Antibiotic Resistance in an Indian Rural Community: A ‘One-Health’ Observational Study on Commensal Coliform from Humans, Animals, and Water
Abstract
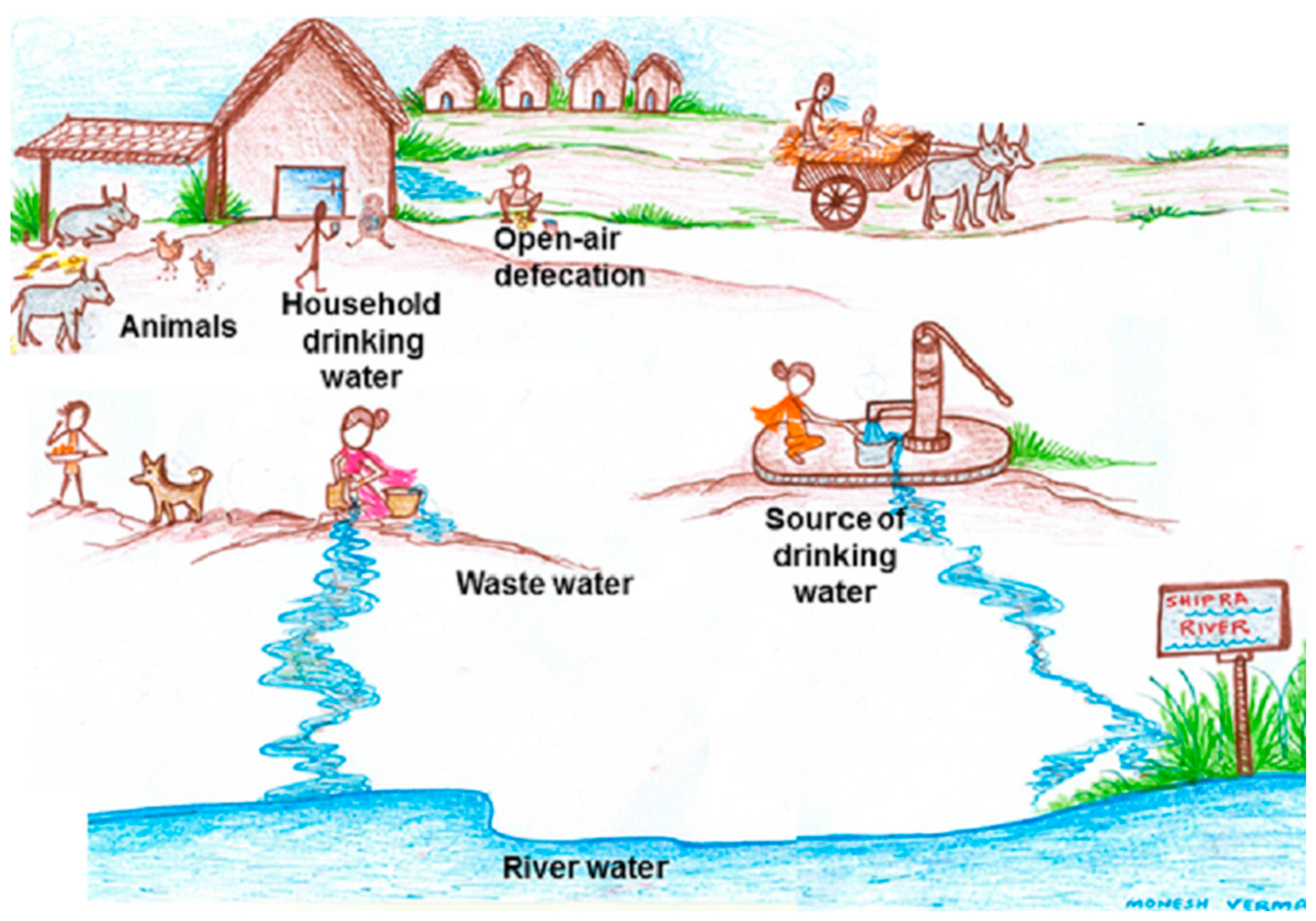
:1. Introduction
2. Materials and Methods
2.1. Study Setting and Sample Collection
2.2. Identification of Coliforms and Confirmation of E. coli
2.3. Antibiotic Susceptibility Testing
2.4. Amplification of Genes
2.5. Data Analysis
2.6. Ethical Consideration
3. Results
3.1. Study Samples
3.2. Antibiotic Resistance Pattern of E. coli in Various Sources
3.3. Antibiotic Resistance Pattern of E. coli to Various Antibiotic Groups
3.4. Antibiotic Resistant Genes in E. coli
3.5. Non-E. coli Coliforms and AST Pattern
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Zhang, T.; Zhang, M.; Fang, H.H.; Cheng, S.P. Characterization and quantification of class 1 integrons and associated gene cassettes in sewage treatment plants. Appl. Microbiol. Biotechnol. 2009, 82, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Zhang, T.; Fang, H.H. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance Draft Global Action Plan on Antimicrobial Resistance. Available online: http://apps.who.int/gb/ebwha/pdf_files/WHA68/A68_20-en.pdf?ua=1 (accessed on 23 December 2015).
- U.S. Food and Drug Administration. National Antimicrobial Resistance Monitoring System—Enteric Bacteria (NARMS): Executive Report; USFDA: Rockville, MD, USA, 2010. [Google Scholar]
- Łuczkiewicz, A.; Jankowska, K.; Fudala-Ksiazek, S.; Olańczuk-Neyman, K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 2010, 44, 5089–5097. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, T.; Kohnen, W.; Jansen, B.; Obst, U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 2003, 43, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Stålsby Lundborg, C.; Diwan, V.; Pathak, A.; Purohit, M.R.; Shah, H.; Sharma, M.; Mahadik, V.K.; Tamhankar, A.J. Protocol: A ‘One health’ two year follow-up, mixed methods study on antibiotic resistance, focusing children under 5 and their environment in rural India. BMC Public Health 2015, 30, 1321–1332. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.F.E.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Iqbal, S.; Robinson, J.; Deere, D.; Saunders, J.R.; Edwards, C.; Porter, J. Efficiency of the polymerase chain reaction amplification of the uid gene for detection of Escherichia coli in contaminated water. Lett. Appl. Microbiol. 1997, 24, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Chandran, S.P.; Diwan, V.; Tamhankar, A.J.; Joseph, B.V.; Rosales-Klintz, S.; Mundayoor, S.; Lundborg, C.S.; Macaden, R. Detection of carbapenem resistance genes and cephalosporin, and quinolone resistance genes along with oqxAB gene in Escherichia coli in hospital wastewater: A matter of concern. J. Appl. Microbiol. 2014, 117, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M.; Heuer, O.E. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin. Infect. Dis. 2009, 48, 916–921. [Google Scholar] [CrossRef] [PubMed]
- Van den Bogaard, A.E.; Bruinsma, N.; Stobberingh, E.E. The effect of banning avoparcin on VRE carriage in The Netherlands. J. Antimicrob. Chemother. 2000, 46, 146–147. [Google Scholar] [CrossRef] [PubMed]
- Abo-State, M.A.; Mahdy, H.M.; Ezzat, S.M.; Abd El Shakour, E.H.; El-Bahnasawy, M.A. Antimicrobial resistance profiles of Enterobacteriaceae isolated from Rosetta branch of River Nile. Egypt. Appl. Sci. J. 2012, 19, 1234–1243. [Google Scholar]
- Sivri, N.; Sandalli, C.; Ozgumus, O.B.; Colakoglu, F.; Dogan, D. Antibiotic Resistance Profiles of Enteric Bacteria Isolated from Kucukcekmece Lagoon (Istanbul–Turkey). Turk. J. Fish. Aquat. Sci. 2012, 12, 699–707. [Google Scholar] [CrossRef]
- Hussain, A.; Ranjan, A.; Nandanwar, N.; Babbar, A.; Jadhav, S.; Ahmed, N. Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob. Agents Chemother. 2014, 58, 7240–7249. [Google Scholar] [CrossRef] [PubMed]
- Sidjabat, H.E.; Paterson, D.L. Multidrug-resistant Escherichia coli in Asia: Epidemiology and management. Expert Rev. Anti-Infect. Ther. 2015, 13, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Schjørring, S.; Krogfelt, K.A. Assessment of bacterial antibiotic resistance transfer in the gut. Int. J. Microbiol. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, S.J.; Bailey, M.; Hansen, L.H.; Kroer, N.; Wuertz, S. Studying plasmid horizontal transfer in situ: A critical review. Nat. Rev. Microbiol. 2005, 3, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Ichor, T.; Umeh, E.U.; Duru, E.E. Microbial Contamination of Surface Water Sources in Rural Areas of Guma Local Government Area of Benue State, Nigeria. Med. Sci. Public Health 2014, 2, 43–51. [Google Scholar]
- Hagedorn, C.; Robinson, S.L.; Filtz, J.R.; Grubbs, S.M.; Angier, T.A.; Reneau, R.B., Jr. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal Streptococci. Appl. Environ. Microbiol. 1999, 65, 5522–5531. [Google Scholar] [PubMed]
- Zhang, Y.; Marrs, C.F.; Simon, C.; Xi, C. Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci. Total Environ. 2009, 407, 3702–3706. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.; Gundry, S.; Conroy, R. Household drinking water in developing countries: A systematic review of microbiological contamination between source and point-of-use. Trop. Med. Int. Health 2004, 9, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D. Extraintestinal Pathogenic Escherichia coli: A Combination of Virulence with Antibiotic Resistance. Front. Microbiol. 2012, 19, 9. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; McGowan, J.E., Jr. Reasons for the emergence of antibiotic resistance. Am. J. Med. Sci. 1996, 311, 9–16. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.; Johnson, S.J.; Stell, A.L.; Doetkott, C.; Johnson, J.R.; Kim, K.S.; Spanjaard, L.; Nolan, L.K. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 2008, 74, 7043–7050. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Saha, S.; Subudhi, E. More Furious Than Ever: Escherichia coli-Acquired Co-resistance toward Colistin and Carbapenems. Clin. Infect. Dis. 2016, 63, 1267–1268. [Google Scholar] [PubMed]
- Gupta, M.; Lakhina, K.; Kamath, A.; Vandana, K.E.; Mukhopadhyay, C.; Vidyasagar, S.; Varma, M. Colistin-resistant Acinetobacter baumannii ventilator-associated pneumonia in a tertiary care hospital: An evolving threat. J. Hosp. Infect. 2016, 94, 72–73. [Google Scholar] [CrossRef] [PubMed]
- Ckakraborty, A.; Adhikari, P.; Shenoy, S.; Baliga, S.; Bhat, G.; Rao, S.; Biranthabail, D.; Saralaya, V. Molecular characterization and clinical significance of New Delhi metallo-beta-lactamases-1 producing Escherichia coli recovered from a South Indian tertiary care hospital. Indian J. Pathol. Microbiol. 2015, 58, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Mittal, G.; Gaind, R.; Kumar, D.; Kaushik, G.; Gupta, K.B.; Verma, P.K.; Deb, M. Risk factors for fecal carriage of carbapenemase producing Enterobacteriaceae among intensive care unit patients from a tertiary care center in India. BMC Microbiol. 2016, 16, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Purohit, M.R.; Chandran, S.P.; Diwan, V.; Harshada, S.; Tamhankar, A.J.; Lundborg, C.S. Detection of carbapenem resistance genes and cephalosporin, and quinolone resistance genes in Escherichia coli in hospital and community wastewater. (Manuscript in preparation).

| Variable | Number (%) |
|---|---|
| Family type | |
| Nuclear family | 6 |
| Joint family | 16 |
| Total number of family members | 162 |
| Male | 84/162 (52) |
| Female | 78/162 (48) |
| Number of children- | |
| Up to five years of age | 46 |
| Male | 25/46 (54) |
| Female | 21/46 (46) |
| Between one and three years of age | 24 |
| Male | 15 |
| Female | 9 |
| Highest education of family member | |
| Primary education (up to 5th grade) | 5 |
| Middle | 11 |
| Secondary | 2 |
| Illiterate | 144 |
| Occupation of head of family | |
| Job | 1 |
| Farmer | 12 |
| Labor/self employed | 6 |
| Unemployed | 3 |
| Type of house | |
| Kuchcha | 11 |
| Pucca/semi-pucca | 1/10 |
| Total number of livestock in all households | 75 |
| Source of drinking water | |
| Piped water into dwelling | 1 |
| In-house tube wells/bore hole | 1 |
| Hand pump | 10 |
| Unprotected dug well | 1 |
| Source of Samples | Number of Samples | Number of E. coli | Number of Non-E. coli |
|---|---|---|---|
| Children stool | 22 | 127 | 67 |
| Dog stool | 1 | 6 | 2 |
| Hen stool | 1 | 6 | 6 |
| Goat stool | 1 | 3 | 0 |
| Horse stool | 1 | 6 | 4 |
| Source-water | 2 | 10 | 14 |
| Waste-water | 2 | 12 | 7 |
| Household drinking water | 22 | 122 | 143 |
| Total | 52 | 292 | 243 |
| Name of Antibiotic Tested | Human Stool (n = 22) | Household Drinking Water (n = 20) | ||
|---|---|---|---|---|
| Resistant E. coli Isolates in Samples (n) * | ||||
| <3 | ≥3 | <3 | ≥3 | |
| Ampicillin | 6 | 12 | 15 | 3 |
| Ceftazidime | 7 | 11 | 13 | 1 |
| Cefotaxime | 7 | 11 | 11 | - |
| Nalidixic acid | 5 | 9 | 6 | 2 |
| Ciprofloxacin | 5 | 6 | 7 | - |
| Nitrofurantoin | 1 | 1 | 2 | - |
| Gentamicin | 2 | 1 | 2 | - |
| Amikacin | 1 | - | 2 | - |
| Tetracycline | 1 | 3 | 8 | - |
| Tigicycline | 3 | 1 | 2 | - |
| Imipenem | - | 1 | - | - |
| Meropenem | 5 | 2 | 1 | - |
| Sulfamethoxazole | 2 | 4 | 4 | 2 |
| Cotrimoxazole | 2 | 4 | 4 | 1 |
| Cephalosporin Resistant Isolates | Cephalosporin Resistance Genes | ||
| CTX-M1 | CTX-M2 | CTX-M9 | |
| HS (n = 73) | 62 | 0 | 0 |
| HDW (n = 26) | 11 | 0 | 0 |
| AS (n = 6) | 4 | 0 | 0 |
| SW (n = 5) | 0 | 0 | 1 |
| WW (n = 5) | 0 | 0 | 5 |
| Quinolone Resistant Isolates | Quinolone Resistance Genes | ||
| qnrA | qnrS | qnrS | |
| HS (n = 72) | 0 | 2 | 23 |
| HDW (n = 13) | 0 | 0 | 3 |
| AS (n = 2) | 0 | 0 | 0 |
| SW (n = 1) | 0 | 0 | 0 |
| WW (n = 8) | 0 | 0 | 0 |
| Phylogenetic Group | A n = 135 | B1 n = 55 | B2 n = 13 | D n = 92 |
|---|---|---|---|---|
| Cephalosporin-Resistant Isolates | ||||
| HS (n = 73) | 35 | 17 | 0 | 21 |
| HDW (n = 26) | 16 | 3 | 0 | 7 |
| AS (n = 6) | 4 | 0 | 1 | 1 |
| SW (n = 5) | 4 | 0 | 1 | 0 |
| WW (n = 5) | 0 | 0 | 0 | 5 |
| Quinolone-Resistant Isolates | ||||
| HS (n = 72) | 28 | 13 | 1 | 30 |
| HDW (n = 13) | 9 | 1 | 0 | 3 |
| AS (n = 2) | 2 | 0 | 0 | 0 |
| SW (n = 1) | 1 | 0 | 0 | 0 |
| WW (n = 8) | 1 | 0 | 0 | 7 |
| Meropenem-Resistant Isolates | ||||
| HS (n = 19) | 10 | 1 | 0 | 8 |
| HDW (n = 8) | 6 | 2 | 0 | 0 |
| AS (n = 1) | 1 | 0 | 0 | 0 |
| SW (n = 1) | 1 | 0 | 0 | 0 |
| WW (n = 0) | 0 | 0 | 0 | 0 |
| Susceptible to All Drugs | ||||
| HS (n = 21) | 2 | 8 | 6 | 5 |
| HDW (n = 9) | 1 | 6 | 1 | 1 |
| AS (n = 2) | 0 | 2 | 0 | 0 |
| SW (n = 3) | 1 | 0 | 2 | 0 |
| WW (n = 1) | 0 | 1 | 0 | 0 |
| Sample * | Total Coliform Count/100 mL | Total E. coli/100 mL n (%) | Total-E. coli = Non-E. coli n (%) (Calculated) |
|---|---|---|---|
| 1 | 1630 | 260 (16) | 1370 (84) |
| 2 | 1400 | 40 (3) | 1360 (97) |
| 3 | 520 | 100 (19) | 400 (81) |
| 4 | 498 | 64 (13) | 434 (87) |
| 5 | 430 | 40 (8) | 390 (92) |
| 6 | 414 | 14 (3) | 400 (97) |
| 7 | 152 | 3 (2) | 149 (98) |
| 8 | 150 | 1 (0.66) | 149 (99.4) |
| 9 | 134 | 34 (25) | 100 (75) |
| 10 | 48 | 20 (41) | 28 (59) |
| 11 | 365,000,000 | 15,000,000 (41) | 215,000,000 (59) |
| 12 | 204,000,000 | 32,000,000 (16) | 1,720,000,000 (84) |
| 13 | 3650 | 150 (4) | 3500 (96) |
| 14 | 3 | 0 | 3 (100) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Purohit, M.R.; Chandran, S.; Shah, H.; Diwan, V.; Tamhankar, A.J.; Stålsby Lundborg, C. Antibiotic Resistance in an Indian Rural Community: A ‘One-Health’ Observational Study on Commensal Coliform from Humans, Animals, and Water. Int. J. Environ. Res. Public Health 2017, 14, 386. https://doi.org/10.3390/ijerph14040386
Purohit MR, Chandran S, Shah H, Diwan V, Tamhankar AJ, Stålsby Lundborg C. Antibiotic Resistance in an Indian Rural Community: A ‘One-Health’ Observational Study on Commensal Coliform from Humans, Animals, and Water. International Journal of Environmental Research and Public Health. 2017; 14(4):386. https://doi.org/10.3390/ijerph14040386
Chicago/Turabian StylePurohit, Manju Raj, Salesh Chandran, Harshada Shah, Vishal Diwan, Ashok J. Tamhankar, and Cecilia Stålsby Lundborg. 2017. "Antibiotic Resistance in an Indian Rural Community: A ‘One-Health’ Observational Study on Commensal Coliform from Humans, Animals, and Water" International Journal of Environmental Research and Public Health 14, no. 4: 386. https://doi.org/10.3390/ijerph14040386
APA StylePurohit, M. R., Chandran, S., Shah, H., Diwan, V., Tamhankar, A. J., & Stålsby Lundborg, C. (2017). Antibiotic Resistance in an Indian Rural Community: A ‘One-Health’ Observational Study on Commensal Coliform from Humans, Animals, and Water. International Journal of Environmental Research and Public Health, 14(4), 386. https://doi.org/10.3390/ijerph14040386






