Ephedra-Treated Donor-Derived Gut Microbiota Transplantation Ameliorates High Fat Diet-Induced Obesity in Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Aqueous Extraction of Ephedra sinica
2.2. High-Performance Liquid Chromatography (HPLC)
2.3. Animals and Experimental Design
2.4. Biochemical Analyses
2.5. Enzyme Immunoassay for Serum Insulin
2.6. Fecal Acetic Acid Quantification by HPLC
2.7. Determination of Fecal Endotoxin
2.8. Real-Time Polymerase Chain Reaction (PCR)
2.9. Pyrosequencing and Bioinformatics Analysis
2.10. Statistical Analysis
3. Results
3.1. Es Extraction Yield and Ephedrine Proportion
3.2. Both Es and Es Altered Gut Microbiota Changed Body Weight (Gain), Fats and Organs Weight and Food Intake
3.3. EsM Ameliorated Hyperglycemia and Hyperinsulinemia, but Did Not Alter Serum Aminotransferases and Lipid Parameters
3.4. Both Es and EsM Ameliorated Enterogenous Endotoxin
3.5. Both Es and EsM Elevated the Fecal Acetic Acid Level
3.6. Both Es and EsM Down-Regulated Gene Expression of Key Proinflammatory Cytokines
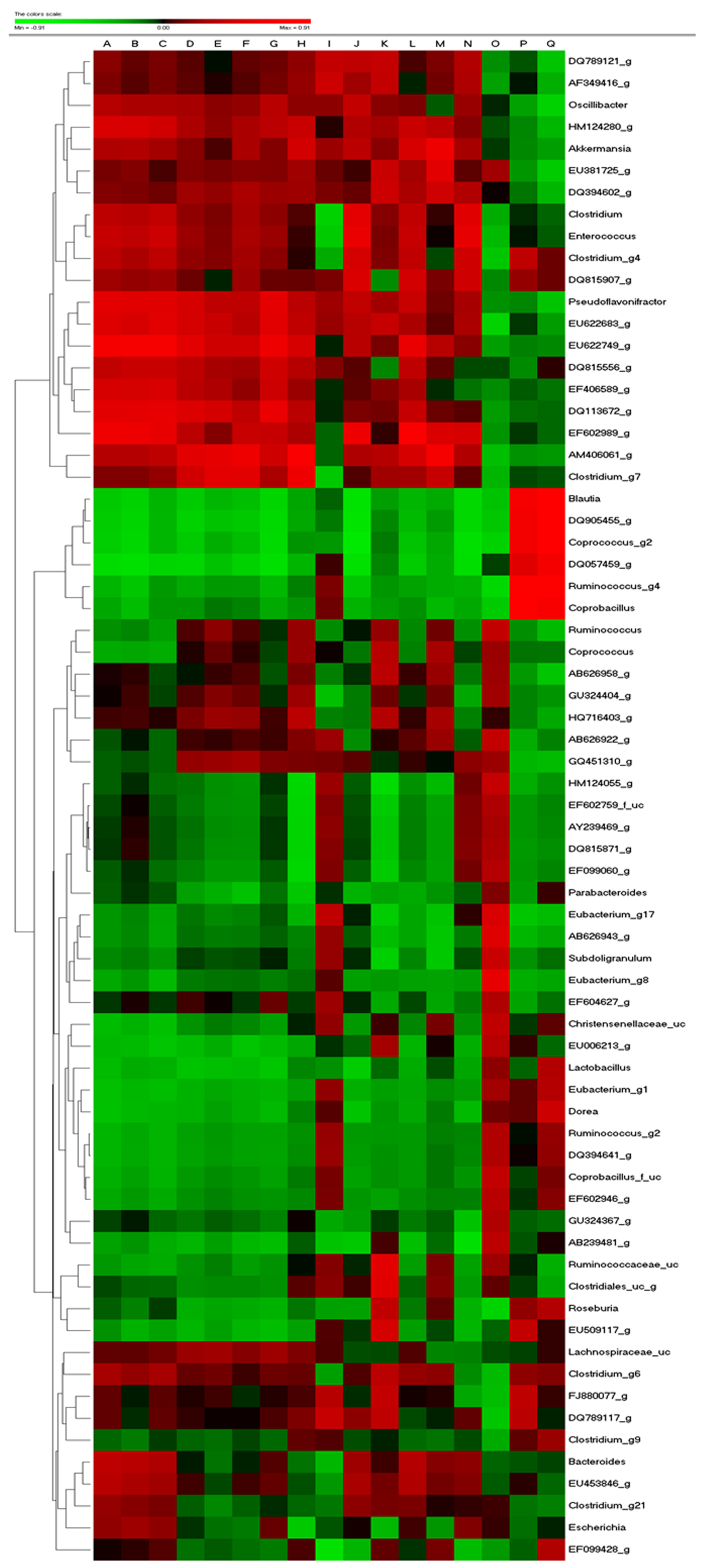
3.7. Gut Microbiota from Donor Evidently Influenced the Host Gut Microbial Community
3.8. Predicted Alteration of Gut Microbiome Function
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stoner, L.; Cornwall, J. Did the American Medical Association make the correct decision classifying obesity as a disease? Australas. Med. J. 2014, 7, 462–464. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Obesity: Preventing and Managing the Global Epidemic: Report of a WHO Consultation; World Health Organization, Division of Noncommunicable Disease, Programme of Nutrition Family and Reproductive Health: Geneva, Switzerland, 2000. [Google Scholar]
- Hill, J.O.; Wyatt, H.R.; Peters, J.C. Energy balance and obesity. Circulation 2012, 126, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Adan, R.A. Mechanisms underlying current and future anti-obesity drugs. Trends Neurosci. 2013, 36, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Mohamed-Ali, V.; Pinkney, J.H.; Coppack, S.W. Adipose tissue as an endocrine and paracrine organ. Int. J. Obes. Relat. Metab. Disord. 1998, 22, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Panee, J. Monocyte Chemoattractant Protein 1 (MCP-1) in obesity and diabetes. Cytokine 2012, 60, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kanda, H.; Tateya, S.; Tamori, Y.; Kotani, K.; Hiasa, K.; Kitazawa, R.; Kitazawa, S.; Miyachi, H.; Maeda, S.; Egashira, K.; et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J. Clin. Investig. 2006, 116, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.; Bouter, L.M.; McQuillan, G.M.; Wener, M.H.; Harris, T.B. Elevated C-reactive protein levels in overweight and obese adults. JAMA 1999, 282, 2131–2135. [Google Scholar] [CrossRef] [PubMed]
- Fain, J.N.; Madan, A.K.; Hiler, M.L.; Cheema, P.; Bahouth, S.W. Comparison of the release of adipokines by adipose tissue, adipose tissue matrix, and adipocytes from visceral and subcutaneous abdominal adipose tissues of obese humans. Endocrinology 2004, 145, 2273–2282. [Google Scholar] [CrossRef] [PubMed]
- DeBoer, M.D. Obesity, systemic inflammation, and increased risk for cardiovascular disease and diabetes among adolescents: A need for screening tools to target interventions. Nutrition 2013, 29, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.A.; Gu, W.; Lee, I.A.; Joh, E.H.; Kim, D.H. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS ONE 2012, 7, e47713. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L. The gut microbiota and obesity: From correlation to causality. Nat. Rev. Microbiol. 2013, 11, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Alang, N.; Kelly, C.R. Weight gain after fecal microbiota transplantation. Open Forum Infect. Dis. 2015, 2, ofv004. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Lund, R.; Laiho, A.; Lundelin, K.; Ley, R.E.; Isolauri, E.; Salminen, S. Gut microbiota as an epigenetic regulator: Pilot study based on whole-genome methylation analysis. mBio 2014, 5, e02113-14. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Bose, S.; Kim, G.C.; Hong, S.U.; Kim, J.H.; Kim, J.E.; Kim, H. Flos Lonicera ameliorates obesity and associated endotoxemia in rats through modulation of gut permeability and intestinal microbiota. PLoS ONE 2014, 9, e86117. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Gu, D.; Li, J.; Cui, K.; Zhang, Y. Effects and action mechanisms of berberine and Rhizoma coptidis on gut microbes and obesity in high-fat diet-fed C57BL/6J mice. PLoS ONE 2011, 6, e24520. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lian, F.; Zhao, L.; Zhao, Y.; Chen, X.; Zhang, X.; Guo, Y.; Zhang, C.; Zhou, Q.; Xue, Z.; et al. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 2014, 9, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Y.; Zhang, M.; Pang, X.; Xu, J.; Kang, C.; Li, M.; Zhang, C.; Zhang, Z.; Zhang, Y.; et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS ONE 2012, 7, e42529. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.K. A comparative study of synthetic and natural ephedrines. J. Pharmacol. Exp. Ther. 1928, 33, 237–258. [Google Scholar]
- Shekelle, P.G.; Hardy, M.L.; Morton, S.C.; Maglione, M.; Mojica, W.A.; Suttorp, M.J.; Rhodes, S.L.; Jungvig, L.; Gagne, J. Efficacy and safety of ephedra and ephedrine for weight loss and athletic performance: A meta-analysis. JAMA 2003, 289, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Song, M.K.; Um, J.Y.; Jang, H.J.; Lee, B.C. Beneficial effect of dietary Ephedra sinica on obesity and glucose intolerance in high-fat diet-fed mice. Exp. Ther. Med. 2012, 3, 707–712. [Google Scholar] [PubMed]
- Kim, B.S.; Song, M.Y.; Kim, H. The anti-obesity effect of Ephedra sinica through modulation of gut microbiota in obese Korean women. J. Ethnopharmacol. 2014, 152, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [PubMed]
- United States Pharmacopeial Convention. General Chapter <85> Bacterial Endotoxins Test; United States Pharmacopeia (USP): Rockville, MD, USA, 2012.
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Abubucker, S.; Segata, N.; Goll, J.; Schubert, A.M.; Izard, J.; Cantarel, B.L.; Rodriguez-Mueller, B.; Zucker, J.; Thiagarajan, M.; Henrissat, B.; et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 2012, 8, e1002358. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef]
- Daniel, H.; Moghaddas Gholami, A.; Berry, D.; Desmarchelier, C.; Hahne, H.; Loh, G.; Mondot, S.; Lepage, P.; Rothballer, M.; Walker, A.; et al. High-fat diet alters gut microbiota physiology in mice. ISME J. 2014, 8, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Woting, A.; Pfeiffer, N.; Loh, G.; Klaus, S.; Blaut, M. Clostridium ramosum promotes high-fat diet-induced obesity in gnotobiotic mouse models. MBio 2014, 5, e01530-14. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Bose, S.; Kim, H.G.; Han, K.S.; Kim, H. Fermented Rhizoma Atractylodis Macrocephalae alleviates high fat diet-induced obesity in association with regulation of intestinal permeability and microbiota in rats. Sci. Rep. 2015, 5, 8391. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Backhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Frost, G.; Sleeth, M.L.; Sahuri-Arisoylu, M.; Lizarbe, B.; Cerdan, S.; Brody, L.; Anastasovska, J.; Ghourab, S.; Hankir, M.; Zhang, S.; et al. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat. Commun. 2014, 5, 3611. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; Van-Hylckama Vlieg, J.E.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Fujisawa, K.; Ito, E.; Idei, S.; Kawaguchi, N.; Kimoto, M.; Hiemori, M.; Tsuji, H. Improvement of obesity and glucose tolerance by acetate in Type 2 diabetic Otsuka Long-Evans Tokushima Fatty (OLETF) rats. Biosci. Biotechnol. Biochem. 2007, 71, 1236–1243. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Lazarevic, V.; Derrien, M.; Girard, M.; Muccioli, G.G.; Neyrinck, A.M.; Possemiers, S.; Van Holle, A.; Francois, P.; De Vos, W.M.; et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 2011, 60, 2775–2786. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Park, Y.J.; Kim, Y.R.; Kim, Y.N.; Ka, S.; Lee, H.Y.; Seong, J.K.; Seok, Y.J.; Kim, J.B. Alteration of gut microbiota by vancomycin and bacitracin improves insulin resistance via glucagon-like peptide 1 in diet-induced obesity. FASEB J. 2015, 29, 2397–2411. [Google Scholar] [CrossRef] [PubMed]
- Nov, O.; Shapiro, H.; Ovadia, H.; Tarnovscki, T.; Dvir, I.; Shemesh, E.; Kovsan, J.; Shelef, I.; Carmi, Y.; Voronov, E.; et al. Interleukin-1β regulates fat-liver crosstalk in obesity by auto-paracrine modulation of adipose tissue inflammation and expandability. PLoS ONE 2013, 8, e53626. [Google Scholar] [CrossRef] [PubMed]
- Huber, J.; Kiefer, F.W.; Zeyda, M.; Ludvik, B.; Silberhumer, G.R.; Prager, G.; Zlabinger, G.J.; Stulnig, T.M. CC chemokine and CC chemokine receptor profiles in visceral and subcutaneous adipose tissue are altered in human obesity. J. Clin. Endocrinol. Metab. 2008, 93, 3215–3221. [Google Scholar] [CrossRef] [PubMed]
- Park, S.K.; Kim, M.S.; Roh, S.W.; Bae, J.W. Blautia stercoris sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2012, 62, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Conterno, L.; Fava, F.; Viola, R.; Tuohy, K.M. Obesity and the gut microbiota: Does up-regulating colonic fermentation protect against obesity and metabolic disease? Genes Nutr. 2011, 6, 241–260. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Tremaroli, V.; Backhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
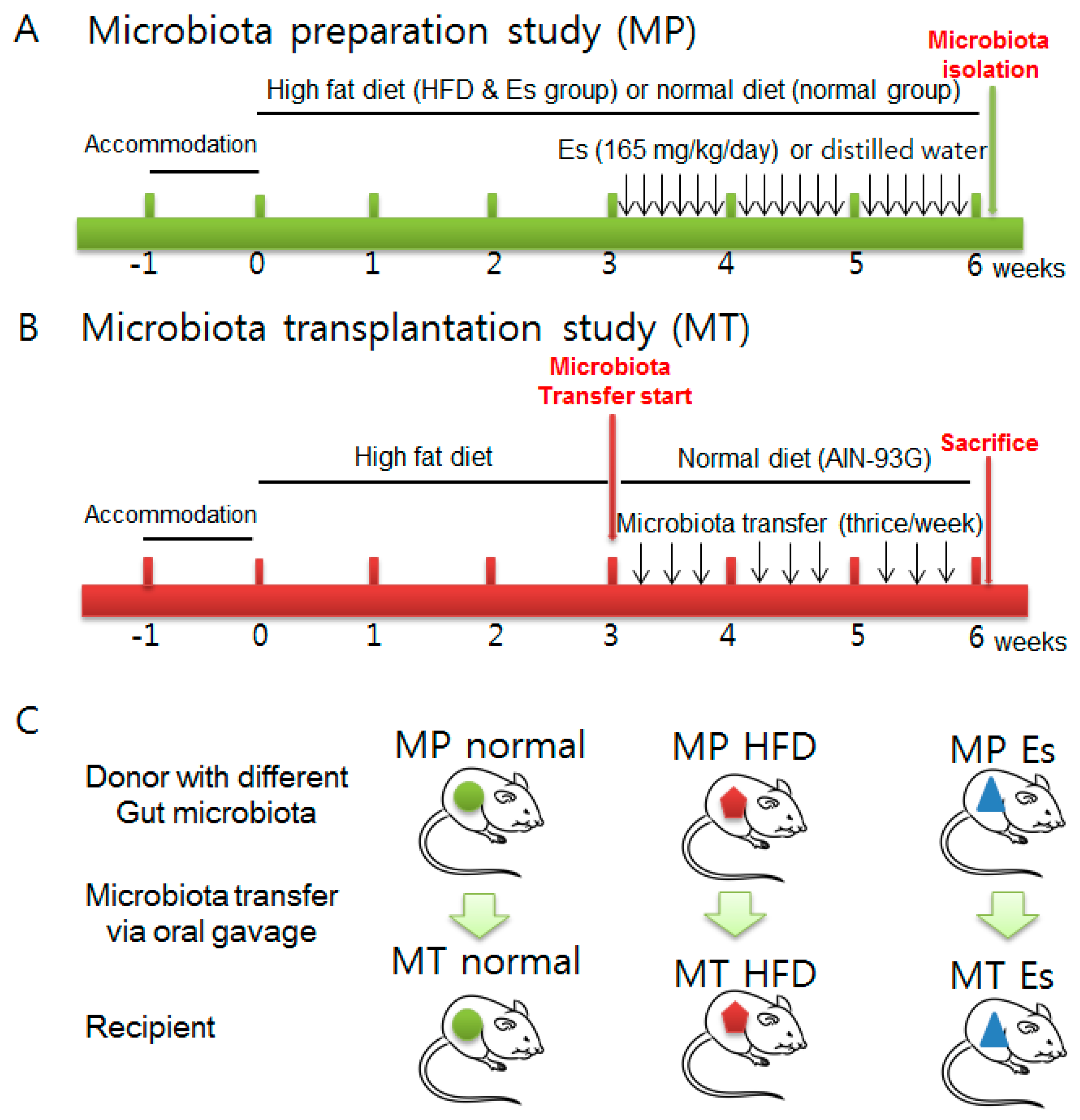

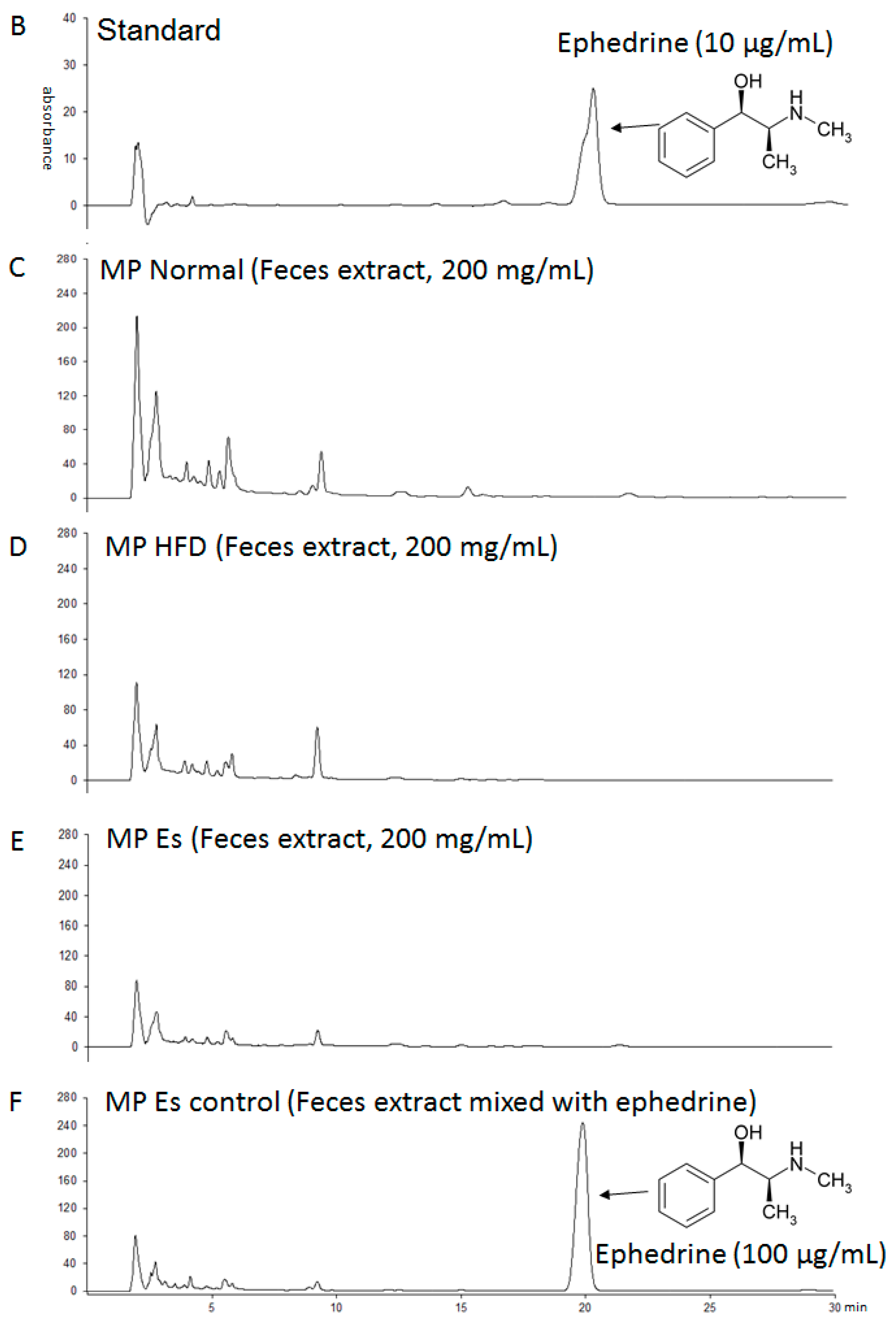
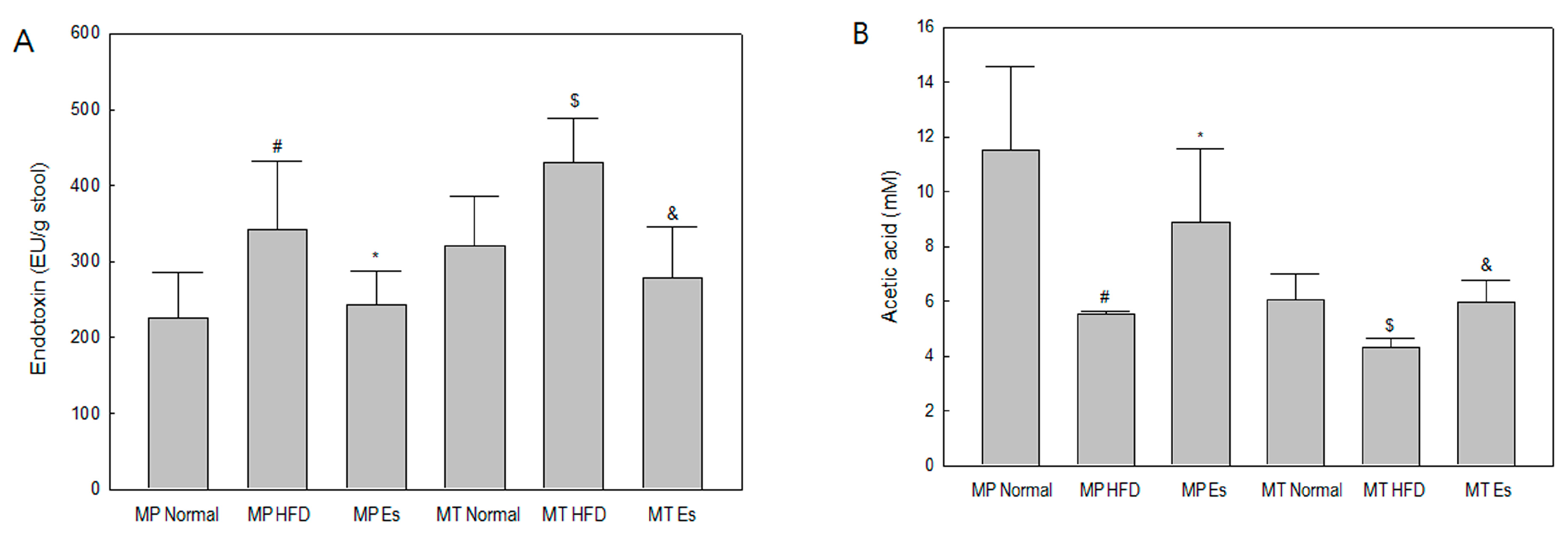

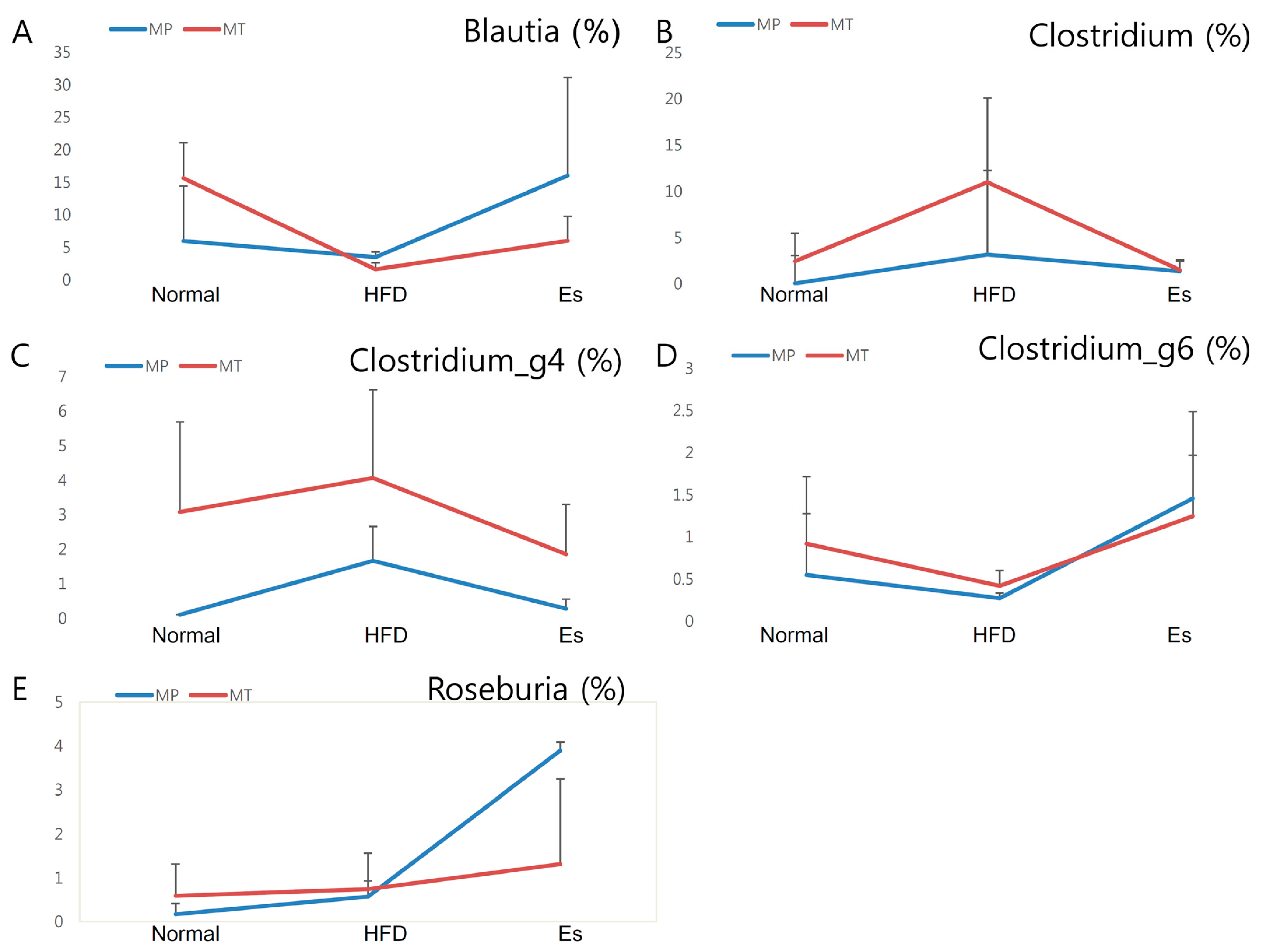
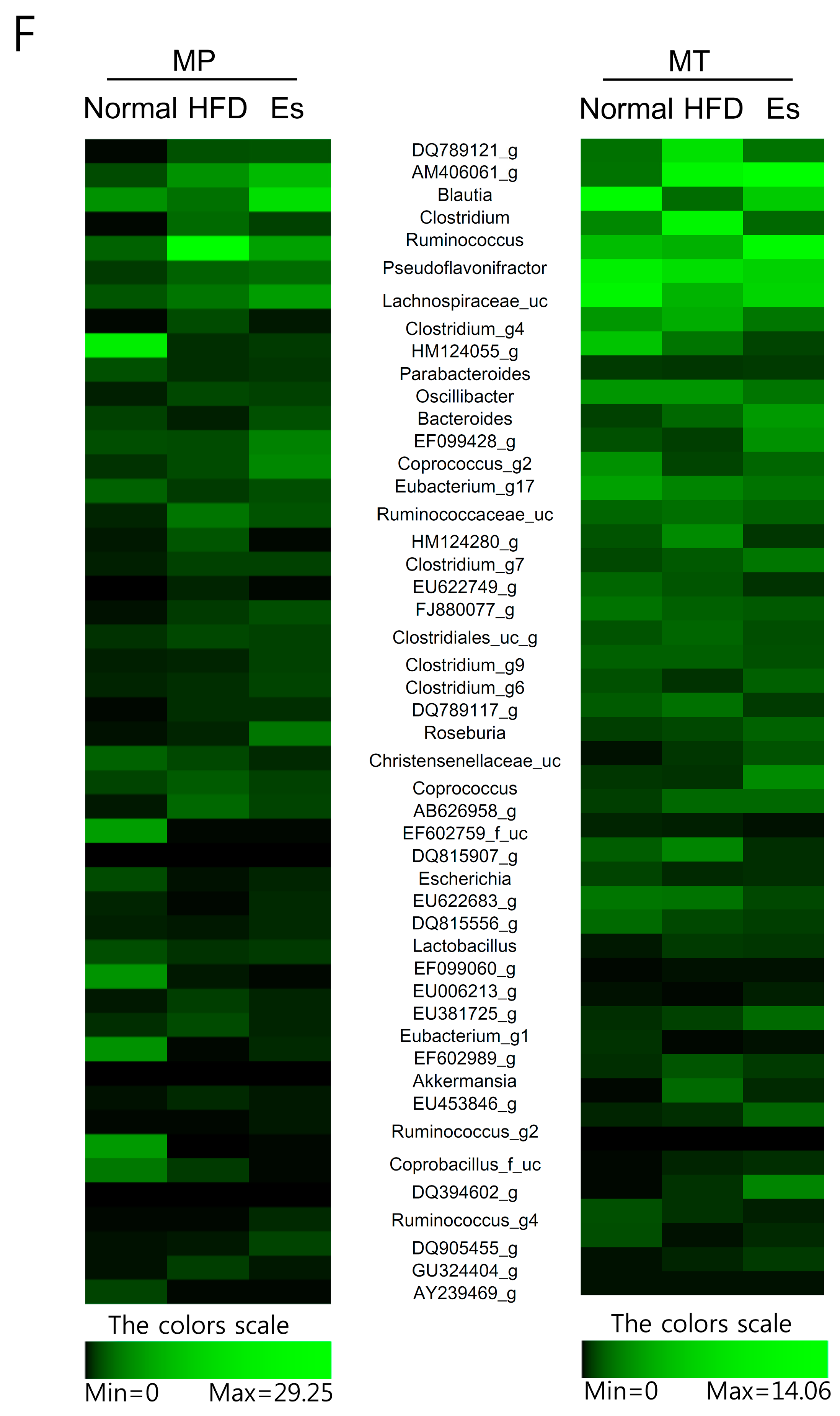
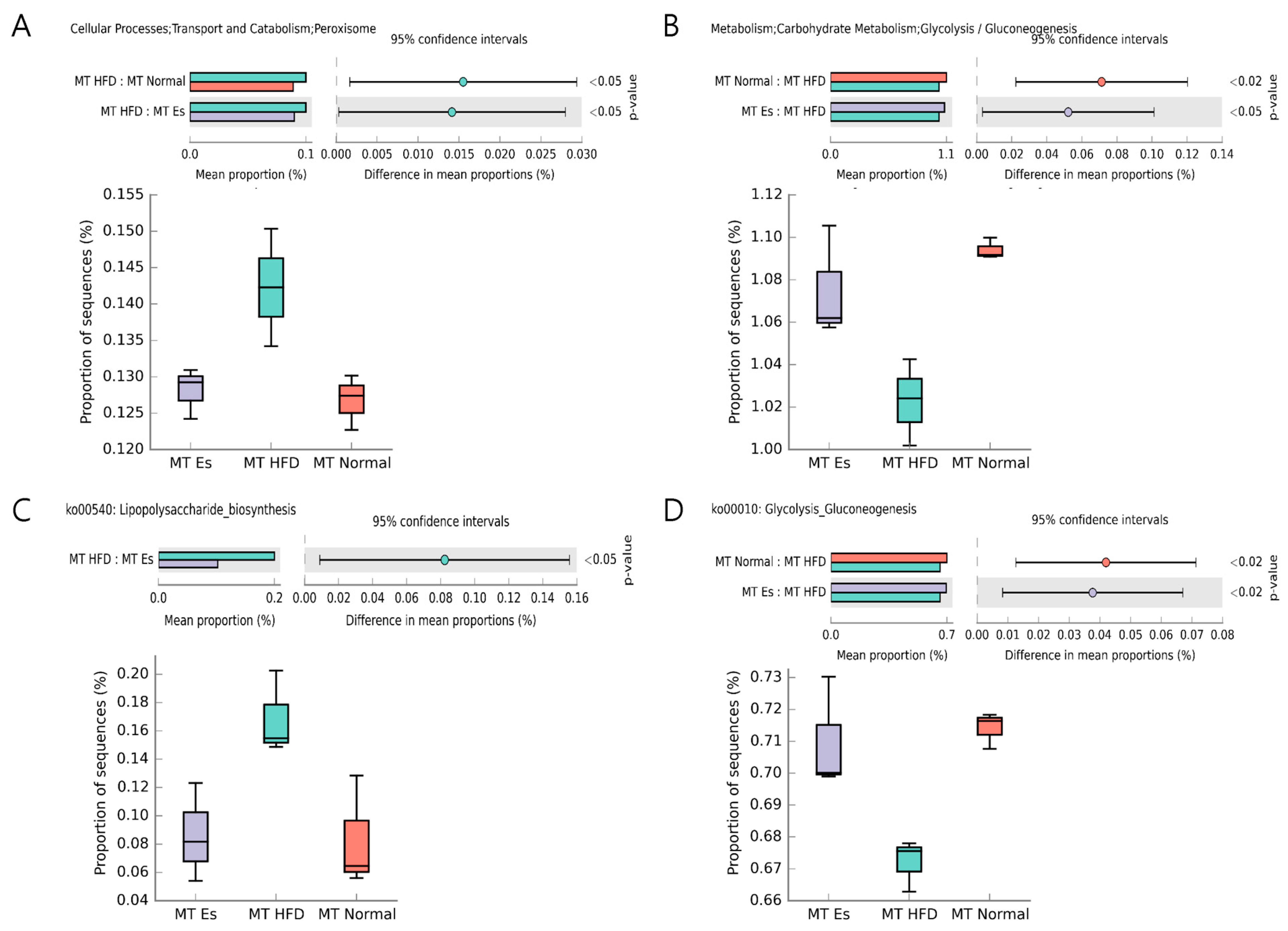
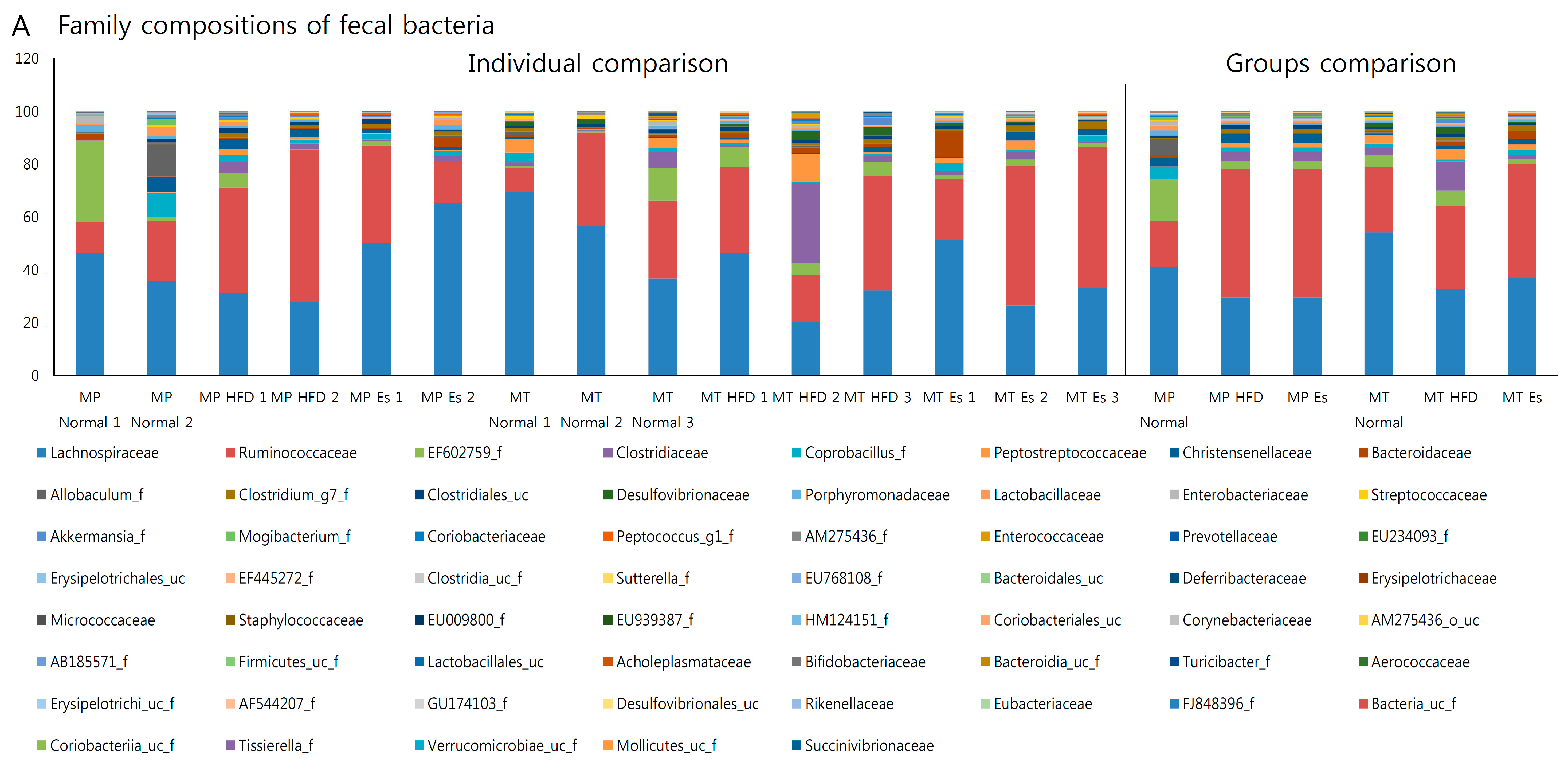
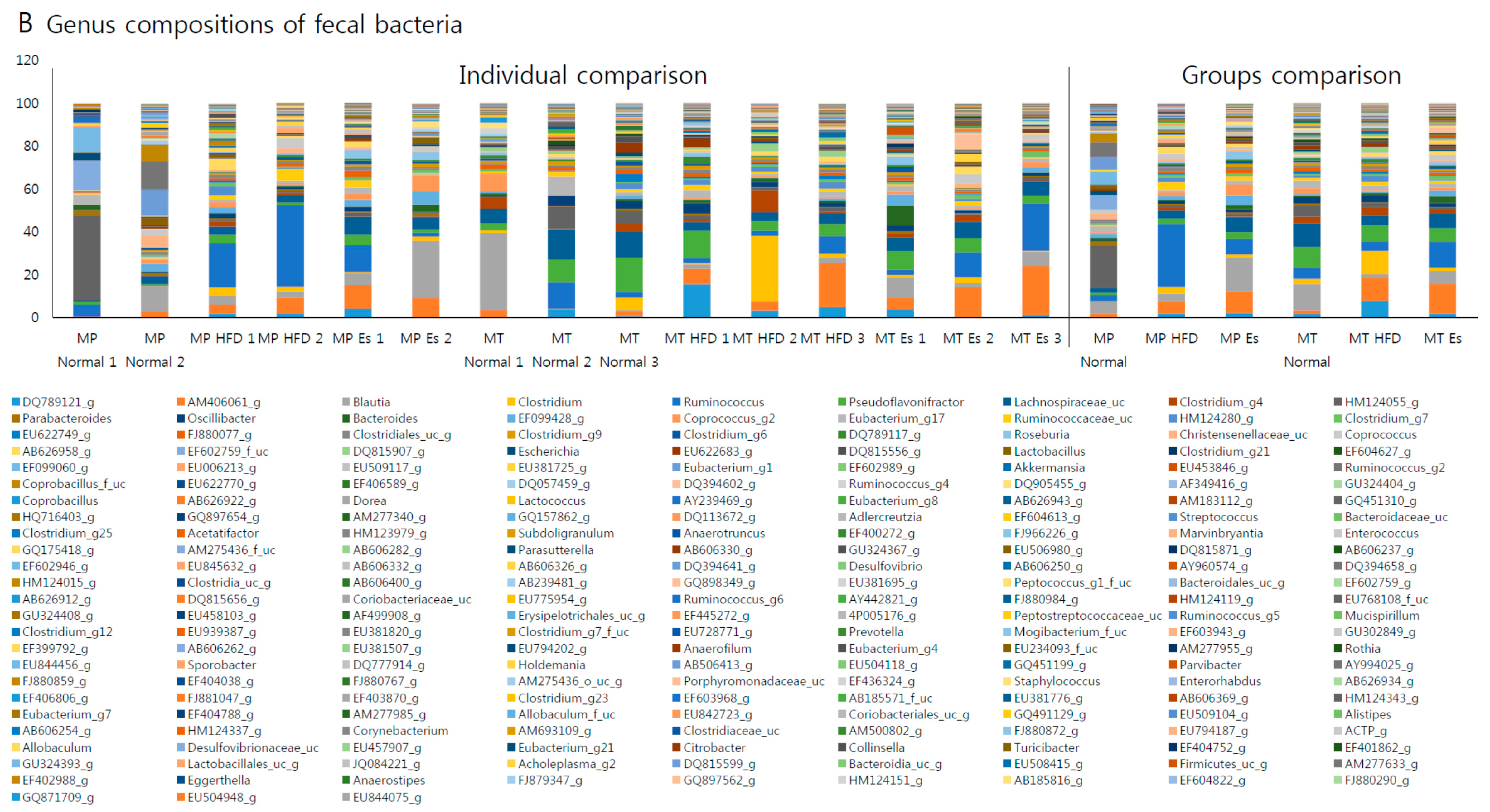
| Formulation | Normal Diet (AIN-93G) | High Fat Diet | ||
|---|---|---|---|---|
| g% | kcal% | g% | kcal% | |
| Protein | 20 | 20 | 27 | 20 |
| Carbohydrate | 64 | 64 | 25 | 20 |
| Fat | 7 | 16 | 36 | 60 |
| kcal/kg | 4000 | 5333 | ||
| Ingredient | g | kcal | g | kcal |
| Casein (from milk) | 200 | 800 | 200 | 800 |
| Corn starch | 397,486 | 1590 | 47,536 | 190 |
| Sucrose | 100 | 400 | 0 | 0 |
| Dextrose | 132 | 528 | 132 | 528 |
| Cellulose | 50 | 0 | 50 | 0 |
| Soybean oil | 70 | 630 | 25 | 225 |
| Lard | 0 | 0 | 245 | 2205 |
| Mineral mixture | 35 | 0 | 35 | 0 |
| Vitamin mixture | 10 | 40 | 10 | 40 |
| TBHQ | 0.014 | 0 | 0.014 | 0 |
| l-Cystine | 3 | 12 | 3 | 12 |
| Choline bitartrate | 2.5 | 0 | 2.5 | 0 |
| Total | 1000 | 4000 | 750.1 | 4000 |
| Microbiota Preparation Experiment (MP) | Microbiota Transfer Experiment (MT) | |||||
|---|---|---|---|---|---|---|
| Groups | MP Normal | MP HFD | MP Es | MT Normal | MT HFD | MT Es |
| Initial body weight (g) | 244.0 ± 9.6 | 248.0 ± 7.7 | 244.9 ± 10.2 | 271.1 ± 3.3 | 268.2 ± 4.9 | 272.9 ± 6.0 |
| Final body weight (g) | 444.9 ± 20.3 | 553.6 ± 61.7 # | 487.9 ± 56.8 * | 524.6 ± 21.5 | 587.3 ± 5.6 & | 532.8 ± 14.1 $ |
| Body weight gain (g) | 200.9 ± 23.8 | 305.6 ± 59.9 # | 249.1 ± 42.9 * | 253.5 ± 18.2 | 319.1 ± 10.3 & | 260.0 ± 12.0 $ |
| Food intake (g/day) | 23.8 ± 2.5 | 22.0 ± 2.8 | 18.1 ± 3.7 * | 25.3 ± 0.8 | 22.1 ± 0.8 & | 18.7 ± 1.2 $ |
| Food efficiency ratio | 0.21 ± 0.02 | 0.35 ± 0.08 # | 0.36 ± 0.11 | 0.24 ± 0.01 | 0.34 ± 0.00 & | 0.33 ± 0.01 |
| Total fat (g) | 46.0 ± 4.9 | 82.3 ± 19.2 # | 63.1 ± 6.7 * | 52.5 ± 9.8 | 83.1 ± 5.9 & | 58.5 ± 2.8 $ |
| Relative of total fat (%) | 7.89 ± 0.66 | 11.75 ± 2.90 # | 10.06 ± 0.73 | 10.0 ± 1.5 | 14.2 ± 1.1 & | 10.9 ± 0.4 $ |
| Abdominal fat (g) | 10.8 ± 1.4 | 17.9 ± 5.1 # | 13.1 ± 2.3 * | 11.3 ± 4.1 | 17.1 ± 1.4 | 16.3 ± 2.0 |
| Epididymal fat (g) | 9.8 ± 0.9 | 17.8 ± 3.9 # | 13.6 ± 2.1 * | 17.3 ± 2.4 | 28.0 ± 1.8 & | 15.0 ± 1.2 $ |
| Perirenal fat (g) | 14.5 ± 2.4 | 28.6 ± 5.9 # | 22.5 ± 3.4 | 23.9 ± 4.0 | 38.0 ± 4.1 & | 27.2 ± 4.5 $ |
| Liver weight (g) | 12.9 ± 1.5 | 16.4 ± 2.1 # | 14.1 ± 2.3 | 15.0 ± 0.8 | 15.2 ± 0.7 | 15.5 ± 1.6 |
| Relative of Liver weight (%) | 2.91 ± 0.34 | 3.00 ± 0.56 | 2.88 ± 0.26 | 2.9 ± 0.1 | 2.6 ± 0.1 & | 2.9 ± 0.3 |
| Cecum weight (g) | 5.6 ± 0.8 | 4.6 ± 0.2 # | 5.0 ± 0.3 * | 4.2 ± 0.1 | 4.6 ± 0.1 & | 5.3 ± 1.3 |
| Relative of Cecum weight (%) | 1.26 ± 0.2 | 0.84 ± 0.08 # | 1.04 ± 0.09 * | 0.80 ± 0.04 | 0.79 ± 0.02 | 0.99 ± 0.27 |
| Cecum contents (g) | 1.90 ± 0.57 | 1.39 ± 0.19 # | 1.69 ± 0.21 * | 1.7 ± 0.2 | 1.1 ± 0.1 & | 1.5 ± 0.2 $ |
| Microbiota Preparation Experiment (MP) | Microbiota Transfer Experiment (MT) | |||||
|---|---|---|---|---|---|---|
| Groups | MP Normal | MP HFD | MP Es | MT Normal | MT HFD | MT Es |
| AST (IU/L) | 76.8 ± 10.8 | 73.9 ± 13.7 | 80.4 ± 14.1 | 79 ± 7.8 | 151.3 ± 41.2 | 113.0 ± 26.0 |
| ALT (IU/L) | 23.3 ± 4.2 | 35.4 ± 5.6 # | 35.1 ± 10.3 | 30.0 ± 6.2 | 32.3 ± 2.9 | 31.7 ± 2.5 |
| TG (mg/dL) | 28.8 ± 1.3 | 59.0 ± 12.1 # | 43.2 ± 6.1 * | 43.0 ± 6.1 | 54.0 ± 11.4 | 39.7 ± 13.6 |
| TC (mg/dL) | 47.9 ± 7.8 | 52.9 ± 5.3 | 59.3 ± 11.9 | 65.7 ± 7.4 | 66.0 ± 4.6 | 59.3 ± 9.5 |
| HDL (mg/dL) | 28.6 ± 11.2 | 24.3 ± 2.0 | 33.1 ± 4.3 * | 35.2 ± 4.1 | 29.7 ± 4.6 | 28.7 ± 5.3 |
| FBG (mg/dL) | 135.8 ± 27.5 | 186.4 ± 29.8 # | 145.1 ± 25.6 * | 158.7 ± 18.5 | 268.7 ± 12.9 & | 190.0 ± 21.4 $ |
| Insulin (ng/mL) | 0.28 ± 0.13 | 0.65 ± 0.14 # | 0.52 ± 0.07 * | 0.24 ± 0.15 | 0.57 ± 0.11 & | 0.28 ± 0.08 $ |
| HOMA-IR (mg/dL) | 0.09 ± 0.04 | 0.29 ± 0.05 # | 0.18 ± 0.04 * | 0.11 ± 0.02 | 0.38 ± 0.09 & | 0.11 ± 0.06 $ |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.-H.; Kim, B.-S.; Han, K.; Kim, H. Ephedra-Treated Donor-Derived Gut Microbiota Transplantation Ameliorates High Fat Diet-Induced Obesity in Rats. Int. J. Environ. Res. Public Health 2017, 14, 555. https://doi.org/10.3390/ijerph14060555
Wang J-H, Kim B-S, Han K, Kim H. Ephedra-Treated Donor-Derived Gut Microbiota Transplantation Ameliorates High Fat Diet-Induced Obesity in Rats. International Journal of Environmental Research and Public Health. 2017; 14(6):555. https://doi.org/10.3390/ijerph14060555
Chicago/Turabian StyleWang, Jing-Hua, Bong-Soo Kim, Kyungsun Han, and Hojun Kim. 2017. "Ephedra-Treated Donor-Derived Gut Microbiota Transplantation Ameliorates High Fat Diet-Induced Obesity in Rats" International Journal of Environmental Research and Public Health 14, no. 6: 555. https://doi.org/10.3390/ijerph14060555
APA StyleWang, J.-H., Kim, B.-S., Han, K., & Kim, H. (2017). Ephedra-Treated Donor-Derived Gut Microbiota Transplantation Ameliorates High Fat Diet-Induced Obesity in Rats. International Journal of Environmental Research and Public Health, 14(6), 555. https://doi.org/10.3390/ijerph14060555








