Prevalence of Integrons and Insertion Sequences in ESBL-Producing E. coli Isolated from Different Sources in Navarra, Spain
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Detection of Integrons
2.3. Detection of Insertion Sequences
2.4. Sequence Analysis
2.5. Statistical Analysis
3. Results and Discussion
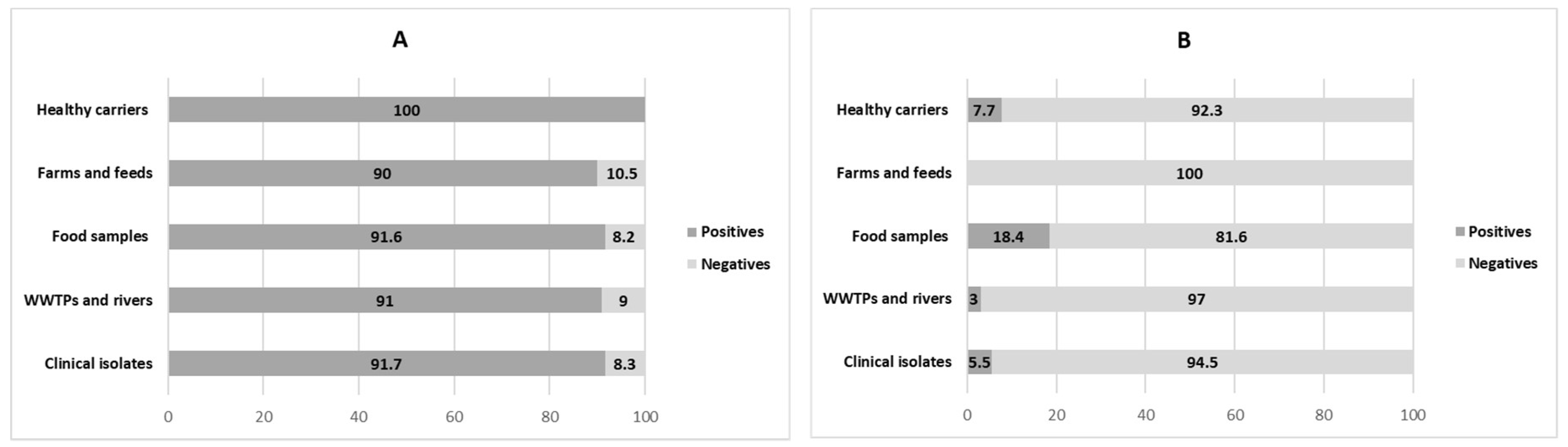
3.1. Distribution of Integrons in ESBL-Producing E. coli
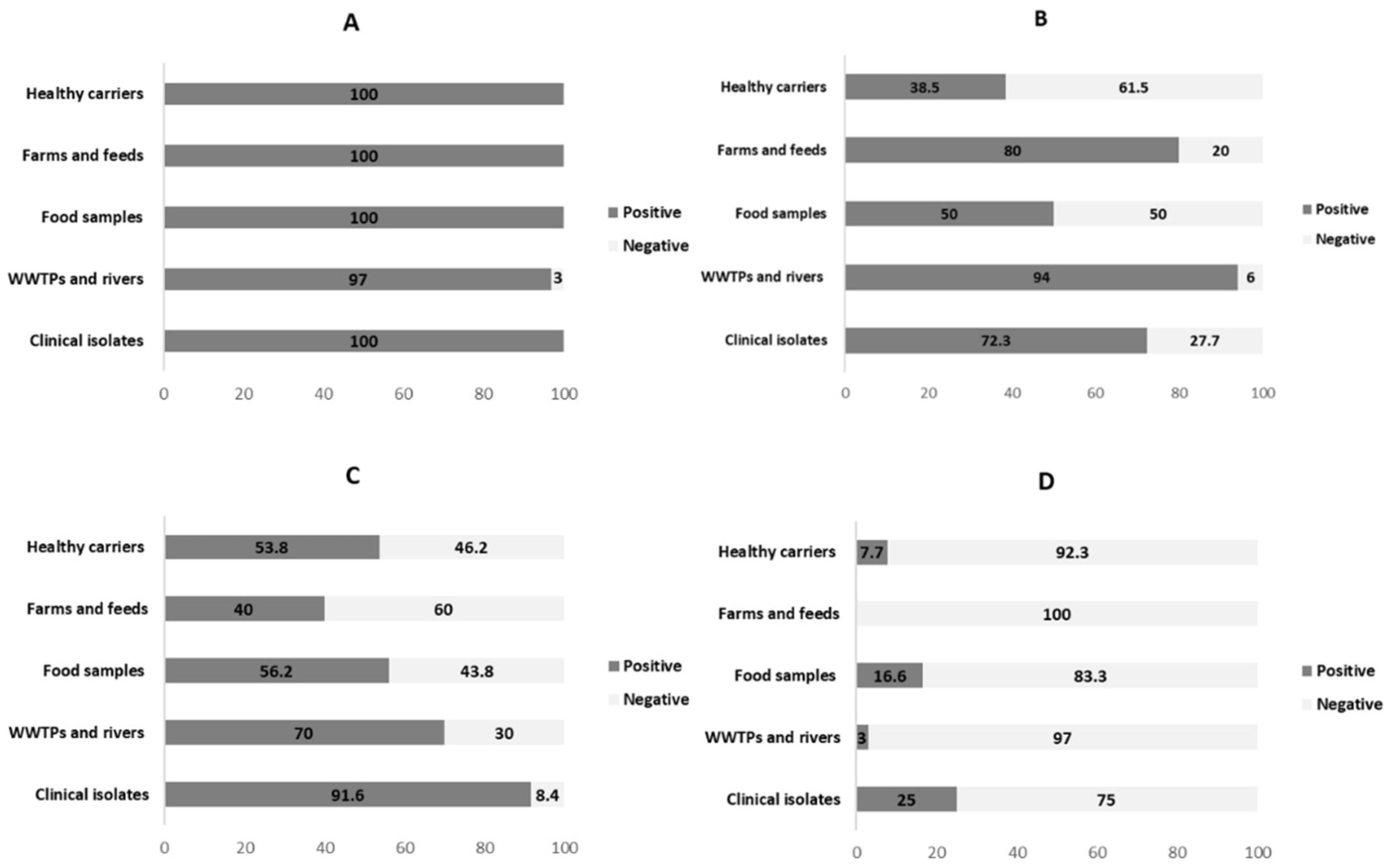
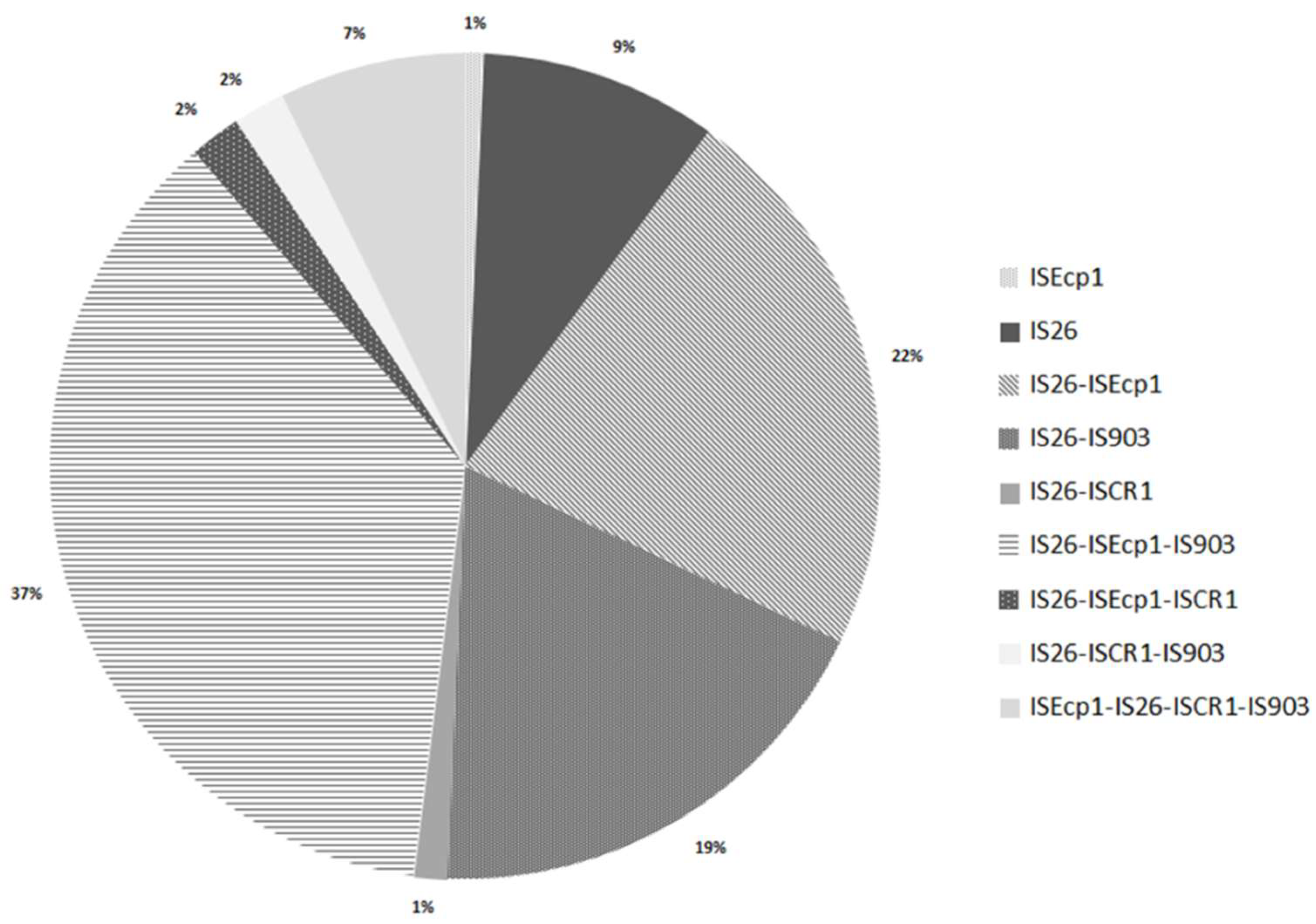
3.2. Analysis of Insertion Sequences
3.3. The Important Role of Horizontal Genetic Elements in the Dissemination of ESBLs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Antimicrobial Resistance. Global Report on Surveillance; WHO: Geneva, Switzerland, 2014; pp. 383–394. [Google Scholar]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.J.; Lyu, Y.; Li, Y.; Xue, F.; Liu, J. Trends in antimicrobial resistance against Enterobacteriaceae strains isolated from blood: A 10-year epidemiological study in mainland China (2004–2014). Chin. Med. J. 2017, 130, 2050–2055. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, S.; Fatima, J.; Shakil, S.; Rizvi, S.M.D.; Kamal, M.A. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J. Biol. Sci. 2015, 22, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Ojer-Usoz, E.; González, D.; Vitas, A.I. Clonal diversity of ESBL-producing Escherichia coli isolated from environmental, human and food samples. Int. J. Environ. Res. Public Health 2017, 14, 676. [Google Scholar] [CrossRef] [PubMed]
- Sikkema, R.; Koopmans, M. One Health training and research activities in Western Europe. Infect. Ecol. Epidemiol. 2016, 6, 33703. [Google Scholar] [CrossRef] [PubMed]
- Medini, D.; Donati, C.; Tettelin, H.; Masignani, V.; Rappuoli, R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005, 15, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Norman, A.; Hansen, L.H.; Sørensen, S.J. Conjugative plasmids: Vessels of the communal gene pool. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Turton, J.F.; Livermore, D.M. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 736–755. [Google Scholar] [CrossRef] [PubMed]
- Stokes, H.W.; Hall, R.M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: Integrons. Mol. Microbiol. 1989, 3, 1669–1683. [Google Scholar] [CrossRef] [PubMed]
- Galani, I.; Souli, M.; Koratzanis, E.; Chryssouli, Z.; Giamarellou, H. Molecular characterization of an Escherichia coli clinical isolate that produces both metallo-β-lactamase VIM-2 and extended-spectrum β-lactamase GES-7: Identification of the In8 integron carrying the blaVIM-2 gene. J. Antimicrob. Chemother. 2006, 58, 432–433. [Google Scholar] [CrossRef] [PubMed]
- Fluit, A.C.; Schmitz, F.J. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 2004, 10, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, M.; Kumar, S.; Kapoor, R.K.; Virdi, J.S.; Gulati, P. Integrons in Enterobacteriaceae: Diversity, distribution and epidemiology. Int. J. Antimicrob. Agents 2018, 51, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Machado, E.; Canto, R.; Baquero, F.; Gala, J.; Coque, T.M. Integron Content of Extended-Spectrum β-Lactamase-Producing Escherichia coli Strains over 12 Years in a Single Hospital in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Odetoyin, B.W.; Labar, A.S.; Lamikanra, A.; Aboderin, A.O.; Okeke, I.N. Classes 1 and 2 integrons in faecal Escherichia coli strains isolated from mother-child pairs in Nigeria. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Saenz, Y.; Brinas, L.; Dominguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of Resistance in Multiple-Antibiotic-Resistant Escherichia coli Strains of Human, Animal, and Food Origins. Antimicrob. Agents Chemother. 2004, 48, 3996–4001. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, Y.; Murakami, M.; Suzuki, K.; Ito, H.; Wacharotayankun, R.; Kato, N.; Ohta, M. A Novel Integron-Like Element Carrying the Metallo-β-Lactamase Gene bla IMP. Microbiology 1995, 39, 1612–1615. [Google Scholar] [CrossRef]
- Correia, M.; Boavida, F.; Grosso, F.; Salgado, M.J.; Lito, L.M.; Cristino, J.M.; Mendo, S.; Duarte, A. Molecular characterization of a new class 3 integron in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2003, 47, 2838–2843. [Google Scholar] [CrossRef] [PubMed]
- Rizk, D.E.; El-Mahdy, A.M. Emergence of class 1 to 3 integrons among members of Enterobacteriaceae in Egypt. Microb. Pathog. 2017, 112, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Mahillon, J.; Chandler, M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998, 62. [Google Scholar] [CrossRef]
- Zhao, W.H.; Hu, Z.Q. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 2013, 39, 79–101. [Google Scholar] [CrossRef] [PubMed]
- Arduino, S.M.; Roy, P.H.; Jacoby, G.A.; Betina, E.; Pineiro, S.A.; Centron, D.; Arduino, S.M.; Roy, P.H.; Jacoby, G.A.; Orman, B.E.; et al. blaCTX-M-2 Is Located in an Unusual Class 1 Integron (In35) Which Includes Orf513 blaCTX-M-2 Is Located in an Unusual Class 1 Integron (In35) Which Includes Orf513. Antimicrob. Agents Chemother. 2002, 46, 2303–2306. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Sun, J.; Zheng, F.; Lu, W.; Yang, Q.; Rui, Y. New structures simultaneously harboring class 1 integron and ISCR1-linked resistance genes in multidrug-resistant Gram-negative bacteria. BMC Microbiol. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Cullik, A.; Pfeifer, Y.; Prager, R.; Von Baum, H.; Witte, W. A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J. Med. Microbiol. 2010, 59, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Diestra, K.; Juan, C.; Curiao, T.; Moya, B.; Miro, E.; Oteo, J.; Coque, T.M.; Perez-Vazquez, M.; Campos, J.; Canton, R.; et al. Characterization of plasmids encoding blaESBL and surrounding genes in Spanish clinical isolates of Escherichia coli and Klebsiella pneumoniae. J. Antimicrob. Chemother. 2008, 63, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Eckert, C.; Gautier, V.; Arlet, G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 2006, 57, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Ojer-Usoz, E.; González, D.; Vitas, A.I.; Leiva, J.; García-Jalón, I.; Febles-Casquero, A.; de la Escolano, M.S. Prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Sci. 2013, 93, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Ojer-Usoz, E.; González, D.; García-Jalón, I.; Vitas, A.I. High dissemination of extended-spectrum β-lactamase-producing Enterobacteriaceae ineffluents from wastewater treatment plants. Water Res. 2014, 56, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Vitas, A.I.; Naik, D.; Pérez-Etayo, L.; González, D. Increased exposure to extended-spectrum β-lactamase-producing multidrug-resistant Enterobacteriaceae through the consumption of chicken and sushi products. Int. J. Food Microbiol. 2018, 269, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Mazel, D.; Dychinco, B.; Webb, V.; Davies, J. Antibiotic resistance in the ECOR collection: Integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 2000, 44, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Novais, Â.; Cantón, R.; Valverde, A.; Machado, E.; Galán, J.C.; Peixe, L.; Carattoli, A.; Baquero, F.; Coque, T.M. Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-α, and IncFI groups. Antimicrob. Agents Chemother. 2006, 50, 2741–2750. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Decousser, J.; Nordmann, P. Insertion Sequence ISEcp1B Is Involved in Expression and Mobilization of a blaCTX-M β-Lactamase Gene. Antimicrob. Agents Chemother. 2003, 47, 2938–2945. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Bao, X.; Ji, L.; Chen, L.; Liu, J.; Miao, J.; Chen, D.; Bian, H.; Li, Y.; Yu, G. Resistance integrons: Class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 2015, 14. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.M.; Collis, C.M. Mobile gene cassettes and integrons: Capture and spread of genes by site-specific recombination. Mol. Microbiol. 1995, 15, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Machado, E.; Coque, T.M.; Cantón, R.; Sousa, J.C.; Peixe, L. Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 2008, 62, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Solberg, O.; Ajiboye, R.; Riley, L. Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J. Clin. Microbiol. 2006, 44, 1347–1351. [Google Scholar] [CrossRef] [PubMed]
- Roe, M.T.; Vega, E.; Pillai, S.D. Antimicrobial Resistance Markers of Class 1 and Class 2 Integron-bearing Escherichia coli from Irrigation Water and Sediments. Emerg. Infect. Dis. 2003, 9, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Ozgumus, O.B.; Sandalli, C.; Sevim, A.; Celik-Sevim, E.; Sivri, N. Class 1 and class 2 integrons and plasmid-mediated antibiotic resistance in coliforms isolated from ten rivers in northern Turkey. J. Microbiol. 2009, 47, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Vinué, L.; Sáenz, Y.; Somalo, S.; Escudero, E.; Moreno, M.Á.; Ruiz-Larrea, F.; Torres, C. Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 2008, 62, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Kargar, M.; Mohammadalipour, Z.; Doosti, A.; Lorzadeh, S.; Japoni-Nejad, A. High prevalence of class 1 to 3 integrons among multidrug-resistant diarrheagenic Escherichia coli in southwest of Iran. Osong Public Health Res. Perspect. 2014, 5, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, C.; Lee, M.D.; Sanchez, S.; Phillips, B.; Register, B.; Grady, M.; Liebert, C.; Summers, A.O.; White, D.G.; Maurer, J.J.; et al. Incidence of Class 1 and 2 Integrases in Clinical and Commensal Bacteria from Livestock, Companion Animals, and Exotics. Antimicrob. Agents Chemother. 2001, 45, 723–726. [Google Scholar] [CrossRef] [PubMed]
- Kor, S.B.; Choo, Q.C.; Chew, C.H. New integron gene arrays from multiresistant clinical isolates of members of the Enterobacteriaceae and Pseudomonas aeruginosa from hospitals in Malaysia. J. Med. Microbiol. 2013, 62, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.; Ur Rahman, S.; Zhang, L.; Shahid, M.; Zhang, S.; Liu, G.; Gao, J.; Han, B. ESBL-producing Escherichia coli from cows suffering mastitis in China contain clinical class 1 integrons with CTX-M linked to ISCR1. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Bae, I.K.; Jeong, S.H.; Chang, C.L.; Lee, C.H.; Lee, K. Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J. Antimicrob. Chemother. 2011, 66, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.P.; Xia, J.; Yang, L.; Li, L.; Sun, J.; Liu, Y.H.; Jiang, H.X. Characterization of CTX-M-14-producing Escherichia coli from food-producing animals. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Bae, I.K.; Lee, Y.H.; Jeong, H.J.; Hong, S.G.; Lee, S.H.; Jeong, S.H. A novel blaCTX-M-14 gene-harboring complex class 1 integron with an In4-like backbone structure from a clinical isolate of Escherichia coli. Diagn. Microbiol. Infect. Dis. 2008, 62, 340–342. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Stephan, R.; Zurfluh, K.; Hächler, H.; Fanning, S. Characterization of the genetic environment of blaESBLgenes, integrons and toxin-antitoxin systems identified on large transferrable plasmids in multi-drug resistant Escherichia coli. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Van Leverstein Hall, M.A.; Dierikx, C.M.; Cohen Stuart, J.; Voets, G.M.; van den Munckhof, M.P.; van Essen-Zandbergen, A.; Platteel, T.; Fluit, A.C.; van de Sande-Bruinsma, N.; Scharinga, J.; et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 2011, 17, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Kiiru, J.; Butaye, P.; Goddeeris, B.M.; Kariuki, S. Analysis for prevalence and physical linkages amongst integrons, ISEcp 1, ISCR 1, Tn 21 and Tn 7 encountered in Escherichia coli strains from hospitalized and non-hospitalized patients in Kenya during a 19-year period (1992–2011). BMC Microbiol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Porse, A.; Schønning, K.; Munck, C.; Sommer, M.O.A. Survival and Evolution of a Large Multidrug Resistance Plasmid in New Clinical Bacterial Hosts. Mol. Biol. Evol. 2016, 33, 2860–2873. [Google Scholar] [CrossRef] [PubMed]
- Billard-Pomares, T.; Fouteau, S.; Jacquet, M.E.; Roche, D.; Barbe, V.; Castellanos, M.; Bouet, J.Y.; Cruveiller, S.; Médigue, C.; Blanco, J.; et al. Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum β-lactamase from an Escherichia coli strain. Antimicrob. Agents Chemother. 2014, 58, 6550–6557. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Hazen, T.H.; Boitano, M.; Tsai, Y.C.; Clark, T.A.; Korlach, J.; Rasko, D.A. Whole-genome assembly of Klebsiella pneumoniae coproducing NDM-1 and OXA-232 carbapenemases using single-molecule, real-time sequencing. Antimicrob. Agents Chemother. 2014, 58, 5947–5953. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zeng, Z.; Chen, S.; Ma, J.; He, L.; Liu, Y.; Deng, Y.; Lei, T.; Zhao, J.; Liu, J.H. High prevalence of blaCTX-M extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 2010, 16, 1475–1481. [Google Scholar] [CrossRef] [PubMed]
- Tamang, M.D.; Nam, H.M.; Gurung, M.; Jang, G.C.; Kim, S.R.; Jung, S.C.; Park, Y.H.; Lim, S.K. Molecular characterization of CTX-M β-lactamase and associated addiction systems in Escherichia coli circulating among cattle, farm workers, and the farm environment. Appl. Environ. Microbiol. 2013, 79, 3898–3905. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Martinez, J.M.; Poirel, L.; Canton, R.; Nordmann, P. Common region CR1 for expression of antibiotic resistance genes. Antimicrob. Agents Chemother. 2006, 50, 2544–2546. [Google Scholar] [CrossRef] [PubMed]
- Toleman, M.A.; Bennett, P.M.; Walsh, T.R. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J. Antimicrob. Chemother. 2006, 58. [Google Scholar] [CrossRef] [PubMed]
- Toleman, M.A.; Bennett, P.M.; Walsh, T.R. ISCR Elements: Novel Gene-Capturing Systems of the 21st Century? Microbiol. Mol. Biol. Rev. 2006, 70, 296–316. [Google Scholar] [CrossRef] [PubMed]
- Millan, B.; Castro, D.; Araque, M.; Ghiglione, B.; Gutkind, G. ISCR1 associated with blaCTX-M-1 y blaCTX-M-2 genes in IncN and IncFIIA plasmids isolated from Klebsiella pneumoniae of nosocomial origin in Mérida, Venezuela. Biomédica 2012, 33, 268–275. [Google Scholar] [CrossRef]
- Woodford, N.; Carattoli, A.; Karisik, E.; Underwood, A.; Ellington, M.J.; Livermore, D.M. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 2009, 53, 4472–4482. [Google Scholar] [CrossRef] [PubMed]
| Sample Origin | Percentages of Detected bla Genes | |||||
|---|---|---|---|---|---|---|
| blaCTX-M-14 | blaCTX-M-15 | blaCTX-M-1 | blaTEM-42 | blaTEML-171 | blaSHV-12 | |
| Hospital inpatients | 41.7 | 61.1 | 8.3 | 11.1 | NA | 5.5 |
| Healthy people | 46.2 | 30.8 | 15.4 | 0 | 46.2 | 0 |
| WWTP and rivers | 33.3 | 30.3 | 18.2 | 6 | NA | 6 |
| Food | 32.7 | 4.1 | 18.3 | 12.3 | 31.8 | 35.6 |
| Farms and feeds | 31.6 | 5.26 | 47.4 | 26.3 | 5 | 21 |
| Primer | Sequence (5′-3′) | Amplicon Size (pb) | T (°C ) 3 | GenBank Accession No | Reference |
|---|---|---|---|---|---|
| intI1-Fw 1 | GGTCAAGGATCTGGATTTCG | 483 | 62 | U49101 | [31] |
| intI1-Rv 2 | ACATGCGTGTAAATCATCGTC | 483 | 62 | U49101 | [31] |
| intI2-Fw 1 | CACGGATATGCGACAAAAAGGT | 789 | 62 | L10818 | [31] |
| intI2-Rv 2 | TAGCAAACGAGTGACGAAATG | 789 | 62 | L10818 | [31] |
| intI3-Fw 1 | AGTGGGTGGCGAATGAGTG | 600 | 60 | D50438 | [31] |
| intI3-Rv 2 | TGTTCTTGTATCGGCAGGTG | 600 | 60 | D50438 | [31] |
| Primer 1 | Sequence (5′-3′) | Amplicon Size (pb) | T (°C) 3 | GenBank Accession No. | Reference |
|---|---|---|---|---|---|
| ISEcp1-Fw 1 | ATCTAACATCAAATGCAGG | 1381 | 60 | AJ972954 | [27] |
| ISEcp1-Rv 2 | AGACTGCTTCTCACACAT | 1381 | 60 | AJ972954 | [27] |
| IS26-Fw 1 | TCACTCCACGATTTACCGCT | 557 | 61 | AF205943 | [27] |
| IS26-Rv 2 | CTTACCAGGCGCATTTCGCC | 557 | 61 | AF205943 | [27] |
| ISCR1-Fw 1 | TCGCTGCGAGGATTGTCATC | 1100 | 60 | AF174129 | [32] |
| ISCR1-Rv 2 | CTCGCTTGAGGCGTTGCAT | 1100 | 60 | AF174129 | [32] |
| IS903-Fw 1 | CATATGAAATCATCTGCGC | 473 | 56 | EU056266 | [33] |
| IS903-Rv 2 | CCGTAGCGGGTTGTGTTTTC | 473 | 56 | EU056266 | [33] |
| bla Genes | IS26 | IS903 | ISEcp1 | ISCR1 | intI1 | intI2 |
|---|---|---|---|---|---|---|
| blaCTX-M | 99.2 | 90.3 | 79 | 11.3 | 94 | 6.5 |
| blaTEM | 100 | 89.9 | 88.5 | 16 | 94 | 7.3 |
| blaOXA-1 | 94.5 | 83.3 | 50 | 5.5 | 100 | 0 |
| blaSHV | 100 | 56.5 | 26 | 0 | 95.7 | 21.8 |
| Number of IS in Each Isolate | N Isolates | N Isolates (%) Producing | |||
|---|---|---|---|---|---|
| 1 ESBL | 2 ESBL | 3 ESBL | 4 ESBL | ||
| 1 | 15 | 46.6 | 40 | 0 | 0 |
| 2 | 63 | 46 | 46 | 8 | 0 |
| 3 | 61 | 49 | 41 | 8.2 | 1.6 |
| 4 | 11 | 0 | 81.8 | 18.2 | 0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Etayo, L.; Berzosa, M.; González, D.; Vitas, A.I. Prevalence of Integrons and Insertion Sequences in ESBL-Producing E. coli Isolated from Different Sources in Navarra, Spain. Int. J. Environ. Res. Public Health 2018, 15, 2308. https://doi.org/10.3390/ijerph15102308
Pérez-Etayo L, Berzosa M, González D, Vitas AI. Prevalence of Integrons and Insertion Sequences in ESBL-Producing E. coli Isolated from Different Sources in Navarra, Spain. International Journal of Environmental Research and Public Health. 2018; 15(10):2308. https://doi.org/10.3390/ijerph15102308
Chicago/Turabian StylePérez-Etayo, Lara, Melibea Berzosa, David González, and Ana Isabel Vitas. 2018. "Prevalence of Integrons and Insertion Sequences in ESBL-Producing E. coli Isolated from Different Sources in Navarra, Spain" International Journal of Environmental Research and Public Health 15, no. 10: 2308. https://doi.org/10.3390/ijerph15102308
APA StylePérez-Etayo, L., Berzosa, M., González, D., & Vitas, A. I. (2018). Prevalence of Integrons and Insertion Sequences in ESBL-Producing E. coli Isolated from Different Sources in Navarra, Spain. International Journal of Environmental Research and Public Health, 15(10), 2308. https://doi.org/10.3390/ijerph15102308





