Whole Genome Transcriptome Analysis of the Association between Obesity and Triple-Negative Breast Cancer in Caucasian Women
Abstract
1. Introduction
2. Material and Methods
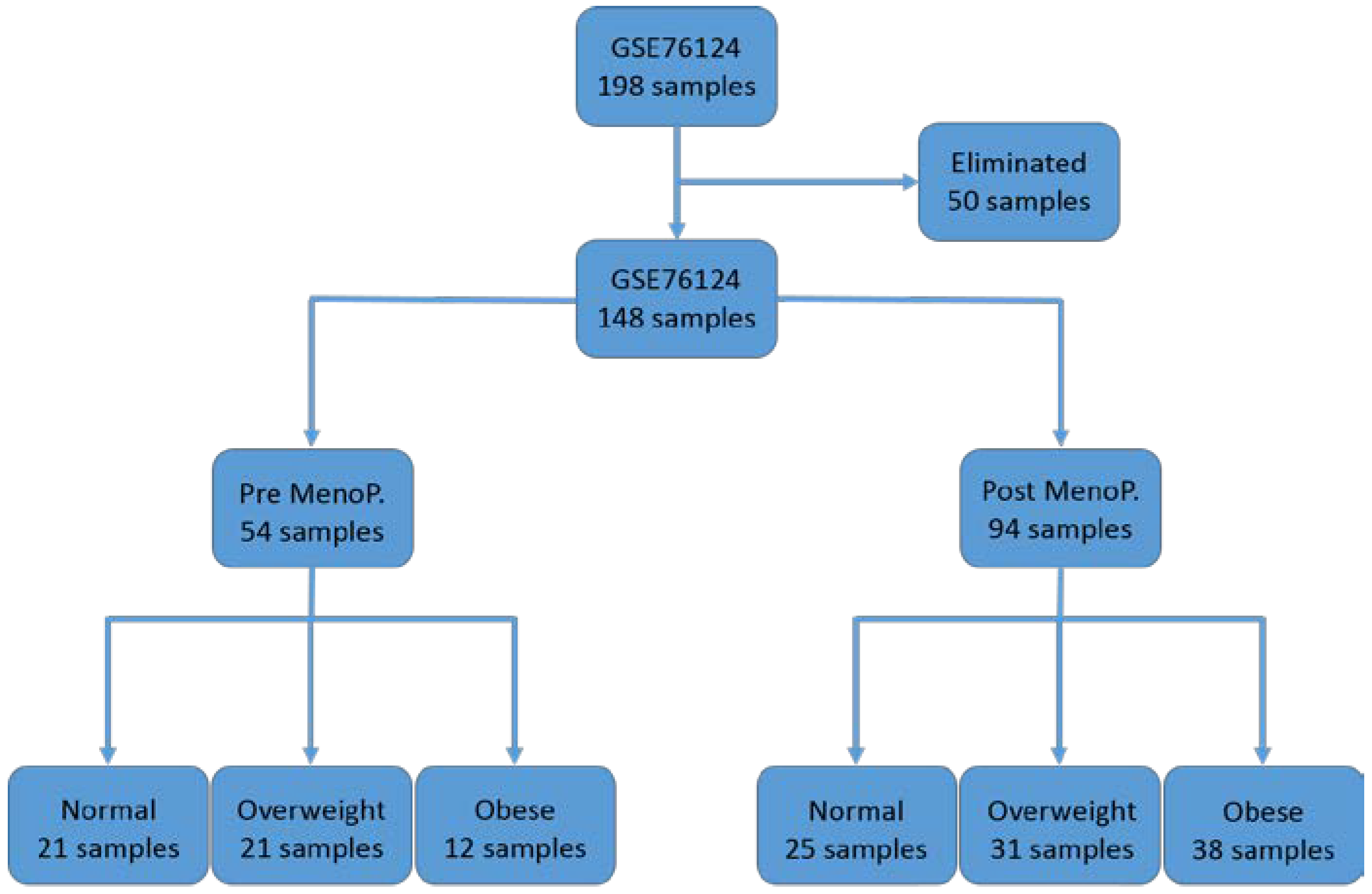
2.1. Research Design and Source of Gene Expression Data
2.2. Data Analysis
3. Results
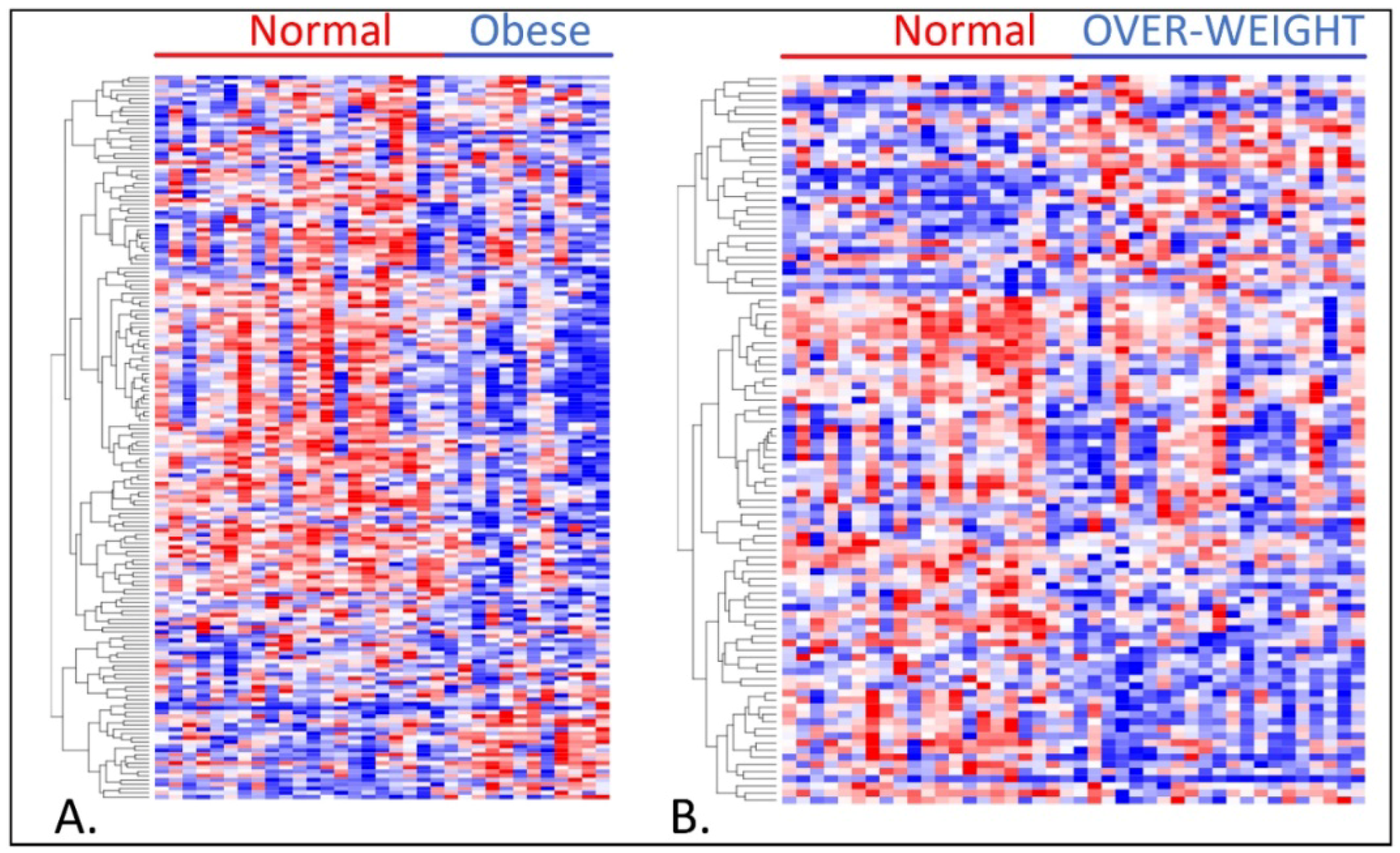
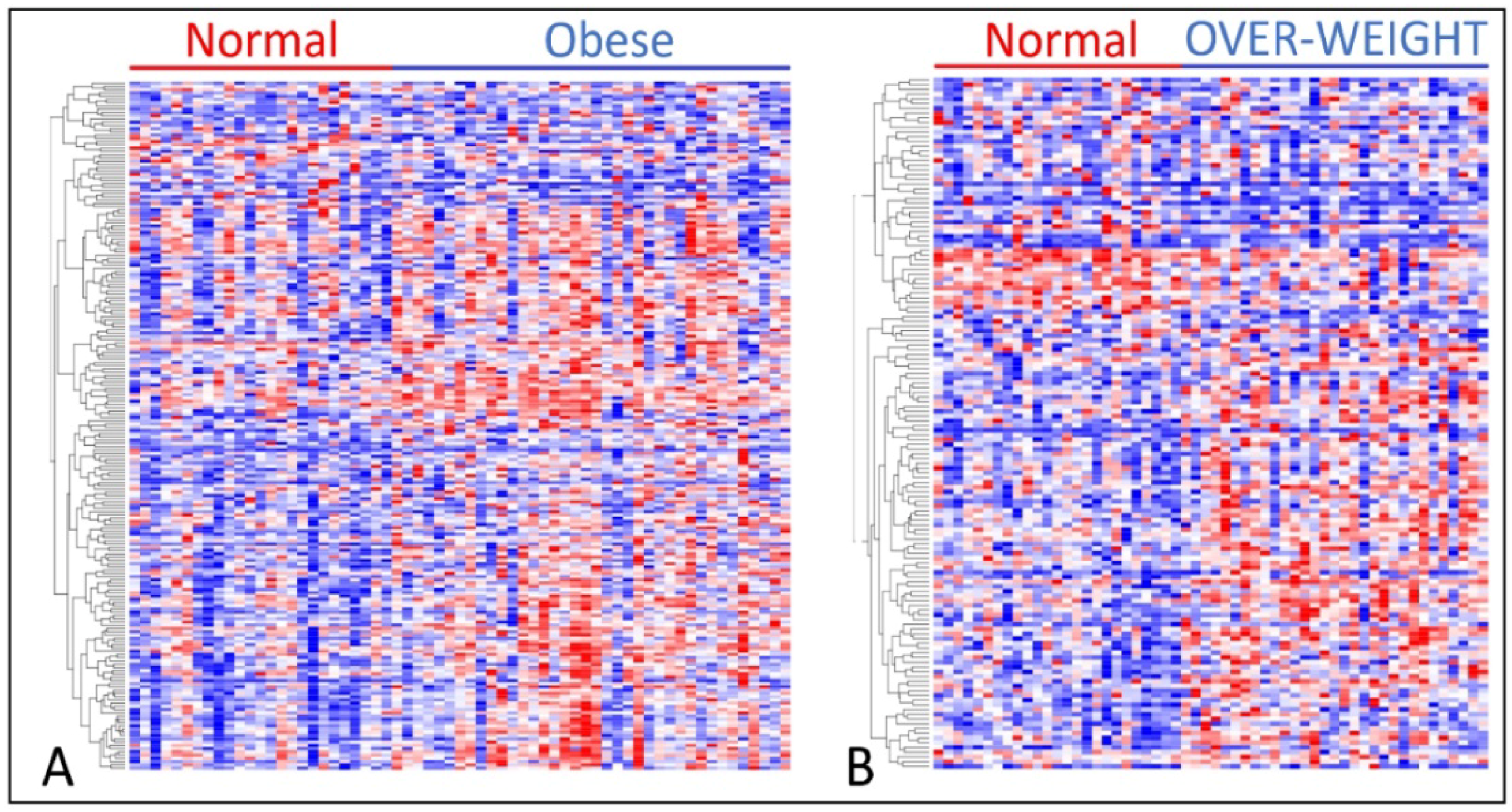
3.1. Differences in Gene Expression Levels among Patient Groups
3.2. Association of Overweight and Obesity with TNBC in Premenopausal Women
3.3. Association of TNBC with Obesity and Overweight in Postmenopausal Women
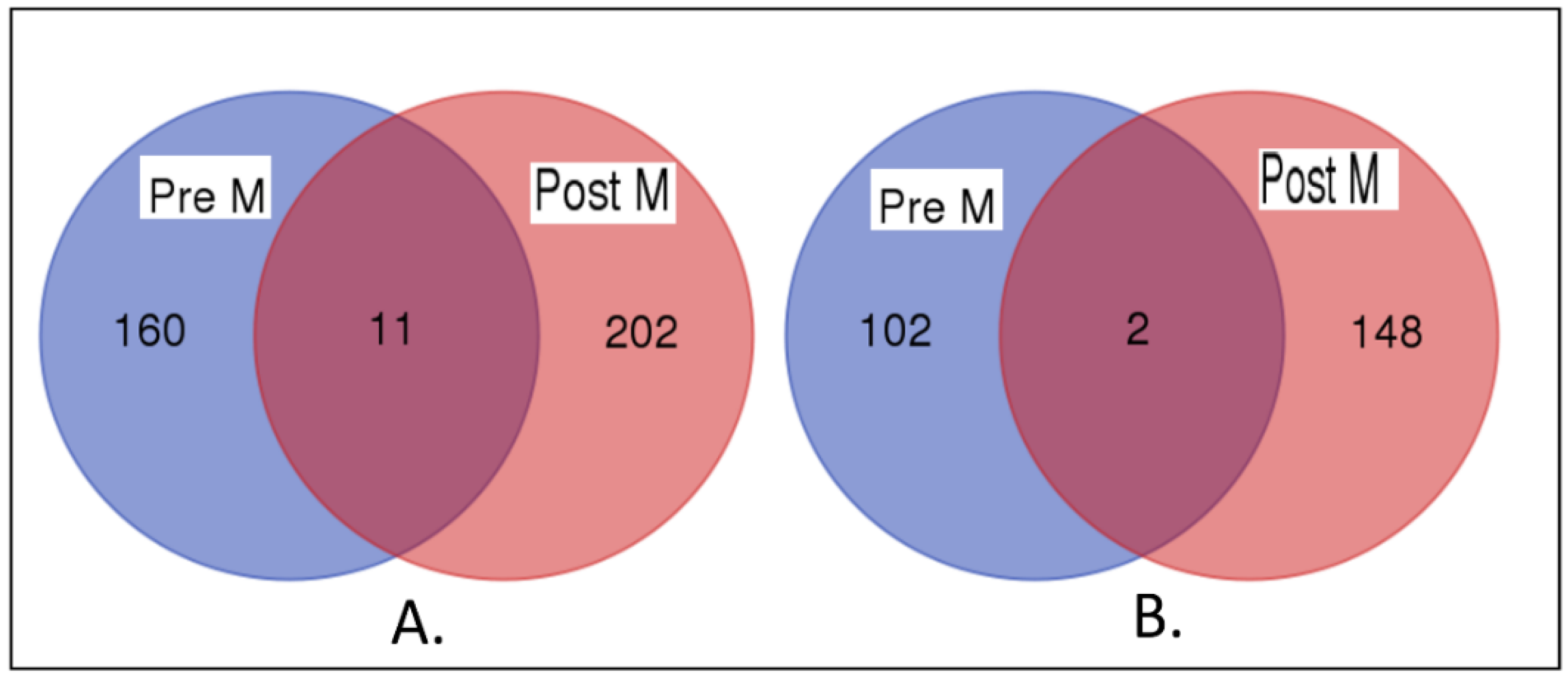
3.4. Premenopausal Versus Postmenopausal
3.5. Similarity in Patterns of Gene Expression Profiles
3.6. Molecular Networks and Biological Pathways Associating Obesity with TNBC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Dietze, E.C.; Sistrunk, C.; Miranda-Carboni, G.; O’regan, R.; Seewaldt, V.L. Triple-negative breast cancer in African-American women: Disparities versus biology. Nat. Rev. Cancer 2015, 15, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.A.; Perou, C.M.; Livasy, C.A.; Dressler, L.G.; Cowan, D.; Conway, K.; Deming, S.L. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006, 295, 2492–2502. [Google Scholar] [CrossRef] [PubMed]
- Turkman, Y.E.; Sakibia Opong, A.; Harris, L.N.; Knobf, M.T. Biologic, demographic, and social factors affecting triple negative breast cancer outcomes. Clin. J. Oncol. Nurs. 2015, 19, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Bradley, C.J.; Given, C.W.; Roberts, C. Race, socioeconomic status, and breast cancer treatment and survival. J. Natl. Cancer Inst. 2002, 94, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Bauer, K.R.; Brown, M.; Cress, R.D.; Parise, C.A.; Caggiano, V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the California cancer Registry. Cancer 2007, 109, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Bao, P.P.; Zhao, G.M.; Shu, X.O.; Peng, P.; Cai, H.; Lu, W.; Zheng, Y. Modifiable lifestyle factors and triple-negative breast cancer survival: A population-based prospective study. Epidemiology 2015, 26, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Pierobon, M.; Frankenfeld, C.L. Obesity as a risk factor for triple-negative breast cancers: A systematic review and meta-analysis. Breast Cancer Res. Treat. 2013, 137, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Turkoz, F.P.; Solak, M.; Petekkaya, I.; Keskin, O.; Kertmen, N.; Sarici, F.; Altundag, K. Association between common risk factors and molecular subtypes in breast cancer patients. Breast 2013, 22, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Al Jarroudi, O.; Abda, N.; Seddik, Y.; Brahmi, S.A.; Afqir, S. Overweight: Is It a Prognostic Factor in Women with Triple-Negative Breast Cancer? Asian Pac. J. Cancer Prev. 2017, 18, 1519–1523. [Google Scholar] [PubMed]
- Hao, S.; Liu, Y.; Yu, K.D.; Chen, S.; Yang, W.T.; Shao, Z.M. Overweight as a prognostic factor for triple-negative breast cancers in Chinese women. PLoS ONE 2015, 10, e0129741. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Ding, A.; Wang, M.L. Impact of central obesity on prognostic outcome of triple negative breast cancer in Chinese women. SpringerPlus 2016, 5, 594. [Google Scholar] [CrossRef] [PubMed]
- Sahin, S.; Erdem, G.U.; Karatas, F.; Aytekin, A.; Sever, A.R.; Ozisik, Y.; Altundag, K. The association between body mass index and immunohistochemical subtypes in breast cancer. Breast 2017, 32, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Cakar, B.; Muslu, U.; Erdogan, A.P.; Ozisik, M.; Ozisik, H.; Dalgic, C.T.; Uslu, R. The role of body mass index in triple negative breast cancer. Oncol. Res. Treat. 2015, 38, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, P.; Friedl, T.W.; Schwentner, L.; DeGregorio, N.; Jaeger, B.; Schramm, A.; Kost, B. The influence of obesity on survival in early, high-risk breast cancer: Results from the randomized SUCCESS A trial. Breast Cancer Res. 2015, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Phipps, A.I.; Malone, K.E.; Porter, P.L.; Daling, J.R.; Li, C.I. Body size and risk of luminal, HER2-overexpressing, and triple-negative breast cancer in postmenopausal women. Cancer Epidemiol. Prev. Biomark. 2008, 17, 2078–2086. [Google Scholar] [CrossRef] [PubMed]
- Tait, S.; Pacheco, J.M.; Gao, F.; Bumb, C.; Ellis, M.J.; Ma, C.X. Body mass index, diabetes, and triple-negative breast cancer prognosis. Breast Cancer Res. Treat. 2014, 146, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Ademuyiwa, F.O.; Groman, A.; O’connor, T.; Ambrosone, C.; Watroba, N.; Edge, S.B. Impact of body mass index on clinical outcomes in triple-negative breast cancer. Cancer 2011, 117, 4132–4140. [Google Scholar] [CrossRef] [PubMed]
- Mowad, R.; Chu, Q.D.; Li, B.D.; Burton, G.V.; Ampil, F.L.; Kim, R.H. Does obesity have an effect on outcomes in triple-negative breast cancer? J. Surg. Res. 2013, 184, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Fontanella, C.; Lederer, B.; Gade, S.; Vanoppen, M.; Blohmer, J.U.; Costa, S.D.; Hilfrich, J. Impact of body mass index on neoadjuvant treatment outcome: A pooled analysis of eight prospective neoadjuvant breast cancer trials. Breast Cancer Res. Treat. 2015, 150, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Karatas, F.; Erdem, G.U.; Sahin, S.; Aytekin, A.; Yuce, D.; Sever, A.R.; Altundag, K. Obesity is an independent prognostic factor of decreased pathological complete response to neoadjuvant chemotherapy in breast cancer patients. Breast 2017, 32, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Bonsang-Kitzis, H.; Chaltier, L.; Belin, L.; Savignoni, A.; Rouzier, R.; Sablin, M.P.; Laé, M. Beyond axillary lymph node metastasis, BMI and menopausal status are prognostic determinants for triple-negative breast cancer treated by neoadjuvant chemotherapy. PLoS ONE 2015, 10, e0144359. [Google Scholar] [CrossRef] [PubMed]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M.; Covington, K.R.; Contreras, A.; Fuqua, S.A.; Mills, G.B. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015, 21, 1688–1698. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M. Molecular stratification of triple-negative breast cancers. Oncologist 2011, 16 (Suppl. 1), 61–70. [Google Scholar] [CrossRef] [PubMed]
- De Onis, M.; Habicht, J.P. Anthropometric reference data for international use: Recommendations from a World Health Organization Expert Committee. Am. J. Clin. Nutr. 1996, 64, 650–658. [Google Scholar] [CrossRef] [PubMed]
- US Department of Health and Human Services, Centers for Disease Control and Prevention. Body Mass Index: Considerations for Practitioners. Available online: https://www.cdc.gov/obesity/downloads/bmiforpactitioners.pdf (accessed on 11 December 2017).
- Morrissey, E.R.; Diaz-Uriarte, R. Pomelo II: Finding differentially expressed genes. Nucleic Acids Res. 2009, 37 (Suppl. 2), W581–W586. [Google Scholar] [CrossRef] [PubMed]
- Radmacher, M.D.; McShane, L.M.; Simon, R. A paradigm for class prediction using gene expression profiles. J. Comput. Biol. 2002, 9, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Morpheus. Available online: https://software.broadinstitute.org/morpheus (accessed on 14 December 2017).
- Ingenuity Pathways Analysis (IPA). Ingenuity Pathways Analysis (IPA) System; Ingenuity Systems, Inc.: Redwood, CA, USA, 2007; Available online: http://www.ingenuity.com/ (accessed on 13 March 2018).
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Creighton, C.J.; Sada, Y.H.; Zhang, Y.; Tsimelzon, A.; Wong, H.; Dave, B.; Landis, M.D.; Bear, H.D.; Rodriguez, A.; Chang, J.C.; et al. A gene transcription signature of obesity in breast cancer. Breast Cancer Res. Treat. 2012, 132, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Toro, A.L.; Costantino, N.S.; Shriver, C.D.; Ellsworth, D.L.; Ellsworth, R.E. Effect of obesity on molecular characteristics of invasive breast tumors: Gene expression analysis in large cohort of female patients. BMC Obes. 2016, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; He, L.; Song, Y.; Lv, Y.; Zhang, L.; Hao, F.; Xu, M. Association between obesity with disease-free survival and overall survival in triple-negative breast cancer. Medicine 2018, 97, e0719. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, L.; Zhou, Q.; Imam, M.U.; Cai, J.; Wang, Y.; Qi, M.; Sun, P.; Ping, Z.; Fu, X. Body mass index had different effects on premenopausal and postmenopausal breast cancer risks: A dose-response meta-analysis with 3,318,796 subjects from 31 cohorts. BMC Public Health 2017, 17, 936. [Google Scholar] [CrossRef] [PubMed]
- Maiti, B.; Kundranda, M.N.; Spiro, T.P.; Daw, H.A. The association of metabolic syndrome with triple-negative breast cancer. Breast Cancer Res. Treat. 2010, 121, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.A.; Kaklamani, V.G. Metabolic syndrome and triple-negative breast cancer: A new paradigm. Int. J. Breast Cancer 2012, 2012, 809291. [Google Scholar] [CrossRef] [PubMed]
- Dietze, E.C.; Chavez, T.A.; Seewaldt, V.L. Obesity and Triple-Negative Breast Cancer: Disparities, Controversies, and Biology. Am. J. Pathol. 2018, 188, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Agresti, R.; Meneghini, E.; Baili, P.; Minicozzi, P.; Turco, A.; Cavallo, I.; Sant, M. Association of adiposity, dysmetabolisms, and inflammation with aggressive breast cancer subtypes: A cross-sectional study. Breast Cancer Res. Treat. 2016, 157, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Vona-Davis, L.; Rose, D.P.; Hazard, H.; Howard-McNatt, M.; Adkins, F.; Partin, J.; Hobbs, G. Triple-negative breast cancer and obesity in a rural Appalachian population. Cancer Epidemiol. Prev. Biomark. 2008, 17, 3319–3324. [Google Scholar] [CrossRef] [PubMed]
- Stead, L.A.; Lash, T.L.; Sobieraj, J.E.; Chi, D.D.; Westrup, J.L.; Charlot, M.; Rosenberg, C.L. Triple-negative breast cancers are increased in black women regardless of age or body mass index. Breast Cancer Res. 2009, 11, R18. [Google Scholar] [CrossRef] [PubMed]
- Tayyari, F.; Gowda, G.N.; Olopade, O.F.; Berg, R.; Yang, H.H.; Lee, M.P.; Mohammed, S.I. Metabolic profiles of triple-negative and luminal A breast cancer subtypes in African-American identify key metabolic differences. Oncotarget 2018, 9, 11677–11690. [Google Scholar] [CrossRef] [PubMed]
- Sturtz, L.A.; Melley, J.; Mamula, K.; Shriver, C.D.; Ellsworth, R.E. Outcome disparities in African American women with triple negative breast cancer: A comparison of epidemiological and molecular factors between African American and Caucasian women with triple negative breast cancer. BMC Cancer 2014, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, J.M.; Gao, F.; Bumb, C.; Ellis, M.J.; Ma, C.X. Racial differences in outcomes of triple-negative breast cancer. Breast Cancer Res. Treat. 2013, 138, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cook, L.S.; Tang, M.T.C.; Porter, P.L.; Hill, D.A.; Wiggins, C.L.; Li, C.I. Body mass index and risk of luminal, HER2-overexpressing, and triple negative breast cancer. Breast Cancer Res. Treat. 2016, 157, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Babu, K.G.; Anand, A.; Lakshmaiah, K.C.; Lokanatha, D.; Jacob, L.A.; Suresh Babu, M.C.; Lokesh, K.N.; Rudresha, H.A.; Rajeev, L.K.; Saldanha, S.C. Correlation of BMI with breast cancer subtype and tumour size. Ecancer 2018, 12, 845. [Google Scholar] [CrossRef] [PubMed]
- Bandera, E.V.; Chandran, U.; Hong, C.C.; Troester, M.A.; Bethea, T.N.; Adams-Campbell, L.L.; Palmer, J.R. Obesity, body fat distribution, and risk of breast cancer subtypes in African American women participating in the AMBER Consortium. Breast Cancer Res. Treat. 2015, 150, 655–666. [Google Scholar] [CrossRef] [PubMed]
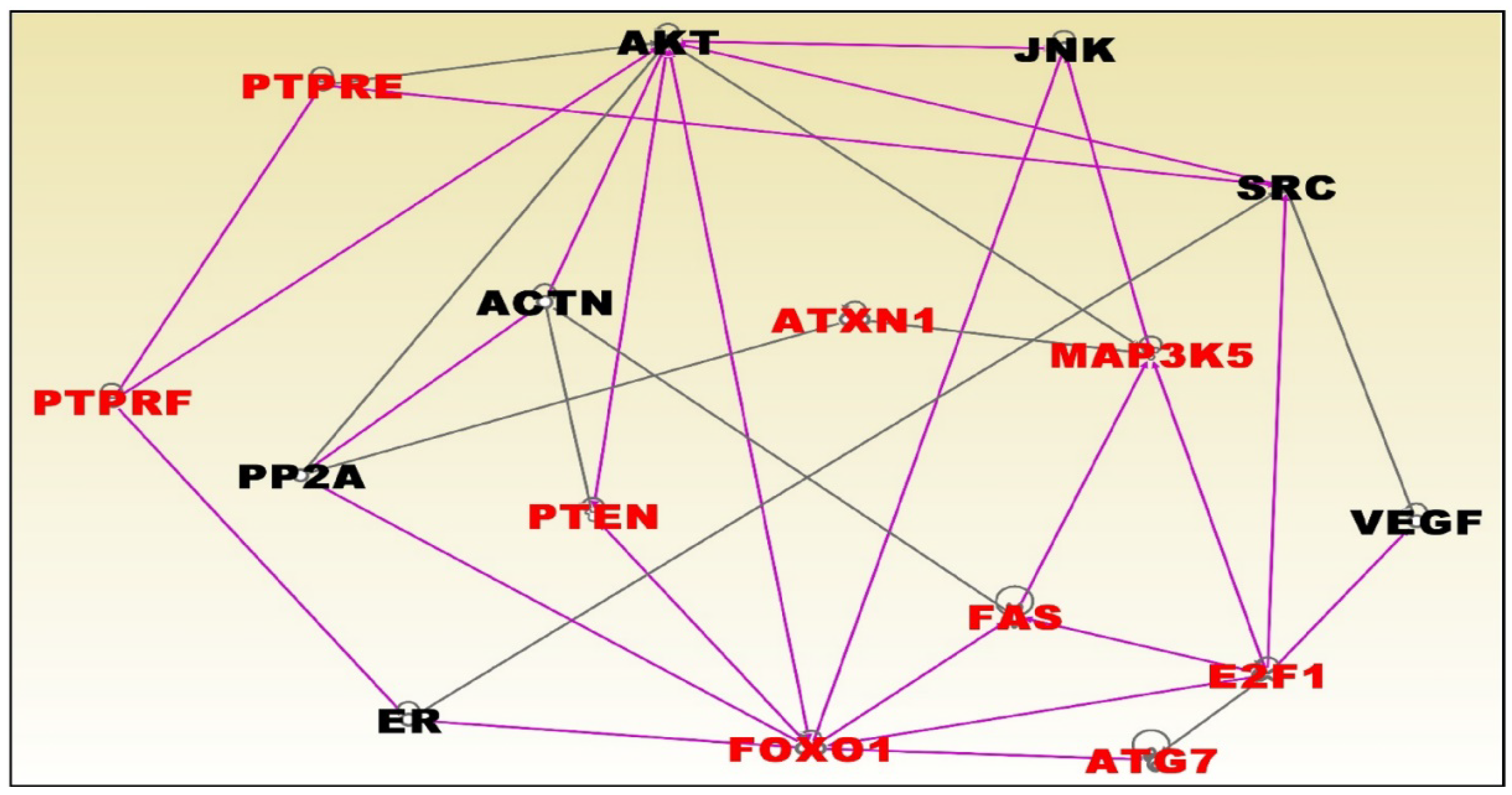
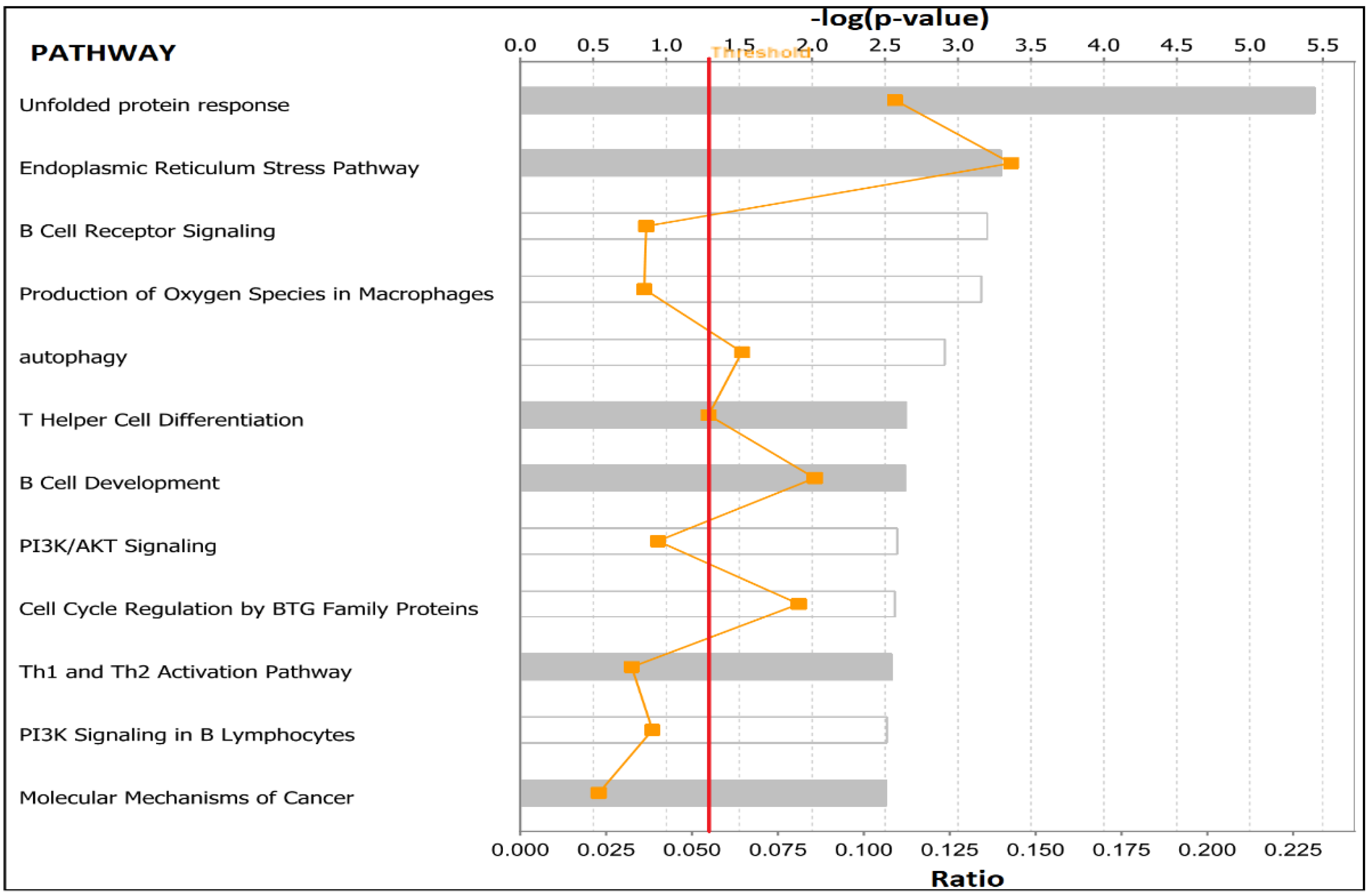
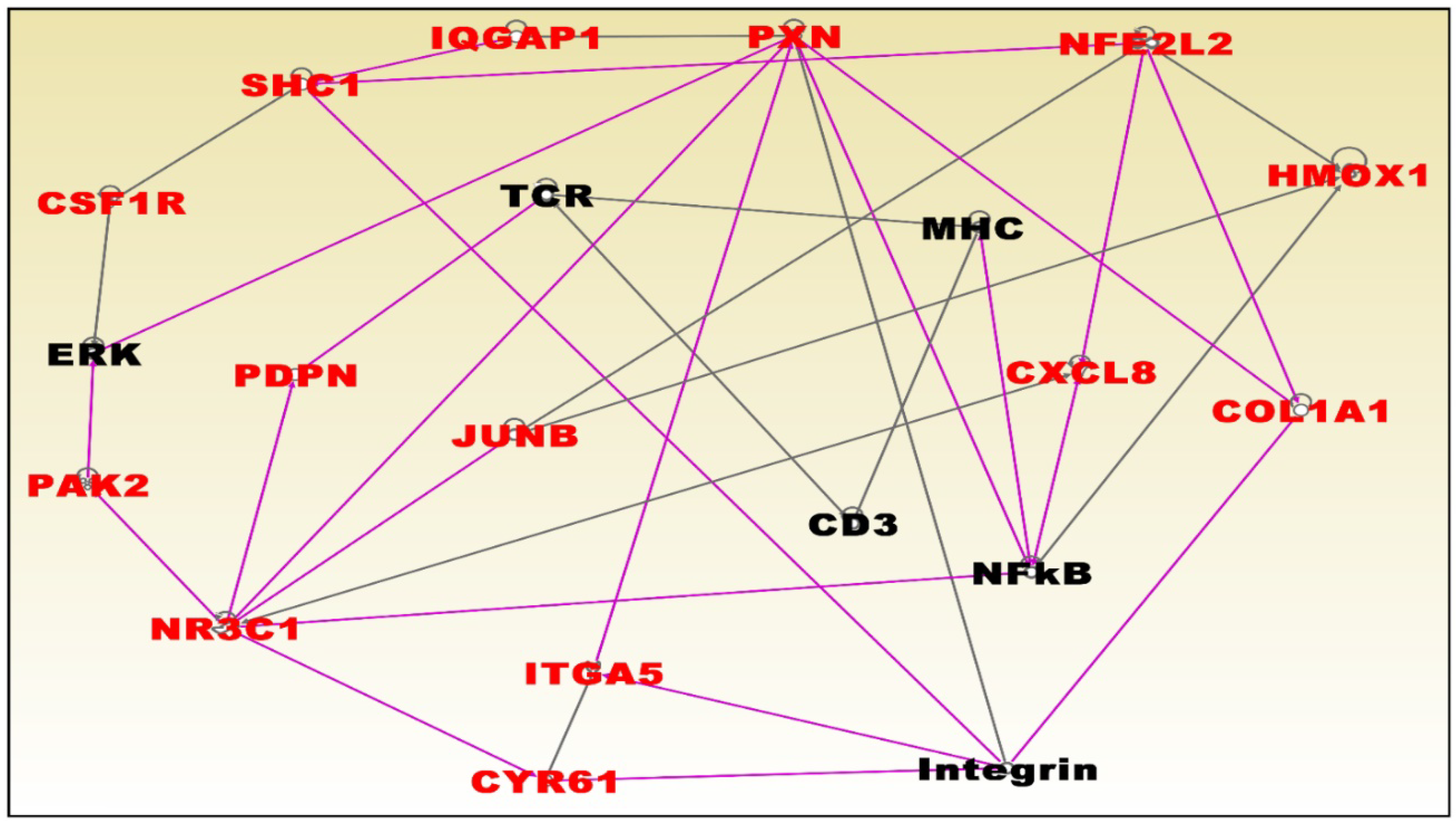

| Normal Weight Versus Overweight | Normal Weight Versus Obese | ||||
|---|---|---|---|---|---|
| Gene Symbol | Cytoband | p-Value | Gene Symbol | Cytoband | p-Value |
| COX10 | 17p12 | 0.000166 | DUXAP8 | 22q11.1 | 0.0000984 |
| DPY19L1 | 7p14.2 | 0.000236 | ABCG1 | 21q22.3 | 0.000115 |
| MYL6B | 12q13.2 | 0.000242 | IFNGR1 | 6q23.3 | 0.000187 |
| CD84 | 1q23.3 | 0.000321 | TPST2 | 22q12.1 | 0.000191 |
| RAPGEF1 | 9q34.13 | 0.000325 | CCDC32 | 15q15.1 | 0.000205 |
| PAWR | 12q21.2 | 0.000396 | SFT2D1 | 6q27 | 0.00024 |
| RPS21 | 20q13.33 | 0.000399 | SMC2 | 9q31.1 | 0.00024 |
| IL13RA1 | Xq24 | 0.000496 | NPC2 | 14q24.3 | 0.000251 |
| GABBR1 | 6p22.1 | 0.000506 | MGAT4A | 2q11.2 | 0.000256 |
| SLF2 | 10q24.31 | 0.000514 | RNASET2 | 6q27 | 0.000261 |
| ZNF621 | 3p22.1 | 0.000586 | WAPL | 10q23.2 | 0.000261 |
| TOP1MT | 8q24.3 | 0.000644 | ATXN1 | 6p22.3 | 0.000267 |
| MMD2 | 7p22.1 | 0.00068 | ALG5 | 13q13.3 | 0.00027 |
| N4BP2L2 | 13q13.1 | 0.000742 | PTEN | 10q23.31 | 0.000358 |
| SNX29 | 16p13.13 | 0.000777 | CREBL2 | 12p13.1 | 0.000429 |
| CCDC34 | 11p14.1 | 0.000803 | SUSD6 | 14q24.1 | 0.000447 |
| EPB41L4A | 5q22.1 | 0.000829 | ZC3H7B | 22q13.2 | 0.000461 |
| MAP3K3 | 17q23.3 | 0.000887 | ITFG1 | 16q12.1 | 0.000497 |
| LYST | 1q42.3 | 0.00093 | FXN | 9q21.11 | 0.000527 |
| STC2 | 5q35.2 | 0.001075 | BLVRA | 7p13 | 0.000542 |
| IQGAP1 | 15q26.1 | 0.001177 | ICAM3 | 19p13.2 | 0.000554 |
| KYNU | 2q22.2 | 0.001258 | XBP1 | 22q12.1 | 0.000671 |
| FBXO28 | 1q42.11 | 0.001358 | CLN5 | 13q22.3 | 0.000682 |
| ARSD | Xp22.3 | 0.001402 | SEL1L | 14q31 | 0.000687 |
| NBN | 8q21.3 | 0.001535 | DNAJC3 | 13q32.1 | 0.000716 |
| IFNGR1 | 6q23.3 | 0.001573 | LAYN | 11q23.1 | 0.000735 |
| CASP9 | 1p36.21 | 0.001591 | GPRIN3 | 4q22.1 | 0.000833 |
| EHD4 | 15q15.1 | 0.00167 | WIPF1 | 2q31.1 | 0.000838 |
| SSH1 | 12q24.11 | 0.001679 | SET | 9q34.11 | 0.000896 |
| SYN2 | 3p25.2 | 0.001725 | FOXN3 | 14q31.3 | 0.00093 |
| ZNF585A | 19q13.13 | 0.001861 | FBXO10 | 9p13.2 | 0.000936 |
| FCGR3A | 1q23. | 0.001938 | SNX29 | 16p13.13 | 0.000962 |
| Gene Symbol | NW vs. Obese | Gene Symbol | NW vs. Overweight | ||
|---|---|---|---|---|---|
| Cytoband | p-Value | Cytoband | p-Value | ||
| MSX1 | 4p16.2 | 0.000002 | TAGLN2 | 1q23.2 | 0.0000002 |
| IL4R | 16p12.1 | 0.0000023 | ZNF92 | 7q11.21 | 0.0000778 |
| STK10 | 5q35.1 | 0.0000194 | KEAP1 | 19p13.11 | 0.0000934 |
| PLK3 | 1p34.1 | 0.0000257 | ZNF253 | 19p13.11 | 0.0000988 |
| EPHA2 | 1p36 | 0.0000278 | YWHAH | 22q12.3 | 0.000138 |
| MUT | 6p12.3 | 0.0000321 | PLEC | 8q24 | 0.000146 |
| SERPINE1 | 7q22.1 | 0.0000412 | TMC6 | 17q25.3 | 0.000161 |
| MVP | 16p11.2 | 0.0000842 | CARHSP1 | 16p13.2 | 0.000196 |
| TNC | 9q33.1 | 0.000134 | YWHAZ | 8q22.3 | 0.000224 |
| OSMR | 5p13.1 | 0.000138 | JRK | 8q24.3 | 0.000255 |
| MESDC1 | 15q25.1 | 0.000141 | PROM2 | 2q11.1 | 0.000269 |
| RBMS1 | 2q24.2 | 0.000196 | SMAD5 | 5q31.1 | 0.000284 |
| PDPN | 1p36.21 | 0.000199 | ZNF138 | 7q11.21 | 0.000293 |
| ZEB2 | 2q22.3 | 0.000218 | MESDC1 | 15q13 | 0.000314 |
| CTDSP2 | 12q14.1 | 0.000243 | ZNF85 | 19p12 | 0.000344 |
| PTPRE | 10q26.2 | 0.000304 | ZNF736 | 7q11.21 | 0.00042 |
| TWIST2 | 2q37.3 | 0.000335 | NADSYN1 | 11q13.4 | 0.00048 |
| PML | 15q24.1 | 0.000349 | SRSF9 | 12q24.31 | 0.000492 |
| C6orf141 | 6p12.3 | 0.000386 | BAHD1 | 15q15.1 | 0.000518 |
| WWTR1-AS1 | 3q25.1 | 0.000441 | ZBTB3 | 11q12.3 | 0.000528 |
| MGAT1 | 5q35.3 | 0.000446 | SRSF7 | 2p22.1 | 0.000554 |
| GCLC | 6p12.1 | 0.000463 | BBS9 | 7p14.3 | 0.000619 |
| PELO | 5q11.2 | 0.000495 | FOSL2 | 2p23.2 | 0.000661 |
| SLC2A3 | 12p13.31 | 0.000506 | ACLY | 17q21.2 | 0.000676 |
| SSH1 | 12q24.11 | 0.000542 | KANSL1L | 2q34 | 0.000728 |
| PRKACA | 19p13.1 | 0.000627 | TFAP2A-AS1 | 6p24.3 | 0.000766 |
| PXN | 12q24.23 | 0.000652 | SMG7 | 1q25.3 | 0.000771 |
| PLEC | 8q24 | 0.000673 | DUOXA1 | 15q21.1 | 0.000772 |
| FAS | 10q23.31 | 0.000737 | HNF4A-AS1 | 20q13.12 | 0.0008 |
| DUSP1 | 5q35.1 | 0.000777 | EPHA2 | 1p36.13 | 0.000811 |
| PTAFR | 1p35.3 | 0.000786 | C11orf54 | 11q23.1 | 0.00097 |
| SCARF1 | 17p13.3 | 0.000807 | ZNF506 | 19p13.11 | 0.000979 |
| KLF7 | 2q3.3 | 0.000839 | C11orf57 | 11q23.1 | 0.001022 |
| FPR1 | 19q13.41 | 0.000848 | HPCAL1 | 2p25.1 | 0.001038 |
| CD163 | 12p13.31 | 0.000862 | KLHDC7B | 22q13.33 | 0.001083 |
| RAP1B | 12q15 | 0.000879 | PLP2 | Xp11.23 | 0.001178 |
| CTSL | 9q21.33 | 0.000896 | ZNF592 | 15q25.2 | 0.001202 |
| GGT5 | 22q11.23 | 0.000929 | TTLL13P | 15q26.1 | 0.001204 |
| GLI2 | 2q14.2 | 0.000951 | RAB34 | 17q11.2 | 0.001223 |
| ITGA5 | 12q13.13 | 0.00097 | ATM | 11q22.3 | 0.001286 |
| RBBP6 | 16p12.1 | 0.000979 | STX8 | 17p13 | 0.001314 |
| MKL1 | 22q13.1 | 0.000982 | PAX8 | 2q14.1 | 0.001328 |
| ST7L | 1p13.2 | 0.000986 | ODF3 | 11p15.5 | 0.001342 |
| ANGPT2 | 8p23.1 | 0.000992 | NEK1 | 4q33 | 0.001358 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamidi, T.K.K.; Wu, J.; Tchounwou, P.B.; Miele, L.; Hicks, C. Whole Genome Transcriptome Analysis of the Association between Obesity and Triple-Negative Breast Cancer in Caucasian Women. Int. J. Environ. Res. Public Health 2018, 15, 2338. https://doi.org/10.3390/ijerph15112338
Mamidi TKK, Wu J, Tchounwou PB, Miele L, Hicks C. Whole Genome Transcriptome Analysis of the Association between Obesity and Triple-Negative Breast Cancer in Caucasian Women. International Journal of Environmental Research and Public Health. 2018; 15(11):2338. https://doi.org/10.3390/ijerph15112338
Chicago/Turabian StyleMamidi, Tarun K. K., Jiande Wu, Paul B. Tchounwou, Lucio Miele, and Chindo Hicks. 2018. "Whole Genome Transcriptome Analysis of the Association between Obesity and Triple-Negative Breast Cancer in Caucasian Women" International Journal of Environmental Research and Public Health 15, no. 11: 2338. https://doi.org/10.3390/ijerph15112338
APA StyleMamidi, T. K. K., Wu, J., Tchounwou, P. B., Miele, L., & Hicks, C. (2018). Whole Genome Transcriptome Analysis of the Association between Obesity and Triple-Negative Breast Cancer in Caucasian Women. International Journal of Environmental Research and Public Health, 15(11), 2338. https://doi.org/10.3390/ijerph15112338







