The State-of-the Art of Environmental Toxicogenomics: Challenges and Perspectives of “Omics” Approaches Directed to Toxicant Mixtures
Abstract
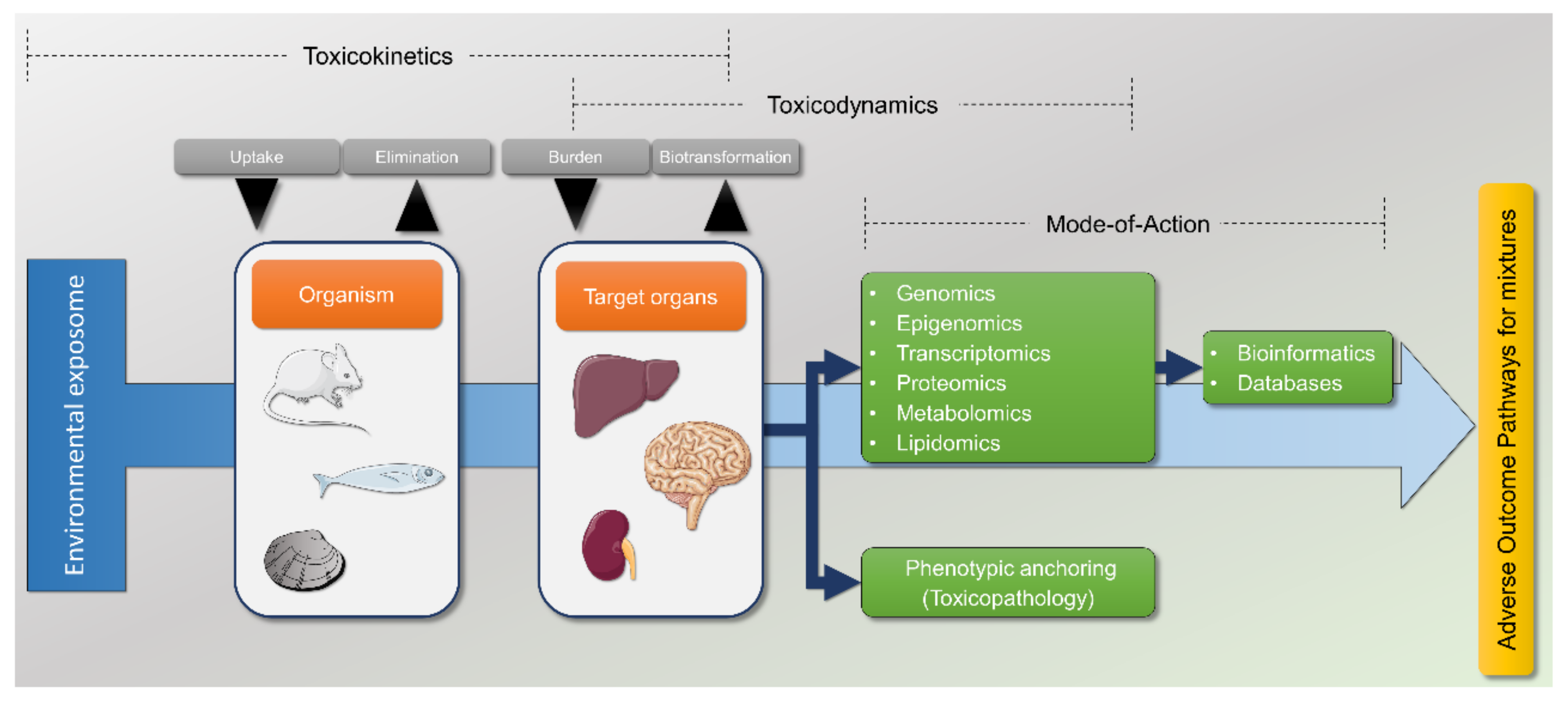
1. Introduction
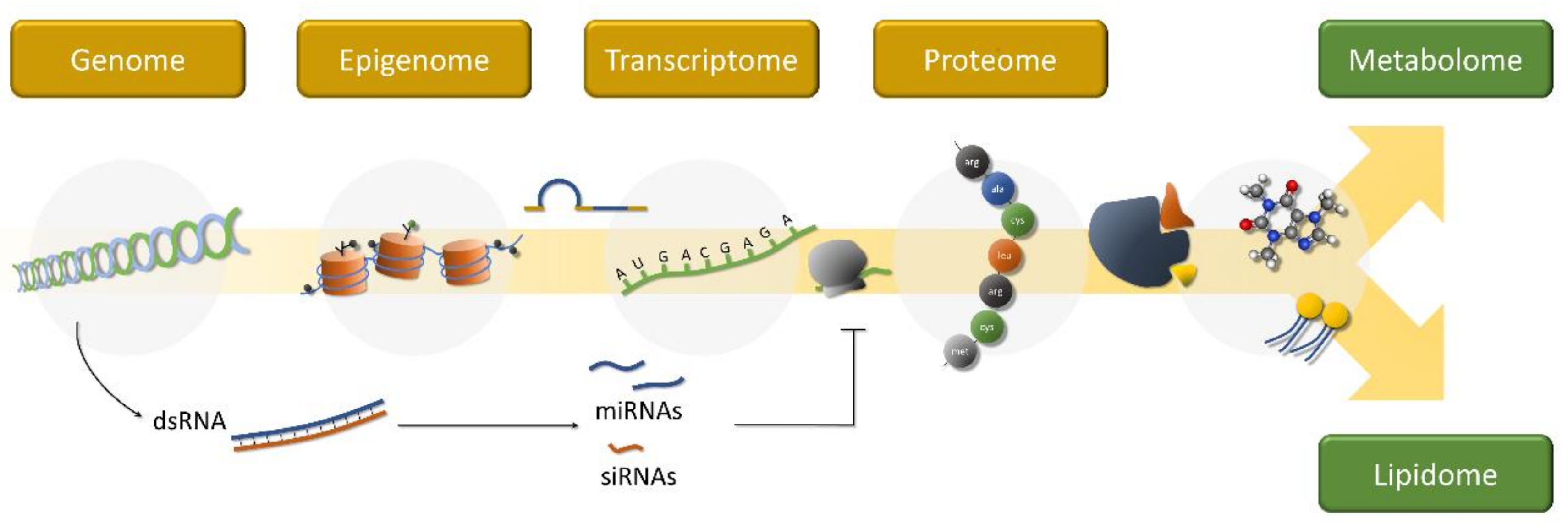
2. What Are “Omics”?
3. Genomics and Epigenomics
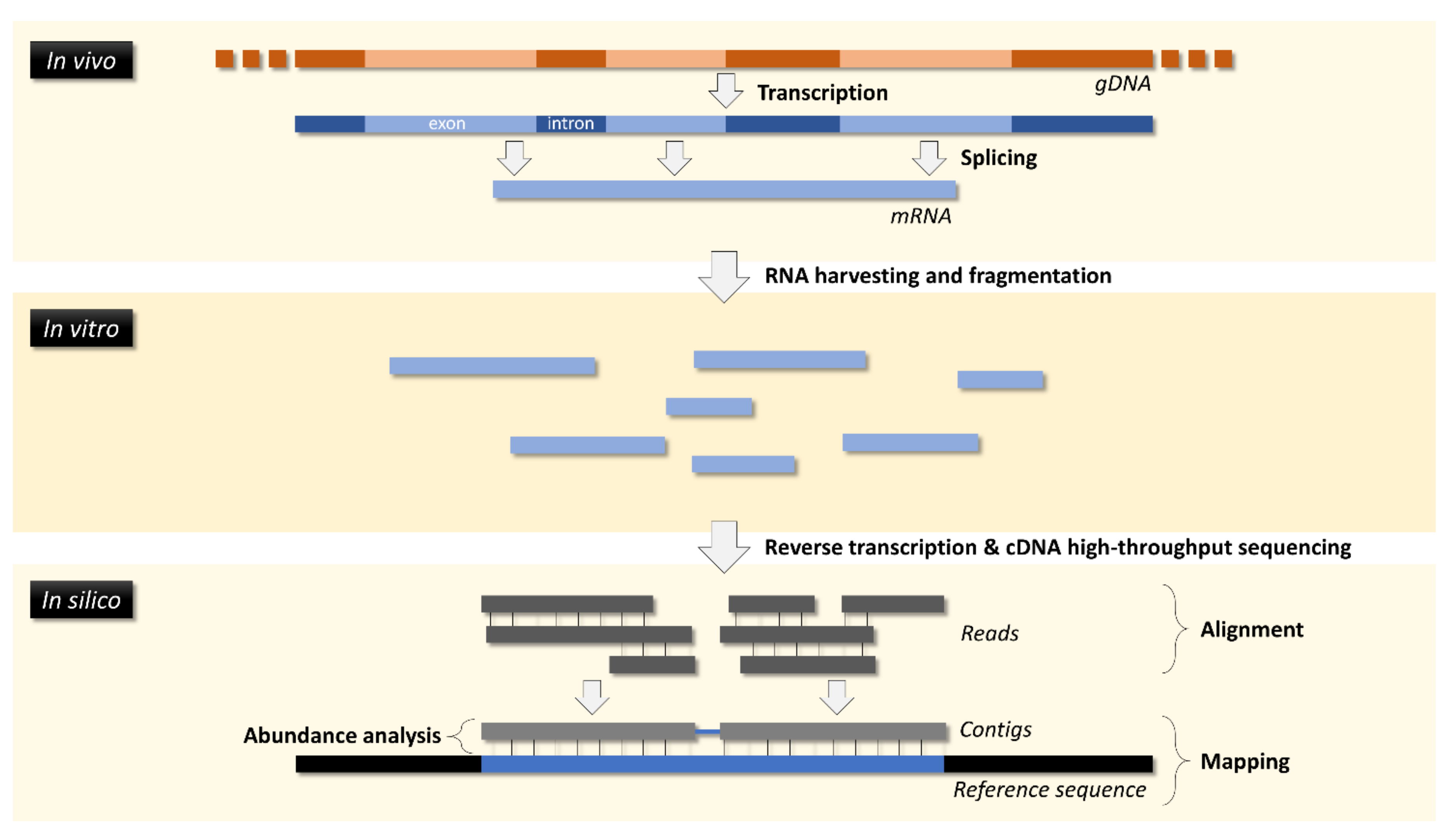
4. Transcriptomics
5. Proteomics
6. Metabolomics and Lipidomics
7. Multi-Omics
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2DE | Two-dimensional gel electrophoresis |
| Ahr | Aryl hydrocarbon receptor |
| AOP | Adverse outcome pathway |
| DEG | Differentially expressed gene |
| EDA | Effects-direct analysis |
| EDC | Endocrine disruptor compound |
| ERA | Environmental risk assessment |
| GC-MS | Gas-chromatography mass spectrometry |
| GWAS | Genome-wide association studies |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MOA | Mode-of-action |
| MS | Mass spectrometry |
| NGS | Next-generation sequencing |
| NMR | Nuclear magnetic resonance |
| PAH | Polycyclic aromatic hydrocarbon |
| PCB | Polychlorinated biphenyl |
| qRT-PCR | Quantitative (real time) reverse-transcription polymerase chain reaction |
| RNA-Seq | Next-generation whole-transcriptome RNA sequencing |
| SNP | Single-nucleotide polymorphism. |
References
- Harada, M. Minamata disease: Methylmercury poisoning in Japan caused by environmental pollution. Crit. Rev. Toxicol. 1995, 25, 1–24. [Google Scholar] [CrossRef]
- Doll, R. Mortality from lung cancer in asbestos workers. Br. J. Ind. Med. 1955, 12, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Hernández, A.F.; Gil, F.; Lacasaña, M. Toxicological interactions of pesticide mixtures: An update. Arch. Toxicol. 2017, 91, 3211–3223. [Google Scholar] [CrossRef] [PubMed]
- Allan, S.E.; Smith, B.W.; Tanguay, R.L.; Anderson, K.A. Bridging environmental mixtures and toxic effects. Environ. Toxicol. Chem. 2012, 31, 2877–2887. [Google Scholar] [CrossRef] [PubMed]
- Perkins, J.T.; Petriello, M.C.; Xu, L.; Stromberg, A.; Hennig, B. An open-sourced statistical application for identifying complex toxicological interactions of environmental pollutants. Rev. Environ. Health 2017, 32, 23–26. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kar, S.; Leszczynski, J. Exploration of computational approaches to predict the toxicity of chemical mixtures. Toxics 2019, 7, 15. [Google Scholar] [CrossRef]
- Macdonald, D.D.; Carr, R.S.; Calder, F.D.; Long, E.R.; Ingersoll, C.G. Development and evaluation of sediment quality guidelines for Florida coastal waters. Ecotoxicology 1996, 5, 253–278. [Google Scholar] [CrossRef]
- Franzellitti, S.; Buratti, S.; Valbonesi, P.; Fabbri, E. The mode of action MOA approach reveals interactive effects of environmental pharmaceuticals on Mytilus galloprovincialis. Aquat. Toxicol. 2013, 140–141, 249–256. [Google Scholar] [CrossRef]
- Kienzler, A.; Bopp, S.K.; van der Linden, S.; Berggren, E.; Worth, A. Regulatory assessment of chemical mixtures: Requirements, current approaches and future perspectives. Regul. Toxicol. Pharmacol. 2016, 80, 321–334. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, M.; Borlak, J.; Tong, W. A decade of toxicogenomic research and its contribution to toxicological science. Toxicol. Sci. 2012, 130, 217–228. [Google Scholar] [CrossRef]
- Schmidt, C.W. Toxicogenomics: An emerging discipline. Environ. Health Perspect. 2002, 110, A750–A755. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hood, L.; Heath, J.R.; Phelps, M.E.; Lin, B. Systems biology and new technologies enable predictive and preventative medicine. Science 2004, 306, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Ankley, G.T.; Bennett, R.S.; Erickson, R.J.; Hoff, D.J.; Hornung, M.W.; Johnson, R.D.; Mount, D.R.; Nichols, J.W.; Russom, C.L.; Schmieder, P.K.; et al. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 2010, 29, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.M.; Fadeel, B. Emerging systems biology approaches in nanotoxicology: Towards a mechanism-based understanding of nanomaterial hazard and risk. Toxicol. Appl. Pharmacol. 2016, 299, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Oziolor, E.M.; Bickham, J.W.; Matson, C.W. Evolutionary toxicology in an omics world. Evol. Appl. 2017, 10, 752–761. [Google Scholar] [CrossRef]
- Altenburger, R.; Scholz, S.; Schmitt-Jansen, M.; Busch, W.; Escher, B.I. Mixture toxicity revisited from a toxicogenomic perspective. Environ. Sci. Technol. 2012, 46, 2508–2522. [Google Scholar] [CrossRef]
- Simbürger, J.M.B.; Dettmer, K.; Oefner, P.J.; Reinders, J. Optimizing the SWATH-MS-workflow for label-free proteomics. J. Proteomics 2016, 145, 137–140. [Google Scholar] [CrossRef]
- Vidova, V.; Spacil, Z. A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition. Anal. Chim. Acta 2017, 964, 7–23. [Google Scholar] [CrossRef]
- Kernaleguen, M.; Daviaud, C.; Shen, Y.; Bonnet, E.; Renault, V.; Deleuze, J.-F.; Mauger, F.; Tost, J. Whole-genome bisulfite sequencing for the analysis of genome-wide DNA methylation and hydroxymethylation patterns at single-nucleotide resolution. In Epigenome Editing: Methods and Protocols, Methods in Molecular Biology 1767; Jeltsch, A., Rots, M.G., Eds.; Humana Press: Totowa, NJ, USA, 2018; pp. 311–350. [Google Scholar] [CrossRef]
- Basu, A.K. DNA damage, mutagenesis and cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef]
- Giacomini, K.M.; Yee, S.W.; Mushiroda, T.; Weinshilboum, R.M.; Ratain, M.J.; Kubo, M. Genome-wide association studies of drug response and toxicity: An oportunity for genome medicine. Nat. Rev. Drug Discov. 2017, 16, 1–8. [Google Scholar] [CrossRef]
- Olden, K.; Guthrie, J. Genomics: Implications for toxicology. Mutat. Res. 2001, 473, 3–10. [Google Scholar] [CrossRef]
- Prokopec, S.D.; Lu, A.; Lee, S.C.S.; Yao, C.Q.; Sun, R.X.; Watson, J.D.; Soliymani, R.; de Borja, R.; Wong, A.; Sam, M.; et al. Comparative toxicoproteogenomics of mouse and rat liver identifies TCDD-resistance genes. Arch. Toxicol. 2019, 93, 2961–2978. [Google Scholar] [CrossRef] [PubMed]
- Maslov, A.Y.; Quispe-Tintaya, W.; Gorbacheva, T.; White, R.R.; Vijg, J. High-throughput sequencing in mutation detection: a new generation of genotoxicity tests? Mutat. Res. 2015, 776, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Pan, B.; Chen, T. Evaluation of chemical mutagenicity using next generation sequencing: A review. J. Environ. Sci. Health Part C Environ. Carcinog. Ecotoxicol. Rev. 2017, 35, 140–158. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Broz, A.K.; Sharbrough, J.; Wu, Z. Detecting rare mutations and DNA damage with sequencing-based methods. Trends Biotechnol. 2018, 36, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Wamucho, A.; Unrine, J.M.; Kieran, T.J.; Glenn, T.C.; Schultz, C.L.; Farman, M.; Svendsen, C.; Spurgeon, D.J.; Tsyusko, O.V. Genomic mutations after multigenerational exposure of Caenorhabditis elegans to pristine and sulfidized silver nanoparticles. Environ. Pollut. 2019, 254, 113078. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, S.; Fujita, Y.; Yamane, M.; Morita, O.; Honda, H. A genome-wide mutation analysis method enabling high-throughput identification of chemical mutagen signatures. Sci. Rep. 2018, 8, 9583. [Google Scholar] [CrossRef]
- Duca, R.-C.; Grova, N.; Ghosh, M.; Do, J.-M.; Hoet, P.H.M.; Vanoirbeek, J.A.J.; Appenzeller, B.M.R.; Godderis, L. Exposure to polycyclic aromatic hydrocarbons leads to non-monotonic modulation of DNA and RNA hydroxymethylation in a rat model. Sci. Rep. 2018, 8, 10577. [Google Scholar] [CrossRef]
- Marczylo, E.L.; Jacobs, M.N.; Gant, T.W. Environmentally induced epigenetic toxicity: Potential public health concerns. Crit. Rev. Toxicol. 2016, 46, 676–700. [Google Scholar] [CrossRef]
- Ding, R.; Jin, Y.; Liu, X.; Zhu, Z.; Zhang, Y.; Wang, T.; Xu, Y. Characteristics of DNA methylation changes induced by traffic-related air pollution. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016, 796, 46–53. [Google Scholar] [CrossRef]
- Desaulniers, D.; Xiao, G.-H.; Lian, H.; Feng, Y.-L.; Zhu, J.; Nakai, J.; Bowers, W.J. Effects of mixtures of polychlorinated biphenyls, methylmercury, and organochlorine pesticides on hepatic DNA methylation in prepubertal female sprague-dawley rats. Int. J. Toxicol. 2009, 28, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.S.; Van Vleet, T.R.; Ciurlionis, R.; Buck, W.R.; Mittelstadt, S.W.; Blomme, E.A.G.; Liguori, M.J. Comparison of RNA-Seq and microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies. Front. Genet. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Evrard, E.; Marchand, J.; Theron, M.; Pichavant-Rafini, K.; Durand, G.; Quiniou, L.; Laroche, J. Impacts of mixtures of herbicides on molecular and physiological responses of the European flounder Platichthys flesus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2010, 152, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Søfteland, L.; Kirwan, J.A.; Hori, T.S.F.; Størseth, T.R.; Sommer, U.; Berntssen, M.H.G.; Viant, M.R.; Rise, M.L.; Waagbø, R.; Torstensen, B.E.; et al. Toxicological effect of single contaminants and contaminant mixtures associated with plant ingredients in novel salmon feeds. Food Chem. Toxicol. 2014, 73, 157–174. [Google Scholar] [CrossRef] [PubMed]
- Tilton, S.C.; Siddens, L.K.; Krueger, S.K.; Larkin, A.J.; Löhr, C.V.; Williams, D.E.; Baird, W.M.; Waters, K.M. Mechanism-based classification of PAH mixtures to predict carcinogenic potential. Toxicol. Sci. 2015, 146, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.K.; Halvorsen, E.; Song, Y.; Hallanger, I.G.; Hansen, E.L.; Brooks, S.J.; Hansen, B.H.; Tollefsen, K.E. Individual and molecular level effects of produced water contaminants on nauplii and adult females of Calanus finmarchicus. J. Toxicol. Environ. Health Part A 2016, 79, 585–601. [Google Scholar] [CrossRef]
- Costa, P.M.; Gosens, I.; Williams, A.; Farcal, L.; Pantano, D.; Brown, D.M.; Stone, V.; Cassee, F.R.; Halappanavar, S.; Fadeel, B. Transcriptional profiling reveals gene expression changes associated with inflammation and cell proliferation following short-term inhalation exposure to copper oxide nanoparticles. J. Appl. Toxicol. 2018, 38, 385–397. [Google Scholar] [CrossRef]
- Zare, A.; Henry, D.; Chua, G.; Gordon, P.; Habibi, H.R. Differential hepatic gene expression profile of male fathead minnows exposed to daily varying dose of environmental contaminants individually and in mixture. Front. Endocrinol. 2018, 9, 749. [Google Scholar] [CrossRef]
- Lichtensteiger, W.; Bassetti-Gaille, C.; Faass, O.; Axelstad, M.; Boberg, J.; Christiansen, S.; Rehrauer, H.; Georgijevic, J.K.; Hass, U.; Kortenkamp, A.; et al. Differential gene expression patterns in developing sexually dimorphic rat brain regions exposed to antiandrogenic, estrogenic, or complex endocrine disruptor mixtures: Glutamatergic synapses as target. Endocrinology 2015, 156, 1477–1493. [Google Scholar] [CrossRef]
- Martínez-Pacheco, M.; Hidalgo-Miranda, A.; Romero-Córdoba, S.; Valverde, M.; Rojas, E. mRNA and miRNA expression patterns associated to pathways linked to metal mixture health effects. Gene 2014, 533, 508–514. [Google Scholar] [CrossRef]
- Curtis, L.R.; Bravo, C.F.; Bayne, C.J.; Tilton, F.; Arkoosh, M.R.; Lambertini, E.; Loge, F.J.; Collier, T.K.; Meador, J.P.; Tilton, S.C. Transcriptional changes in innate immunity genes in head kidneys from Aeromonas salmonicida challenged rainbow trout fed a mixture of polycyclic aromatic hydrocarbons. Ecotoxicol. Environ. Saf. 2017, 142, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Ezerskyte, M.; Paredes, J.A.; Malvezzi, S.; Burns, J.A.; Margison, G.P.; Olsson, M.; Scicchitano, D.A.; Dreij, K. O6-methylguanine-induced transcriptional mutagenesis reduces p53 tumor-suppressor function. Proc. Natl. Acad. Sci. USA 2018, 115, 4731–4736. [Google Scholar] [CrossRef] [PubMed]
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Yadetie, F.; Zhang, X.; Hanna, E.M.; Aranguren-Abadía, L.; Eide, M.; Blaser, N.; Brun, M.; Jonassen, I.; Goksøyr, A.; Karlsen, O.A. RNA-Seq analysis of transcriptome responses in Atlantic cod Gadus morhua precision-cut liver slices exposed to benzo[a]pyrene and 17Α-ethynylestradiol. Aquat. Toxicol. 2018, 201, 174–186. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; Von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Chen, H.; Feng, W.; Chen, K.; Qiu, X.; Xu, H.; Mao, G.; Zhao, T.; Ding, Y.; Wu, X. Transcriptomic analysis reveals potential mechanisms of toxicity in a combined exposure to dibutyl phthalate and diisobutyl phthalate in zebrafish Danio rerio ovary. Aquat. Toxicol. 2019, 216, 105290. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.D., III; Dhammi, A.; Wallace, A.; Hodgson, E.; Roe, R.M. Impact of environmental chemicals on the transcriptome of primary human hepatocytes: Potential for Health Effects. J. Biochem. Mol. Toxicol. 2016, 30, 375–394. [Google Scholar] [CrossRef] [PubMed]
- Titz, B.; Elamin, A.; Martin, F.; Schneider, T.; Dijon, S.; Ivanov, N.V.; Hoeng, J.; Peitsch, M.C. Proteomics for systems toxicology. Comput. Struct. Biotechnol. J. 2014, 11, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Romero-Ruiz, A.; Carrascal, M.; Alhama, J.; Gómez-Ariza, J.L.; Abian, J.; López-Barea, J. Utility of proteomics to assess pollutant response of clams from the Doñana bank of Guadalquivir Estuary SW Spain. Proteomics 2006, 6, S245–S255. [Google Scholar] [CrossRef]
- Costa, P.M.; Chicano-Gálvez, E.; Caeiro, S.; Lobo, J.; Martins, M.; Ferreira, A.M.; Caetano, M.; Vale, C.; Alhama-Carmona, J.; Lopez-Barea, J.; et al. Hepatic proteome changes in Solea senegalensis exposed to contaminated estuarine sediments: A laboratory and in situ survey. Ecotoxicology 2012, 21, 1194–1207. [Google Scholar] [CrossRef]
- Costa, P.M.; Chicano-Gálvez, E.; López Barea, J.; Delvalls, T.A.; Costa, M.H. Alterations to proteome and tissue recovery responses in fish liver caused by a short-term combination treatment with cadmium and benzo[a]pyrene. Environ. Pollut. 2010, 158, 3338–3346. [Google Scholar] [CrossRef]
- Maria, V.L.; Gomes, T.; Barreira, L.; Bebianno, M.J. Impact of benzoapyrene, Cu and their mixture on the proteomic response of Mytilus galloprovincialis. Aquat. Toxicol. 2013, 144–145, 284–295. [Google Scholar] [CrossRef]
- Galland, C.; Dupuy, C.; Loizeau, V.; Danion, M.; Auffret, M.; Quiniou, L.; Laroche, J.; Pichereau, V. Proteomic analysis of the European flounder Platichthys flesus response to experimental PAH-PCB contamination. Mar. Pollut. Bull. 2015, 95, 646–657. [Google Scholar] [CrossRef]
- Hooven, L.A.; Baird, W.M. Proteomic analysis of MCF-7 cells treated with benzo[a]pyrene, dibenzo[a,l]pyrene, coal tar extract, and diesel exhaust extract. Toxicology 2008, 249, 1–10. [Google Scholar] [CrossRef]
- Yin, X.; Wang, H.; Zhang, Y.; Dahlgren, R.A.; Zhang, H.; Shi, M.; Gao, M.; Wang, X. Toxicological assessment of trace β-diketone antibiotic mixtures on zebrafish Danio rerio by proteomic analysis. PLoS ONE 2014, 9, 1–12. [Google Scholar] [CrossRef]
- Robertson, D.G. Metabonomics in toxicology: A review. Toxicol. Sci. 2005, 85, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Jordan, J.; Zare, A.; Jackson, L.J.; Habibi, H.R.; Weljie, A.M. Environmental contaminant mixtures at ambient concentrations invoke a metabolic stress response in goldfish not predicted from exposure to individual compounds alone. J. Proteome Res. 2012, 11, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, W.X. NMR-based metabolomic studies on the toxicological effects of cadmium and copper on green mussels Perna viridis. Aquat. Toxicol. 2010, 100, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Melvin, S.D.; Habener, L.J.; Leusch, F.D.L.; Carroll, A.R. 1H NMR-based metabolomics reveals sub-lethal toxicity of a mixture of diabetic and lipid-regulating pharmaceuticals on amphibian larvae. Aquat. Toxicol. 2017, 184, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.Y.; Wang, P.; Sun, Y.J.; Wu, Y.J. Metabolomic analysis for combined hepatotoxicity of chlorpyrifos and cadmium in rats. Toxicology 2017, 384, 50–58. [Google Scholar] [CrossRef]
- David, A.; Lange, A.; Abdul-Sada, A.; Tyler, C.R.; Hill, E.M. Disruption of the prostaglandin Mmtabolome and characterization of the pharmaceutical exposome in fish exposed to wastewater treatment works effluent as revealed by nanoflow-nanospray Mass spectrometry-based metabolomics. Environ. Sci. Technol. 2017, 51, 616–624. [Google Scholar] [CrossRef]
- Tufi, S.; Wassenaar, P.N.H.; Osorio, V.; De Boer, J.; Leonards, P.E.G.; Lamoree, M.H. Pesticide mixture toxicity in surface water extracts in Snails Lymnaea stagnalis by an in vitro acetylcholinesterase inhibition assay and metabolomics. Environ. Sci. Technol. 2016, 50, 3937–3944. [Google Scholar] [CrossRef]
- Chiappini, F.; Coilly, A.; Kadar, H.; Gual, P.; Tran, A.; Desterke, C.; Samuel, D.; Duclos-Vallée, J.-C.; Touboul, D.; Bertrand-Michel, J.; et al. Metabolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef]
- Phillips, B.; Titz, B.; Kogel, U.; Sharma, D.; Leroy, P.; Xiang, Y.; Vuillaume, G.; Lebrun, S.; Sciuscio, D.; Ho, J.; et al. Toxicity of the main electronic cigarette components, propylene glycol, glycerin, and nicotine, in Sprague-Dawley rats in a 90-day OECD inhalation study complemented by molecular endpoints. Food Chem. Toxicol. 2017, 109, 315–332. [Google Scholar] [CrossRef]
- Borgert, C.J. Predicting interactions from mechanistic information: Can omic data validate theories? Toxicol. Appl. Pharmacol. 2007, 223, 114–120. [Google Scholar] [CrossRef]
- Song, Q.; Chen, H.; Li, Y.; Zhou, H.; Han, Q.; Diao, X. Toxicological effects of benzoapyrene, DDT and their mixture on the green mussel Perna viridis revealed by proteomic and metabolomic approaches. Chemosphere 2016, 144, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Zhou, H.; Han, Q.; Diao, X. Toxic responses of Perna viridis hepatopancreas exposed to DDT, benzoapyrene and their mixture uncovered by iTRAQ-based proteomics and NMR-based metabolomics. Aquat. Toxicol. 2017, 192, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.D.; Davies, I.M.; Wu, H.; Diab, A.M.; Webster, L.; Viant, M.R.; Chipman, J.K.; Leaver, M.J.; George, S.G.; Moffat, C.F.; et al. Molecular responses of European flounder Platichthys flesus chronically exposed to contaminated estuarine sediments. Chemosphere 2014, 108, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Vandenbrouck, T.; Soetaert, A.; van der Ven, K.; Blust, R.; De Coen, W. Nickel and binary metal mixture responses in Daphnia magna: Molecular fingerprints and suborganismal effects. Aquat. Toxicol. 2009, 92, 18–29. [Google Scholar] [CrossRef]
- Dondero, F.; Negri, A.; Boatti, L.; Marsano, F.; Mignone, F.; Viarengo, A. Transcriptomic and proteomic effects of a neonicotinoid insecticide mixture in the marine mussel Mytilus galloprovincialis, Lam. Sci. Total Environ. 2010, 408, 3775–3786. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, K.; Deng, Y.; Zhao, Y.; Wu, B.; Xu, K.; Ren, H. Evaluation of the toxic effects of municipal wastewater effluent on mice using omic approaches. Environ. Sci. Technol. 2013, 47, 9470–9477. [Google Scholar] [CrossRef]
- Zhang, Y.; Deng, Y.; Zhao, Y.; Ren, H. Using combined bio-omics methods to evaluate the complicated toxic effects of mixed chemical wastewater and its treated effluent. J. Hazard. Mater. 2014, 272, 52–58. [Google Scholar] [CrossRef]
- Seeger, B.; Mentz, A.; Knebel, C.; Schmidt, F.; Bednarz, H.; Niehaus, K.; Albaum, S.; Kalinowski, J.; Noll, T.; Steinberg, P.; et al. Assessment of mixture toxicity of triazoles and their hepatotoxic effects in vitro by means of omics technologies. Arch. Toxicol. 2019, 93, 2321–2333. [Google Scholar] [CrossRef]
| “Omics” | Toxicants | Model | Organ/Tissue | Exposure | Exposure Range | Molecular Alterations | Reference |
|---|---|---|---|---|---|---|---|
| Transcriptomics (microarray) Metabolomics (NMR, GC-MS) | Ni, Cd, Pb | Daphnia magna | Whole-body | 96 h | Ni2+ (0.5 mg/L), Pb2+ (0.5 mg/L), Cd2+ (0.05 mg/L) | Genes involved in carbohydrate catabolic processes and proteolysis; genes coding for: mannanase precursor, chymotrypsin-like serine proteases, cellulases, carboxypeptidase, amylase. | Vandenbrouck et al. [75] |
| Transcriptomics (microarray) Proteomics (2DE, MS) | Imidacloprid, thiacloprid | Mytilus galloprovincialis | Digestive gland | 4 days | 0.1 mg/L; 1 mg/L; 10 mg/L | Protein polymerization; microtubule based movement, and GTPase activity. | Dondero et al. [76] |
| Transcriptomics (microarray) Metabolomics (NMR) | Wastewater effluents: semi volatile organic compounds | Mus musculus | Liver, blood serum and urine | 90 days | - | Alterations of lipid, nucleotide, amino acid, and energy metabolism. Disruption of signal transduction processes, hepatotoxicity- and nephrotoxicity-related pathways. | Zhang et al. [77] |
| Transcriptomics (microarray) Metabolomics (NMR) | Marine sediments: metals, PAHs, organochlorines, butyltins | Platichthys flesus | Blood, liver | 7 months | - | Xenobiotic metabolism, immune response and apoptosis. | Williams et al. [74] |
| Transcriptomics (microarray) Metabolomics (NMR) Lipidomics (FT-ICR 1 MS) | Benzo(a)pyrene, phenanthrene, Chlorpyrifos, endosulfan | Hepatocytes (Salmo salar) | - | 24 h | 1 µM, 50.5 µM, 100 µM | Suppression of unsaturated fatty acids and steroid biosynthesis. Alterations in linoleic acid metabolism. | Søfteland et al. [35] |
| Transcriptomics (RNA-seq) Metabolomics (NMR) | Wastewater: PAHs, PAEs, OCCs | Mus musculus | Liver and blood serum | 90 days | 0.1 to 2 ng/L | Molecular pathways related to lipid metabolism and hepatotoxicity | Zhang et al. [78] |
| Proteomics (2DE, MS/MS) Metabolomics (NMR) | DDT, Benzo(a)pyrene | Perna viridis | Gills | 7 days | 10 μg/L | Impact on of proteins related to oxidative stress, cytoskeleton and cell structure, protein biosynthesis and modification, energy metabolism, cell growth and apoptosis. | Song et al. [72] |
| Proteomics (RPLC 1– MS/MS) Metabolomics (NMR) | DDT, Benzo(a)pyrene | Perna viridis | Digestive gland | 7 days | 10 µg/L | Effects on proteins related to cytoskeleton, gene expression, energy balance, reproduction, development, stress response, signal transduction and apoptosis. | Song et al. [73] |
| Transcriptomics (microarray) Metabolomics (GC-MS) | (Tri)azoles | Primary hepatocytes (human and rat) | - | 24 h | µM range | Activation of pathways related to drug and porphyrin metabolism, peroxisome proliferator-activated receptor (PPAR) signaling pathway and others. | Seeger et al. [79] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, C.; Dreij, K.; Costa, P.M. The State-of-the Art of Environmental Toxicogenomics: Challenges and Perspectives of “Omics” Approaches Directed to Toxicant Mixtures. Int. J. Environ. Res. Public Health 2019, 16, 4718. https://doi.org/10.3390/ijerph16234718
Martins C, Dreij K, Costa PM. The State-of-the Art of Environmental Toxicogenomics: Challenges and Perspectives of “Omics” Approaches Directed to Toxicant Mixtures. International Journal of Environmental Research and Public Health. 2019; 16(23):4718. https://doi.org/10.3390/ijerph16234718
Chicago/Turabian StyleMartins, Carla, Kristian Dreij, and Pedro M. Costa. 2019. "The State-of-the Art of Environmental Toxicogenomics: Challenges and Perspectives of “Omics” Approaches Directed to Toxicant Mixtures" International Journal of Environmental Research and Public Health 16, no. 23: 4718. https://doi.org/10.3390/ijerph16234718
APA StyleMartins, C., Dreij, K., & Costa, P. M. (2019). The State-of-the Art of Environmental Toxicogenomics: Challenges and Perspectives of “Omics” Approaches Directed to Toxicant Mixtures. International Journal of Environmental Research and Public Health, 16(23), 4718. https://doi.org/10.3390/ijerph16234718





