Knowledge Discovery from Posts in Online Health Communities Using Unified Medical Language System
Abstract
1. Introduction
2. Materials and Methods
2.1. Framework of Domain Knowledge Support
2.2. Method for Knowledge Discovery from Patient-Reported Posts
| Algorithm 1 Knowledge-involved topic modeling method |
| Input: source={(id_field, label_field, text_field)} Output: KI-TM
|
| Algorithm 2 Domain discovery algorithm |
| Input: labels={l1, l2, …, lm} Output: domain labels of medical background
|
| Algorithm 3 Latent knowledge discovery algorithm |
| Input: knowledge graphs of explicit terms within text, the number of latent layers Output: knowledge graphs of latent terms
|
2.3. Evaluation Metrics
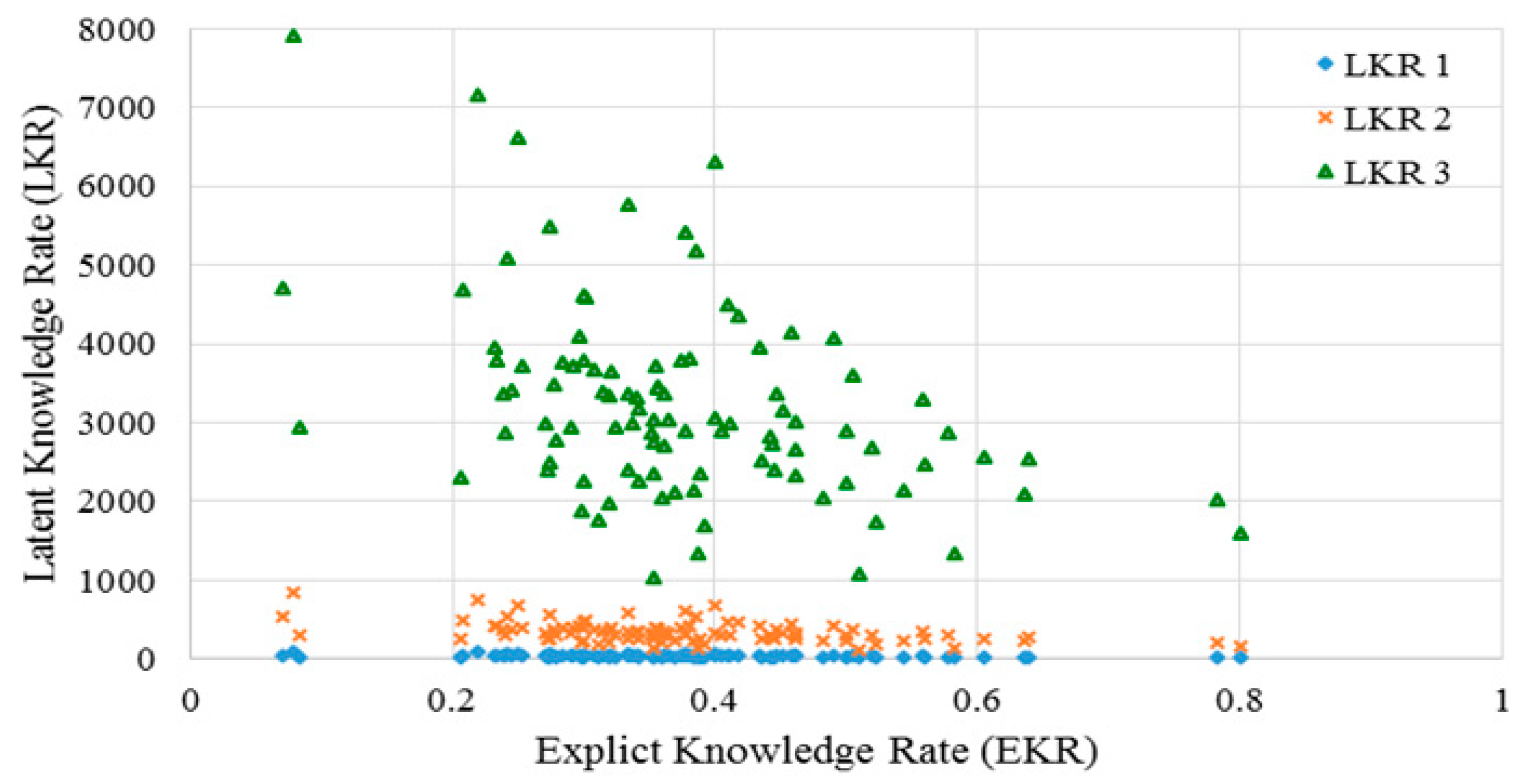
2.3.1. Explicit Knowledge Rate
2.3.2. Latent Knowledge Rate
2.3.3. Knowledge Correlation Rate
2.3.4. Perplexity of KI-TM
3. Results
3.1. Datasets
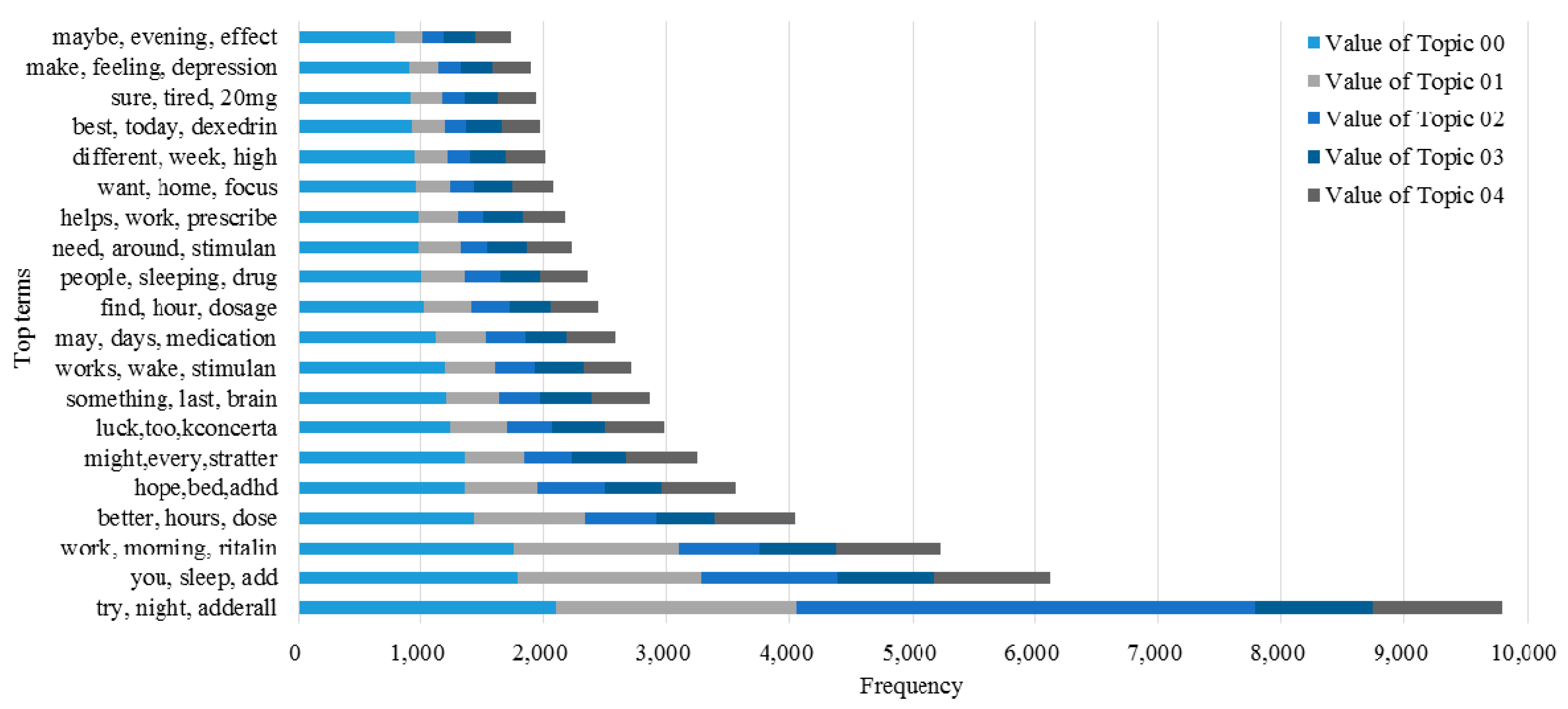
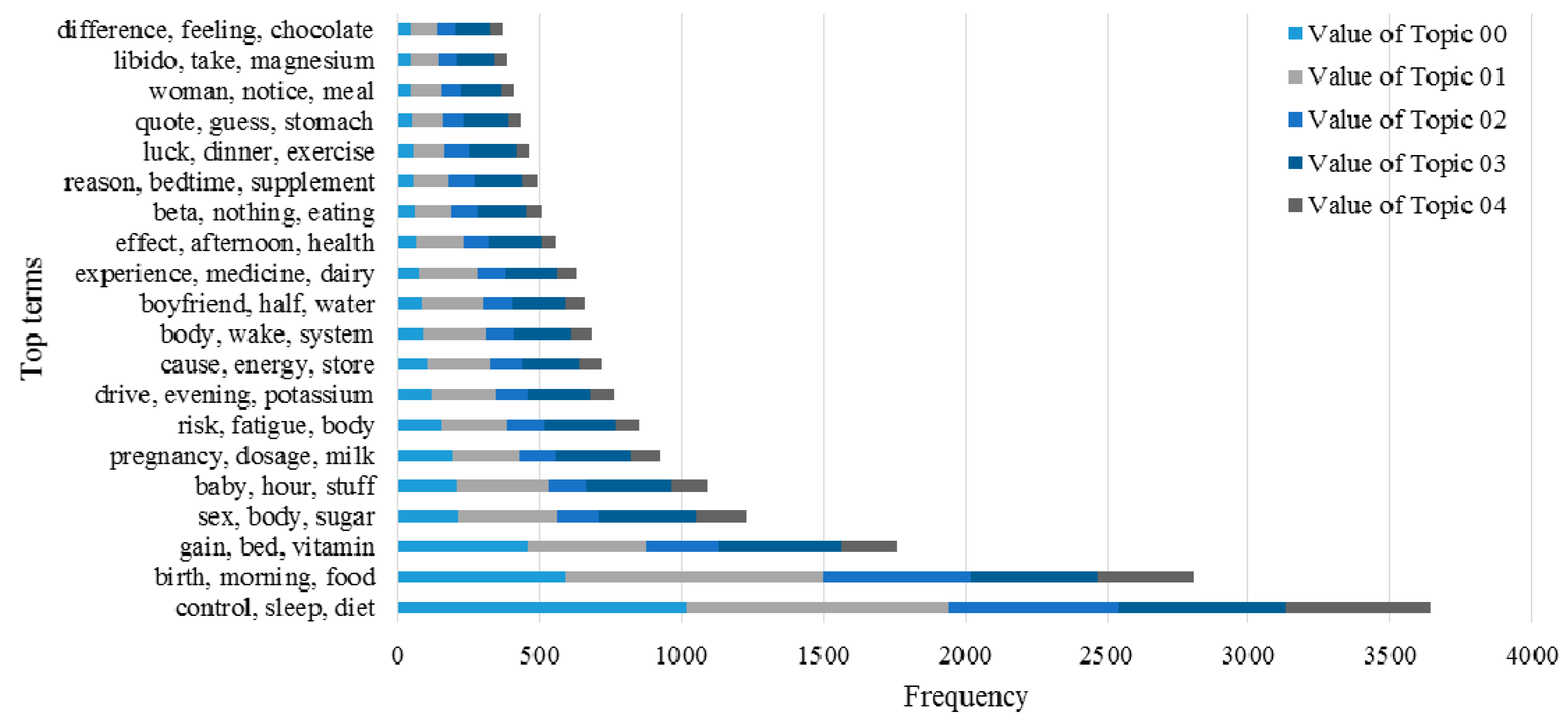
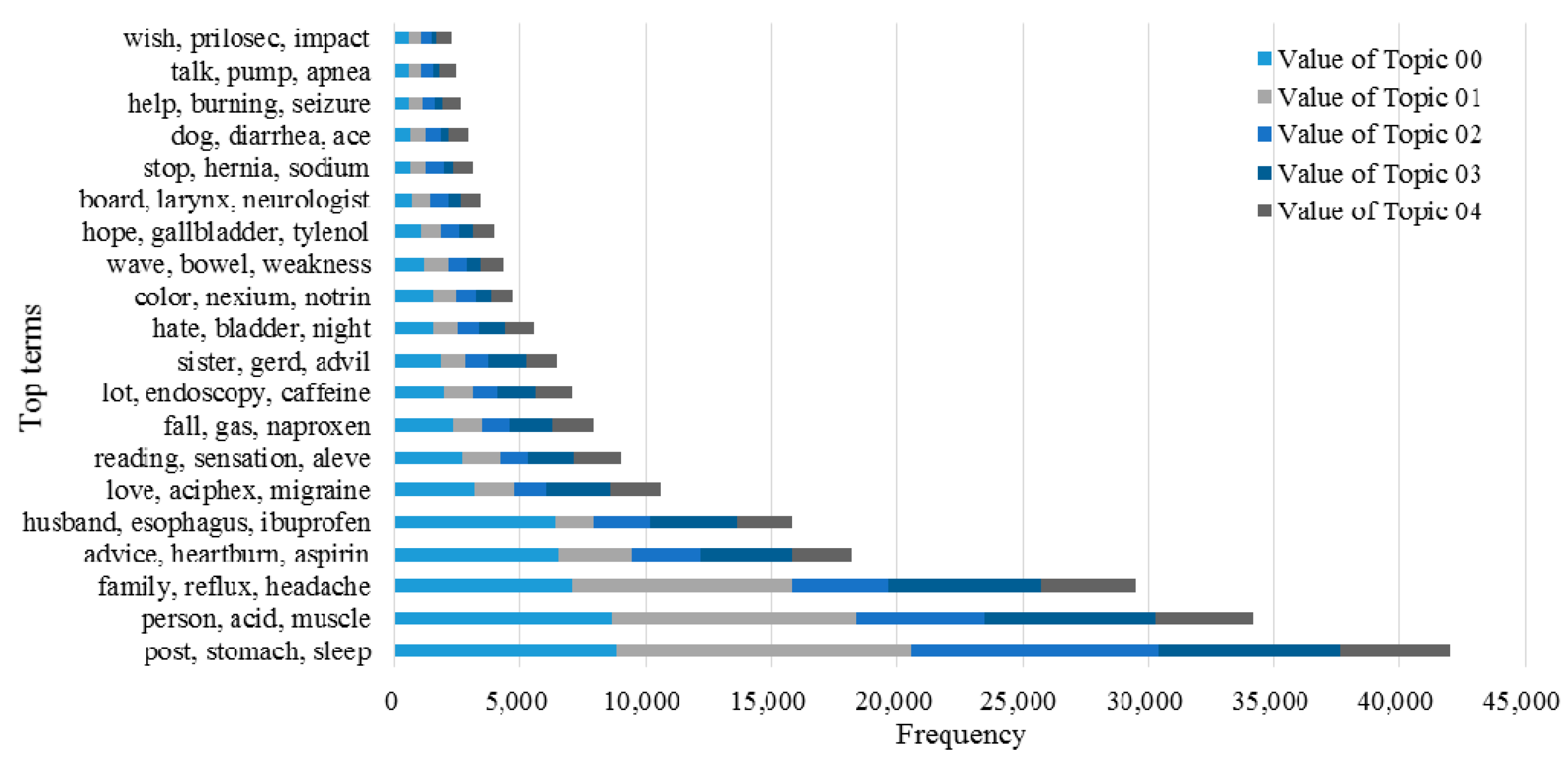
3.2. Knowledge Support Provided by the Proposed Method
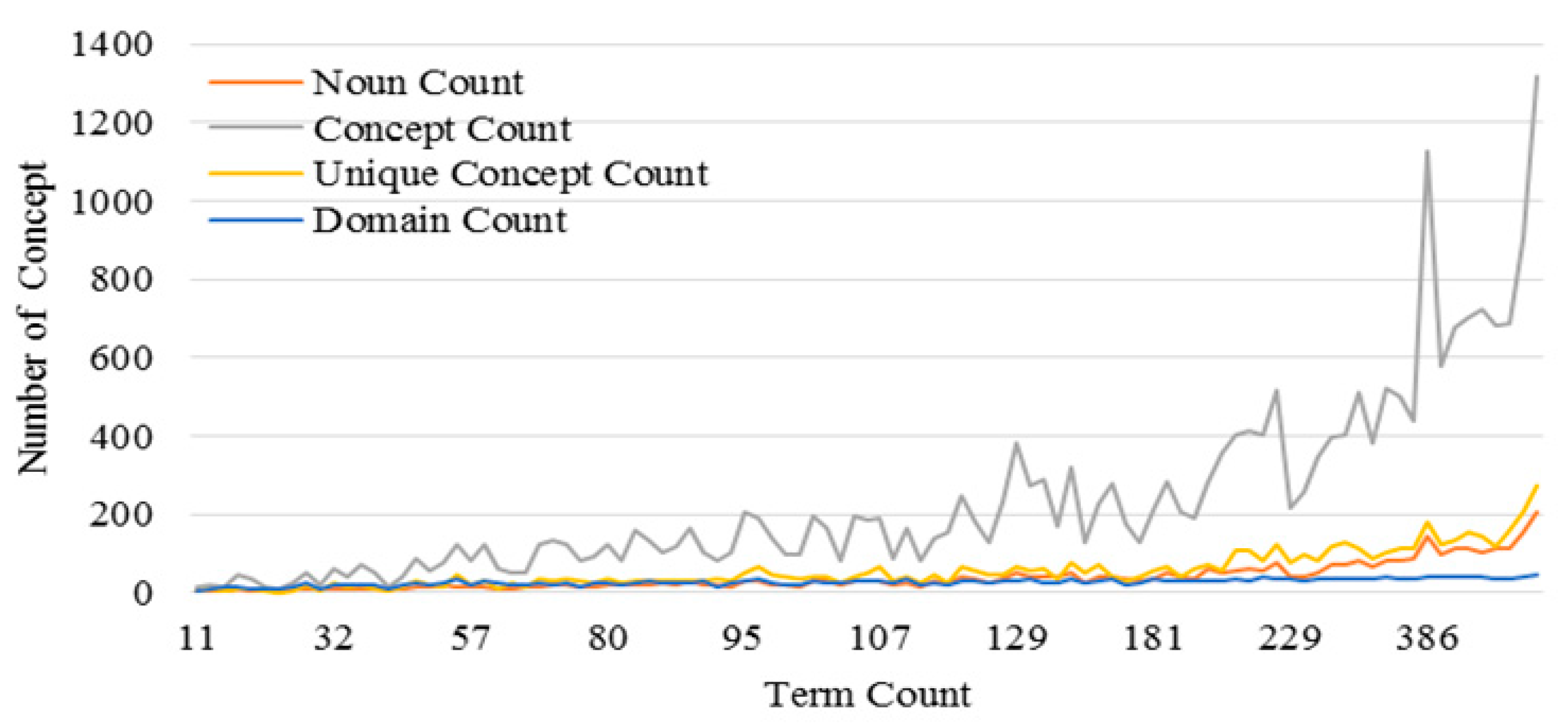
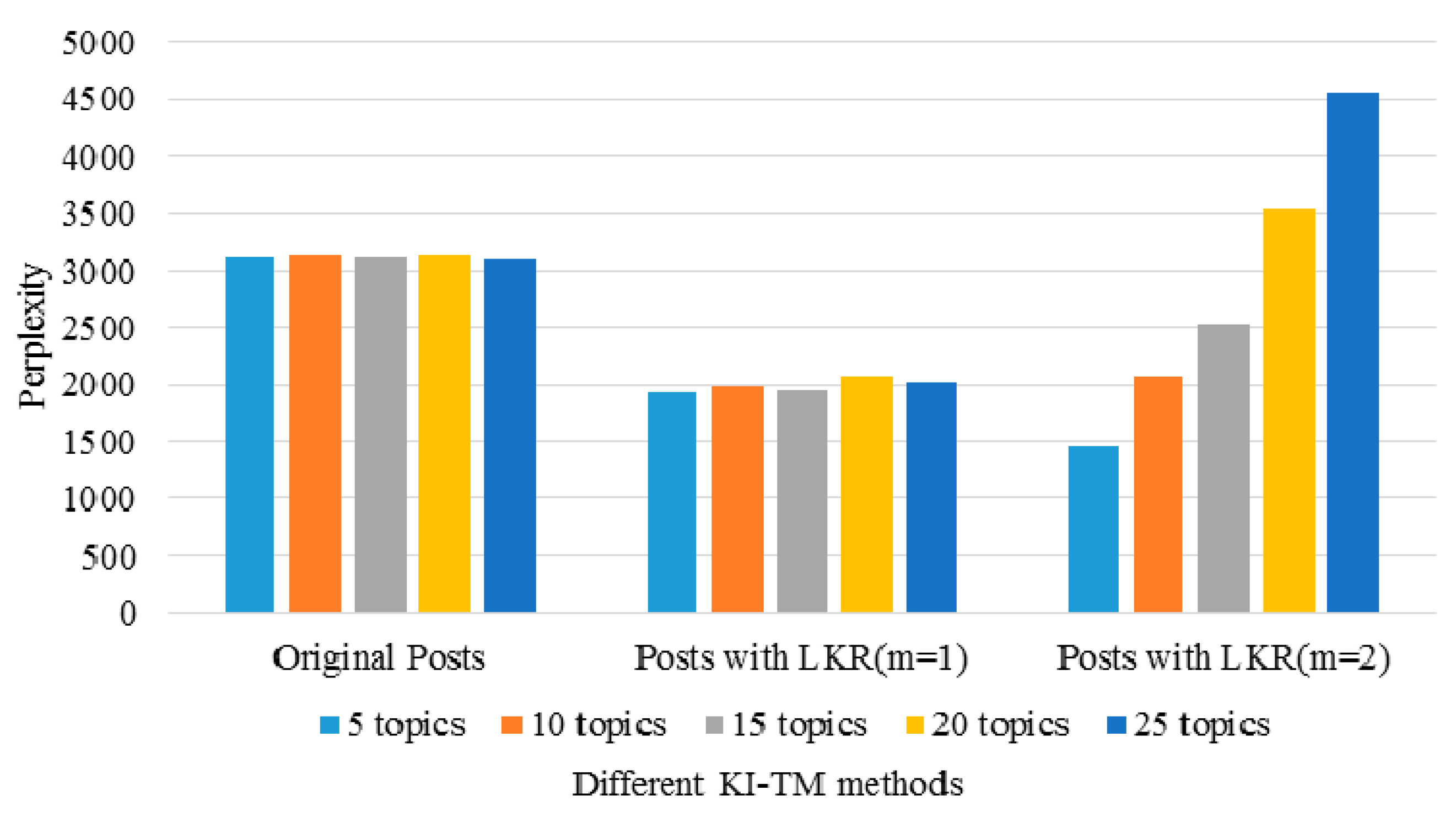
3.3. Performance of the KI-TM Method
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yan, Z.; Wang, T.; Chen, Y.; Zhang, H. Knowledge sharing in online health communities: A social exchange theory perspective. Inform. Manag. 2016, 53, 643–653. [Google Scholar] [CrossRef]
- Willis, E.; Royne, M.B. Online health communities and chronic disease self-management. Health Comm. 2016, 32, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Corley, C.D.; Cook, D.J.; Mikler, A.R.; Singh, K.P. Text and Structural Data Mining of Influenza Mentions in Web and Social Media. Int. J. Environ. Res. Public Health 2010, 7, 596–651. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.W. Trending now: Using social media to predict and track disease outbreaks. Environ. Health Perspect. 2012, 120, a30–a33. [Google Scholar] [CrossRef] [PubMed]
- Nath, C.; Huh, J.; Adupa, A.K.; Jonnalagadda, S.R. Website sharing in online health communities: A descriptive analysis. J. Med. Int. Res. 2016, 18, e11. [Google Scholar] [CrossRef] [PubMed]
- Palomino, M.; Taylor, T.; Göker, A.; Isaacs, J.; Warber, S. The Online Dissemination of Nature-Health Concepts: Lessons from Sentiment Analysis of Social Media Relating to “Nature-Deficit Disorder”. Int. J. Environ. Res. Public Health 2016, 13, 142. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, S.; Yoon, H.J.; Tourassi, G. Extracting Patient Demographics and Personal Medical Information from Online Health Forums. AMIA Annu. Symp. Proc. 2014, 2014, 1825–1834. [Google Scholar] [PubMed]
- Foster, D. ‘Keep complaining til someone listens’: Exchanges of tacit healthcare knowledge in online illness communities. Soc. Sci. Med. 2016, 166, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Gurulingappa, H.; Rajput, A.M.; Roberts, A.; Fluck, J.; Holmann-Apitius, M. Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J. Biomed. Inform. 2012, 45, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Van Mulligen, E.M.; Fourrier-Reglat, A.; Gurwitz, D.; Molokhia, M.; Nieto, M.; Trifiro, G.; Kors, J.A.; Furlong, L.I. The EU-ADR corpus: Annotated drugs, diseases, targets, and their relationships. J. Biomed. Inform. 2012, 45, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Rubrichi, S.; Quaglini, S. Summary of Product Characteristics content extraction for a safe drugs usage. J. Biomed. Inform. 2012, 45, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Dobkin, P.L.; Boothroyd, L.J. Organizing Health Services for Patients with Chronic Pain: When There Is a Will There Is a Way. Pain Med. 2008, 9, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Cornet, R.; Keizer, N.D. Forty years of SNOMED: A literature review. BMC Med. Inform. Decis. Mak. 2008, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Van der Eijk, M.; Faber, M.J.; Aarts, J.W.; Kremer, J.A.; Munneke, M.; Bloem, B.R. Using Online Health Communities to Deliver Patient-Centered Care to People with Chronic Conditions. J. Med. Int. Res. 2013, 15, e115. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Lai, A.M.; Mukhopadhyay, A.; Machiraju, R.; Huang, K. Effectively processing medical term queries on the UMLS Metathesaurus by layered dynamic programming. BMC Med. Genom. 2014, 7, S11. [Google Scholar] [CrossRef] [PubMed]
- Alonso, I.; Contreras, D. Evaluation of semantic similarity metrics applied to the automatic retrieval of medical documents: An UMLS approach. Exp. Syst. Appl. 2015, 44, 386–399. [Google Scholar] [CrossRef]
- Albin, A.; Ji, X.; Borlawsky, T.B.; Ye, Z.; Lin, S.; Payne, P.R.O.; Huang, K.; Xiang, Y. Enabling online studies of conceptual relationships between medical terms: Developing an efficient web platform. Int. J. Med. Inform. 2014, 2, 914–925. [Google Scholar] [CrossRef] [PubMed]
- Kallinikos, J.; Tempini, N. Patient Data as Medical Facts: Social Media Practices as a Foundation for Medical Knowledge Creation. Inform. Syst. Res. 2014, 25, 817–833. [Google Scholar] [CrossRef]
- Marco-Ruiz, L.; Pedrinaci, C.; Maldonado, J.A.; Panziera, L.; Chen, R.; Bellika, J.G. Publication, Discovery and Interoperability of Clinical Decision Support Systems: A Linked Data Approach. J. Biomed. Inform. 2016, 62, 243–264. [Google Scholar] [CrossRef] [PubMed]
- Scuba, W.; Tharp, M.; Mowery, D.; Tseytlin, E.; Liu, Y.; Drews, F.A.; Chapman, W.W. Knowledge Author: Facilitating user-driven, domain content development to support clinical information extraction. J. Biomed. Semant. 2016, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Ravorie, S.; Lang, M.; Perrin, E. Advantages and limitations of online communities of patients for research on health products. Therapie 2017, 72, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Vandam, C.; Kanthawala, S.; Pratt, W.; Chai, J.; Hub, J. Detecting clinically related content in online patient posts. J. Biomed. Inform. 2017, 75, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.C.; Lee, A.J.; Kuo, S.C. Mining Health Social Media with Sentiment Analysis. J. Med. Syst. 2016, 40, 236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, H. Methods for Linking EHR Notes to Education Materials. Inform. Retriev. J. 2015, 19, 174–188. [Google Scholar] [CrossRef]
- Distante, D.; Fermandex, A.; Cerulo, L.; Visagio, A. Enhancing Online Discussion Forums with Topic-Driven Content Search and Assisted Posting. In International Joint Conference on Knowledge Discovery, Knowledge Engineering, and Knowledge Management; Springer International Publishing: Cham, Switzerland, 2015; pp. 161–180. [Google Scholar]
- Hanbury, A. How users search and what they search for in the medical domain. Inform. Retriev. J. 2016, 19, 189–224. [Google Scholar]
- Alecu, I.; Bousquet, C.; Jaulent, M.C. A case report: Using SNOMED CT for grouping Adverse Drug Reactions Terms. BMC Med. Inform. Deci. Mak. 2008, 8, S4. [Google Scholar] [CrossRef] [PubMed]
- Cherichi, S.; Faiz, R. Analyzing the Behavior and Text Posted by Users to Extract Knowledge. In Proceedings of the International Conference on Computational Collective Intelligence, Seoul, Korea, 24–26 September 2014; pp. 524–533. [Google Scholar]
- Huang, L.; Ma, J.; Chen, C. Topic Detection from Microblogs Using T-LDA and Perplexity. In Proceedings of the 24th Asia-Pacific Software Engineering Conference Workshops (APSECW), Nanjing, China, 4–8 December 2017; pp. 71–77. [Google Scholar]
- Albert, P.; Hartzler, A.L.; Jina, H.; Mcdonald, D.W.; Wanda, P. Automatically Detecting Failures in Natural Language Processing Tools for Online Community Text. J. Med. Inter. Res. 2015, 17, e212. [Google Scholar]
- Ramage, D.; Hall, D.; Nallapati, R.; Manning, C.D. Labeled LDA: A supervised topic model for credit attribution in multi-labeled corpora. In Proceedings of the 2009 Conference on Empirical Methods in Natural Language Processing, Singapore, 6–7 August 2009; pp. 248–256. [Google Scholar]
- Ramage, D.; Manning, C.D.; Dumais, S. Partially labeled topic models for interpretable text mining. In Proceedings of the 17th ACM SIGKDD Conference on Knowledge Discovery and Data Mining, San Diego, CA, USA, 21–24 August 2011; pp. 457–465. [Google Scholar]
- Tang, B.; Cao, H.; Wu, Y.; Jiang, M.; Xu, H. Recognizing clinical entities in hospital discharge summaries using Structural Support Vector Machines with word representation features. BMC Med. Inform. Decis. Mak. 2013, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dasmahapatra, P.; Raja, P.; Gilbert, J.; Wicks, P. Clinical trials from the patient perspective: Survey in an online patient community. BMC Serv. Res. 2017, 17, 166. [Google Scholar] [CrossRef] [PubMed]
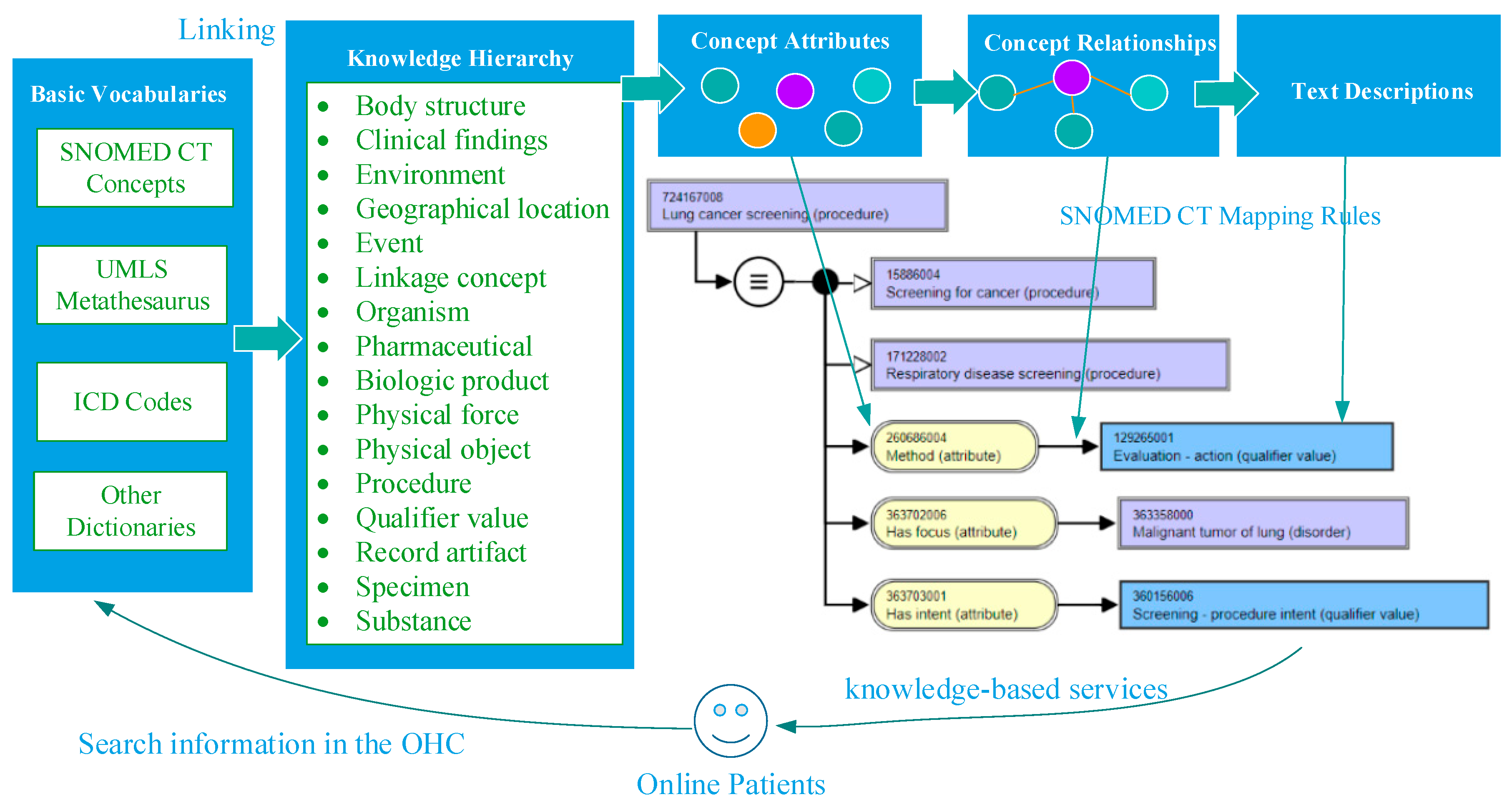
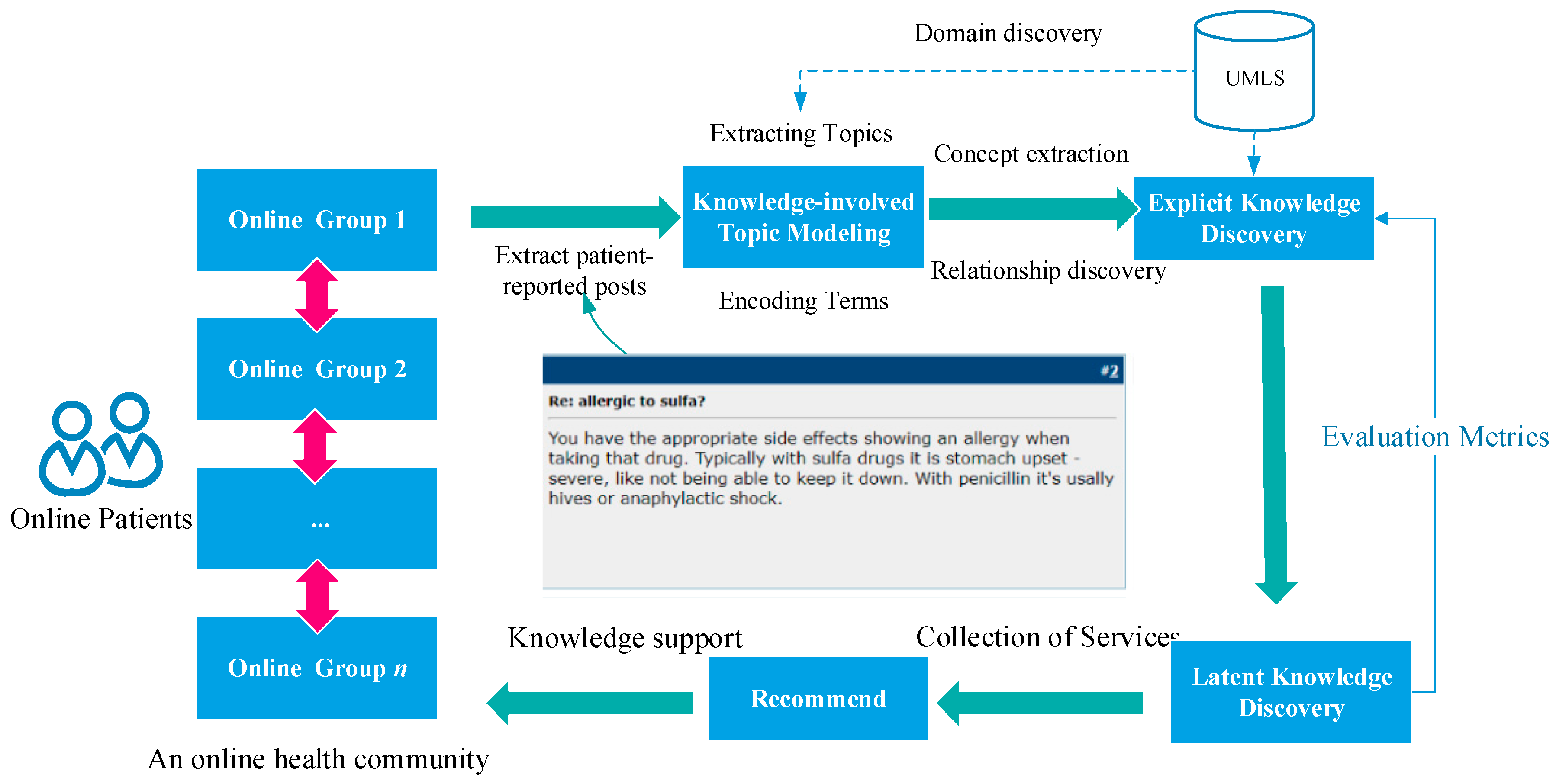
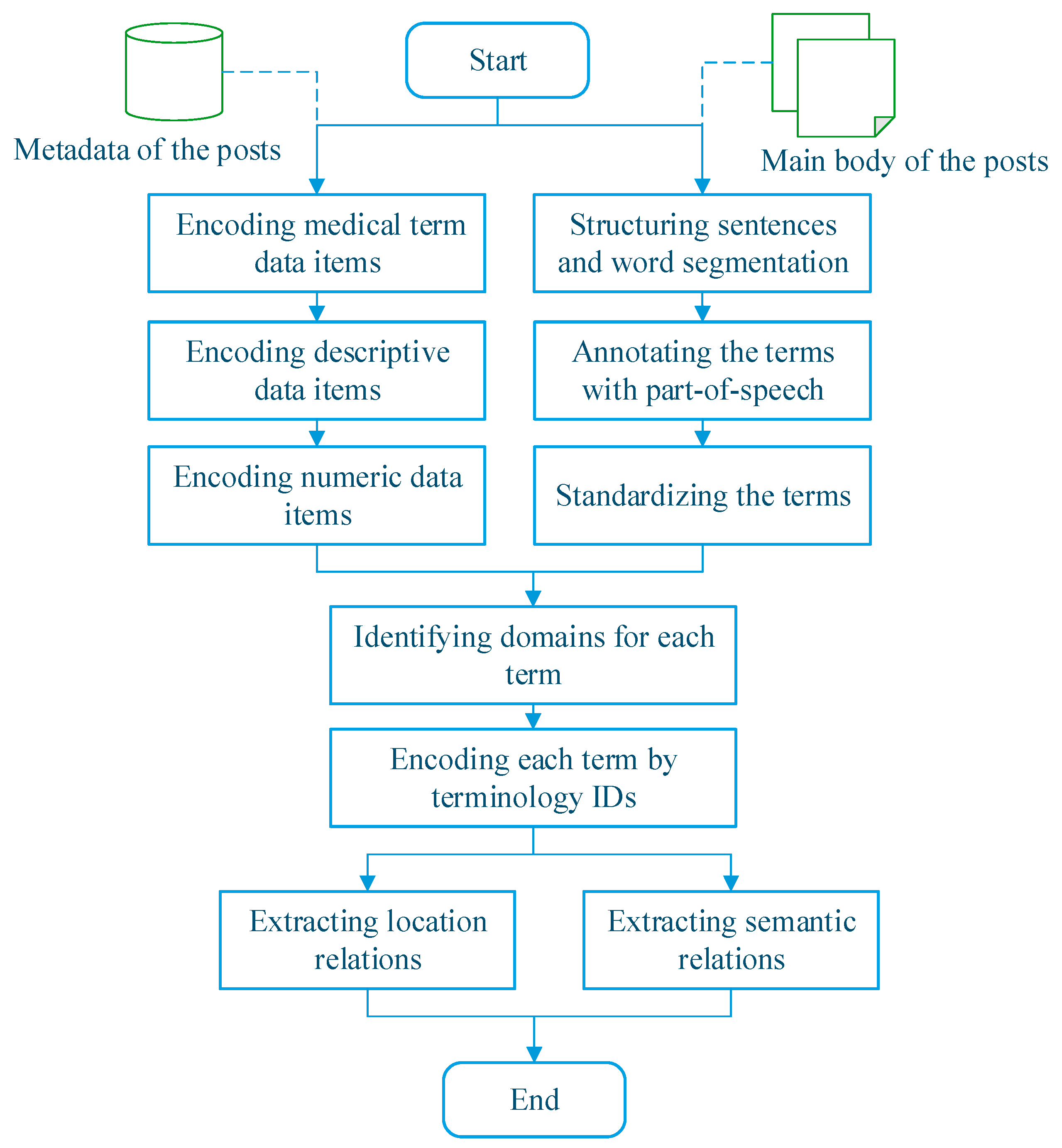
| Element Type of UMLS | Element Type of Our DKFS |
|---|---|
| Concept | Term |
| Concept names | Term names (standardized terms) |
| Relationships | Concept relationships |
| Attribute | Concept attribute (part-of-speech) |
| Source vocabularies | Domains (medical meanings) |
| String identifiers | Entity in narrative text |
| Lexical identifiers | Semantic relationships |
| Knowledge hierarchy | Knowledge hierarchy |
| Name of Drug | Caption of Online Post | Main Body of Online Post |
|---|---|---|
| Atenolol | Low Libido | Wow..........is atenolol the answer? Bookish, I hope you get this resolved.... Sincerely, Oleander. |
| Diovan | Stopped Diovan—Hurrah!! | Hello, I take Diovan. I missed why you wanted to get off it? Bad side effects? |
| Tazorac | Should I Give Retin-A Micro the Boot | Tazorac is basically the same thing as Retin A accept it’s supposed to be more potent. |
| Trazodone | Generic Amb ien!? | trazodone—nonaddictive, no grogginess and something that I’d suggest to anyone. |
| Wellbutrin | I am Going to Quit Smoking Soon....but I have Panic Disorder | Wellbutrin really worked for me. I wish I had tried it years ago. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.; Zhang, R.; Liu, K.; Hou, L. Knowledge Discovery from Posts in Online Health Communities Using Unified Medical Language System. Int. J. Environ. Res. Public Health 2018, 15, 1291. https://doi.org/10.3390/ijerph15061291
Chen D, Zhang R, Liu K, Hou L. Knowledge Discovery from Posts in Online Health Communities Using Unified Medical Language System. International Journal of Environmental Research and Public Health. 2018; 15(6):1291. https://doi.org/10.3390/ijerph15061291
Chicago/Turabian StyleChen, Donghua, Runtong Zhang, Kecheng Liu, and Lei Hou. 2018. "Knowledge Discovery from Posts in Online Health Communities Using Unified Medical Language System" International Journal of Environmental Research and Public Health 15, no. 6: 1291. https://doi.org/10.3390/ijerph15061291
APA StyleChen, D., Zhang, R., Liu, K., & Hou, L. (2018). Knowledge Discovery from Posts in Online Health Communities Using Unified Medical Language System. International Journal of Environmental Research and Public Health, 15(6), 1291. https://doi.org/10.3390/ijerph15061291





