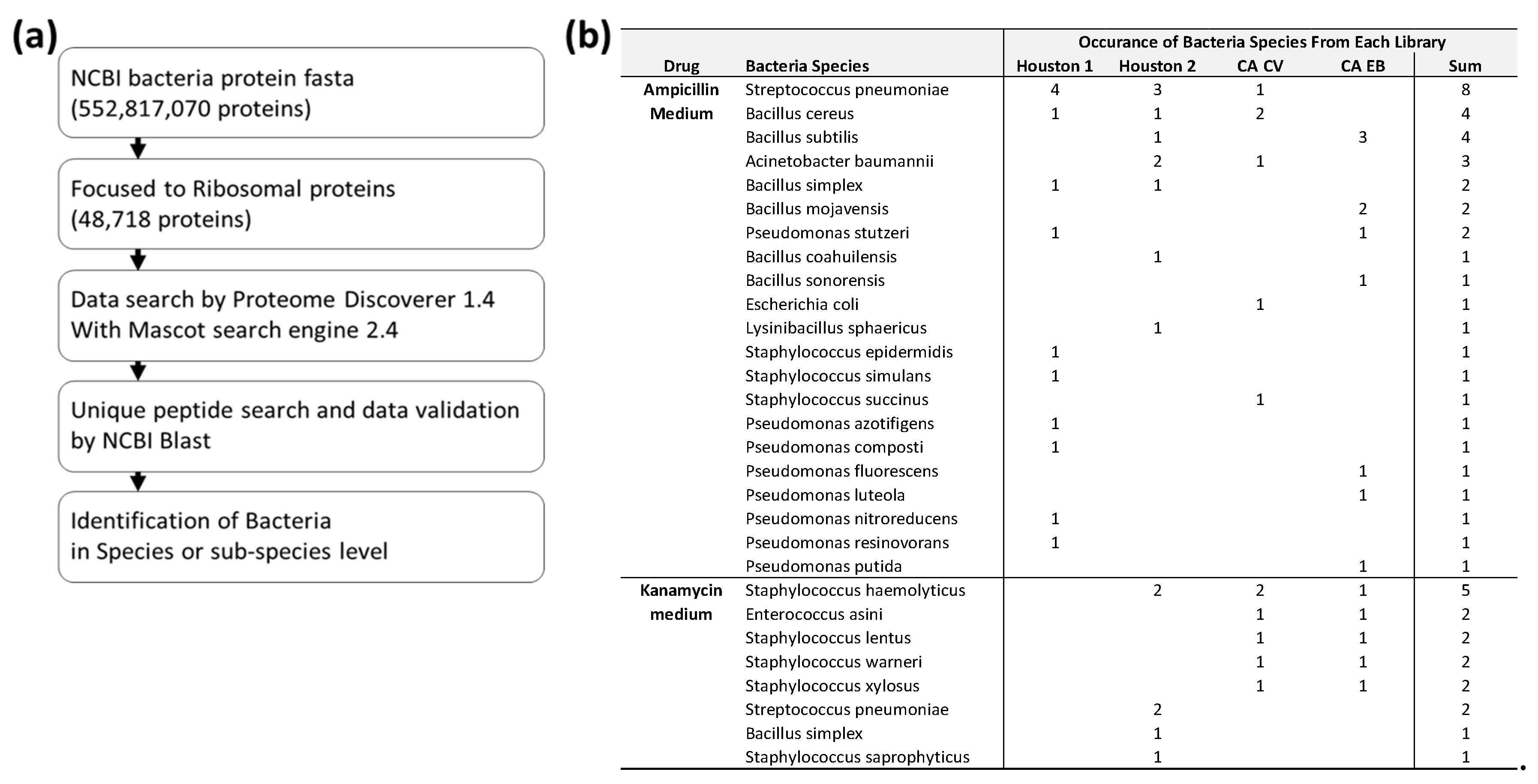
2.4. Data Analysis with Commercial and In-House Computer Software
2.5. Data Availability
3. Result
3.1. Growth of Bactria in Antibiotic Medium
3.2. Identified Bacteria Grown in Ampicillin or Kanamycin-Containing Medium
3.3. Identified of Bacteria at Species Level
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- McClary, A. Beware the deadly books: A forgotten episode in library history. J. Libr. Hist. 1985, 20, 427–433. [Google Scholar] [PubMed]
- Brook, S.J.; Brook, I. Are public library books contaminated by bacteria? J. Clin. Epidemiol. 1994, 47, 1173–1174. [Google Scholar] [CrossRef]
- Rafiei, H.; Chadeganipour, M.; Ojaghi, R.; Maracy, M.R.; Nouri, R. The comparison of printed resources bacterial contamination in libraries of Al-Zahra Hospital and Sciences Faculty of Isfahan University and the determination of their antibiotic sensitivity pattern. J. Educ. Health Promot. 2017, 6, 19. [Google Scholar]
- Gambale, W.; Croce, J.; Costa-Manso, E.; Croce, M.; Sales, M. Library fungi at the University of Sao Paulo and their relationship with respiratory allergy. J. Investig. Allergol. Clin. Immunol. 1993, 3, 45–50. [Google Scholar] [PubMed]
- Research Team Seeks Answers on How Dirty the Library Is. Available online: https://universe.byu.edu/2011/12/04/dirty-library-books/ (accessed on 24 January 2019).
- Library Books Could Come with a Side of Germs. Available online: https://www.sciencenewsforstudents.org/blog/eureka-lab/library-books-could-come-side-germs (accessed on 24 January 2019).
- Bradley, J.S.; Byington, C.L.; Shah, S.S.; Alverson, B.; Carter, E.R.; Harrison, C.; Kaplan, S.L.; Mace, S.E.; McCracken, G.H., Jr.; Moore, M.R.; et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin. Infect. Dis. 2011, 53, e25–e76. [Google Scholar] [CrossRef]
- Simmonds, P.G. Whole microorganisms studied by pyrolysis-gas chromatography-mass spectrometry: Significance for extraterrestrial life detection experiments. Appl. Microbiol. 1970, 20, 567–572. [Google Scholar]
- Mohamed, H.; Miloud, B.; Zohra, F.; Garcia-Arenzana, J.M.; Veloso, A.; Rodriguez-Couto, S. Isolation and Characterization of actinobacteria from Algerian Sahara Soils with antimicrobial activities. Int. J. Mol. Cell Med. 2017, 6, 109–120. [Google Scholar] [PubMed]
- Avanzi, I.R.; Gracioso, L.H.; Baltazar, M.D.; Karolski, B.; Perpetuo, E.A.; Do Nascimento, C.A. Rapid bacteria identification from environmental mining samples using MALDI-TOF MS analysis. Environ. Sci. Pollut. Res. Int. 2017, 24, 3717–3726. [Google Scholar] [CrossRef]
- Nacef, M.; Chevalier, M.; Chollet, S.; Drider, D.; Flahaut, C. MALDI-TOF mass spectrometry for the identification of lactic acid bacteria isolated from a French cheese: The Maroilles. Int. J. Food Microbiol. 2017, 247, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.P.; Reddy, P.M. Identification of pathogens by mass spectrometry. Clin. Chem. 2010, 56, 525–536. [Google Scholar] [CrossRef]
- Sauer, S.; Freiwald, A.; Maier, T.; Kube, M.; Reinhardt, R.; Kostrzewa, M.; Geider, K. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS ONE 2008, 3, e2843. [Google Scholar] [CrossRef]
- Krasny, L.; Rohlova, E.; Ruzickova, H.; Santrucek, J.; Hynek, R.; Hochel, I. Differentiation of Cronobacter spp. by tryptic digestion of the cell suspension followed by MALDI-TOF MS analysis. J. Microbiol. Methods 2014, 98, 105–113. [Google Scholar] [CrossRef]
- He, Y.; Li, H.; Lu, X.; Stratton, C.W.; Tang, Y.W. Mass spectrometry biotyper system identifies enteric bacterial pathogens directly from colonies grown on selective stool culture media. J. Clin. Microbiol. 2010, 48, 3888–3892. [Google Scholar] [CrossRef]
- Richter, S.S.; Sercia, L.; Branda, J.A.; Burnham, C.A.; Bythrow, M.; Ferraro, M.J.; Garner, O.B.; Ginocchio, C.C.; Jennemann, R.; Lewinski, M.A.; et al. Identification of Enterobacteriaceae by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using the VITEK MS system. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1571–1578. [Google Scholar] [CrossRef]
- Clark, C.G.; Kruczkiewicz, P.; Guan, C.; McCorrister, S.J.; Chong, P.; Wylie, J.; Van Caeseele, P.; Tabor, H.A.; Snarr, P.; Gilmour, M.W.; et al. Evaluation of MALDI-TOF mass spectroscopy methods for determination of Escherichia coli pathotypes. J. Microbiol. Methods 2013, 94, 180–191. [Google Scholar] [CrossRef]
- Angeletti, S.; Dicuonzo, G.; D’Agostino, A.; Avola, A.; Crea, F.; Palazzo, C.; Dedej, E.; De Florio, L. Turnaround time of positive blood cultures after the introduction of matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. New Microbiol. 2015, 38, 379–386. [Google Scholar]
- Curtoni, A.; Cipriani, R.; Marra, E.S.; Barbui, A.M.; Cavallo, R.; Costa, C. Rapid identification of microorganisms from positive blood culture by MALDI-TOF MS after short-term incubation on solid medium. Curr. Microbiol. 2017, 74, 97–102. [Google Scholar] [CrossRef]
- Sloan, A.; Wang, G.; Cheng, K. Traditional approaches versus mass spectrometry in bacterial identification and typing. Clin. Chim. Acta 2017, 473, 180–185. [Google Scholar] [CrossRef]
- Afshinnekoo, E.; Meydan, C.; Chowdhury, S.; Jaroudi, D.; Boyer, C.; Bernstein, N.; Maritz, J.M.; Reeves, D.; Gandara, J.; Chhangawala, S.; et al. Geospatial resolution of human and bacterial diversity with city-scale metagenomics. Cell Syst. 2015, 1, 97. [Google Scholar] [CrossRef]
- Jung, S.Y.; Choi, J.M.; Rousseaux, M.W.; Malovannaya, A.; Kim, J.J.; Kutzera, J.; Wang, Y.; Huang, Y.; Zhu, W.; Maity, S.; et al. An anatomically resolved mouse brain proteome reveals parkinson disease-relevant pathways. Mol. Cell Proteom. 2017, 16, 581–593. [Google Scholar] [CrossRef]
- Goloborodko, A.A.; Levitsky, L.I.; Ivanov, M.V.; Gorshkov, M.V. Pyteomics–A Python framework for exploratory data analysis and rapid software prototyping in proteomics. J. Am. Soc. Mass Spectrom. 2013, 24, 301–304. [Google Scholar] [CrossRef]
- Suarez, S.; Ferroni, A.; Lotz, A.; Jolley, K.A.; Guérin, P.; Leto, J.; Dauphin, B.; Jamet, A.; Maiden, M.C.; Nassif, X.; et al. Ribosomal proteins as biomarkers for bacterial identification by mass spectrometry in the clinical microbiology laboratory. J. Microbiol. Methods 2013, 94, 390–396. [Google Scholar] [CrossRef]
- Pineda, F.J.; Antoine, M.D.; Demirev, P.A.; Feldman, A.B.; Jackman, J.; Longenecker, M.; Lin, J.S. Microorganism identification by matrix-assisted laser/desorption ionization mass spectrometry and model-derived ribosomal protein biomarkers. Anal. Chem. 2003, 75, 3817–3822. [Google Scholar] [CrossRef]
- Ryzhov, V.; Fenselau, C. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 2001, 73, 746–750. [Google Scholar] [CrossRef]
- Teramoto, K.; Sato, H.; Sun, L.; Torimura, M.; Tao, H.; Yoshikawa, H.; Hotta, Y.; Hosoda, A.; Tamura, H. Phylogenetic classification of Pseudomonas putida strains by MALDI-MS using ribosomal subunit proteins as biomarkers. Anal. Chem. 2007, 79, 8712–8719. [Google Scholar] [CrossRef]
- Teramoto, K.; Sato, H.; Sun, L.; Torimura, M.; Tao, H. A simple intact protein analysis by MALDI-MS for characterization of ribosomal proteins of two genome-sequenced lactic acid bacteria and verification of their amino acid sequences. J. Proteome. Res. 2007, 6, 3899–3907. [Google Scholar] [CrossRef]
- Falcone, M.; Campanile, F.; Giannella, M.; Borbone, S.; Stefani, S.; Venditti, M. Staphylococcus haemolyticus endocarditis: Clinical and microbiologic analysis of 4 cases. Diagn. Microbiol. Infect. Dis. 2007, 57, 325–331. [Google Scholar] [CrossRef]
- Barros, E.M.; Ceotto, H.; Bastos, M.C.; Dos Santos, K.R.; Giambiagi-Demarval, M. Staphylococcus haemolyticus as an important hospital pathogen and carrier of methicillin resistance genes. J. Clin. Microbiol. 2012, 50, 166–168. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. A framework for human microbiome research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Ortqvist, A.; Hedlund, J.; Kalin, M. Streptococcus pneumoniae: Epidemiology, risk factors, and clinical features. Semin. Respir. Crit Care Med. 2005, 26, 563–574. [Google Scholar] [CrossRef]
- Torres, A.; Blasi, F.; Dartois, N.; Akova, M. Which individuals are at increased risk of pneumococcal disease and why? Impact of COPD, asthma, smoking, diabetes, and/or chronic heart disease on community-acquired pneumonia and invasive pneumococcal disease. Thorax 2015, 70, 984–989. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Montefour, K.; Frieden, J.; Hurst, S.; Helmich, C.; Headley, D.; Martin, M.; Boyle, D.A. Acinetobacter baumannii: An emerging multidrug-resistant pathogen in critical care. Crit Care Nurse 2008, 28, 15–25. [Google Scholar]
- Sebeny, P.J.; Riddle, M.S.; Petersen, K. Acinetobacter baumannii skin and soft-tissue infection associated with war trauma. Clin. Infect. Dis. 2008, 47, 444–449. [Google Scholar] [CrossRef]
- Cerqueira, G.M.; Peleg, A.Y. Insights into Acinetobacter baumannii pathogenicity. IUBMB Life 2011, 63, 1055–1060. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Spencer, R.C. Bacillus anthracis. J. Clin. Pathol. 2003, 56, 182–187. [Google Scholar] [CrossRef]
- Kotiranta, A.; Lounatmaa, K.; Haapasalo, M. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2000, 2, 189–198. [Google Scholar] [CrossRef]
- Palleroni, N.J. The Pseudomonas story. Environ. Microbiol. 2010, 12, 1377–1383. [Google Scholar] [CrossRef]
- Zubarev, R.A.; Makarov, A. Orbitrap mass spectrometry. Anal. Chem. 2013, 85, 5288–5296. [Google Scholar] [CrossRef]
- Hessling, B.; Buttner, K.; Hecker, M.; Becher, D. Global relative quantification with liquid chromatography-matrix-assisted laser desorption ionization time-of-flight (LC-MALDI-TOF)—Cross-validation with LTQ-Orbitrap proves reliability and reveals complementary ionization preferences. Mol. Cell Proteom. 2013, 12, 2911–2920. [Google Scholar] [CrossRef]
- Balazova, T.; Makovcova, J.; Sedo, O.; Slany, M.; Faldyna, M.; Zdrahal, Z. The influence of culture conditions on the identification of Mycobacterium species by MALDI-TOF MS profiling. FEMS Microbiol. Lett. 2014, 353, 77–84. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, R.H.; Kim, M.; Bhatt, B.; Choi, J.M.; Roh, J.H. Identification of Pathogenic Bacteria from Public Libraries via Proteomics Analysis. Int. J. Environ. Res. Public Health 2019, 16, 912. https://doi.org/10.3390/ijerph16060912
Jung RH, Kim M, Bhatt B, Choi JM, Roh JH. Identification of Pathogenic Bacteria from Public Libraries via Proteomics Analysis. International Journal of Environmental Research and Public Health. 2019; 16(6):912. https://doi.org/10.3390/ijerph16060912
Chicago/Turabian StyleJung, Ryan Hyunjae, Minzae Kim, Bhoomi Bhatt, Jong Min Choi, and Jung H. Roh. 2019. "Identification of Pathogenic Bacteria from Public Libraries via Proteomics Analysis" International Journal of Environmental Research and Public Health 16, no. 6: 912. https://doi.org/10.3390/ijerph16060912
APA StyleJung, R. H., Kim, M., Bhatt, B., Choi, J. M., & Roh, J. H. (2019). Identification of Pathogenic Bacteria from Public Libraries via Proteomics Analysis. International Journal of Environmental Research and Public Health, 16(6), 912. https://doi.org/10.3390/ijerph16060912




