Leptin Receptor Gene Polymorphism and the Risk of Cardiovascular Disease: A Systemic Review and Meta-Analysis
Abstract
1. Introduction
2. Methods
2.1. Literature Search
2.2. Selection Criteria and Data Extraction
2.3. Quality Assessment
2.4. Statistical Analysis
3. Results
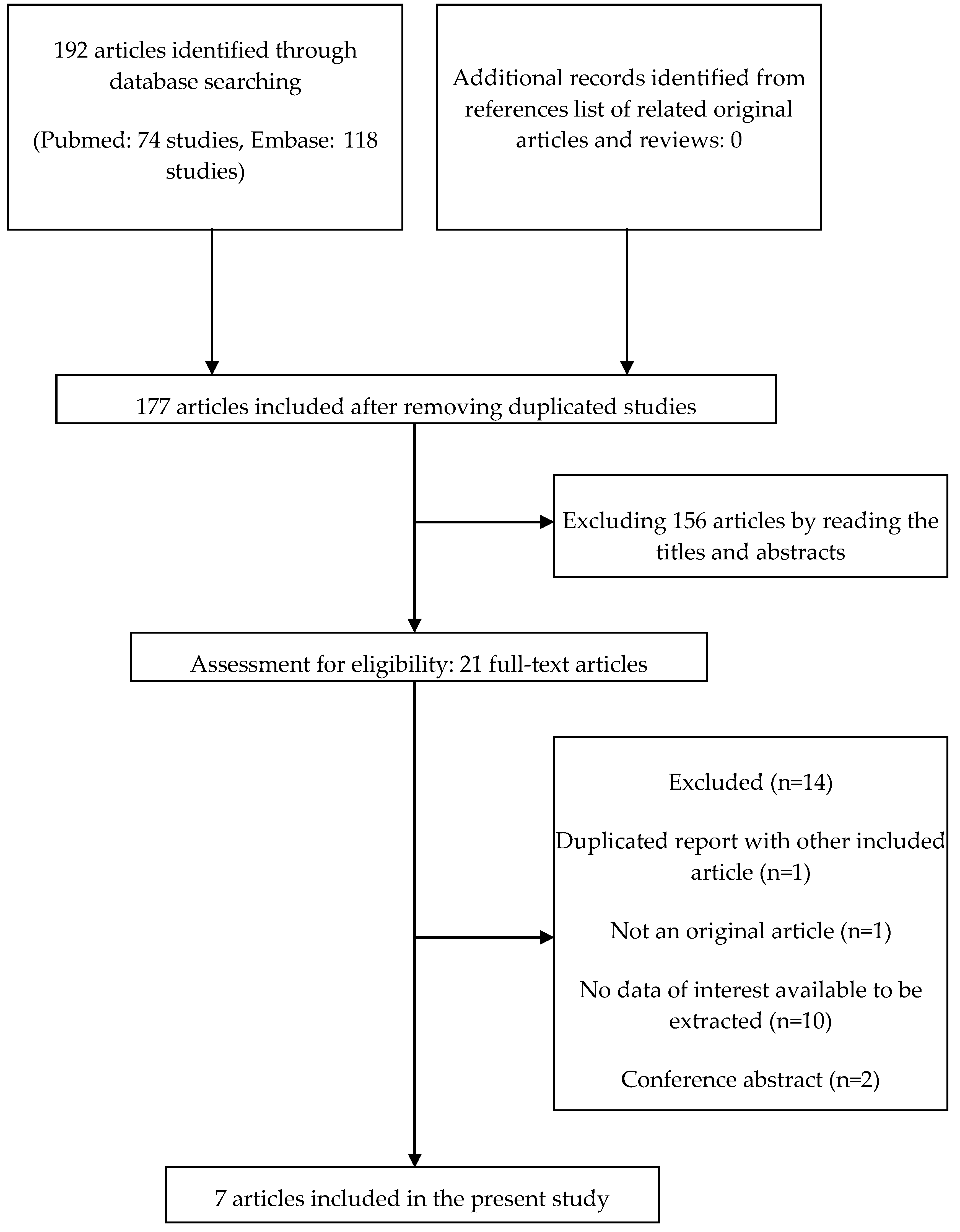
3.1. Study Identification and Selection
3.2. Study Characteristics
3.3. Quality Assessment
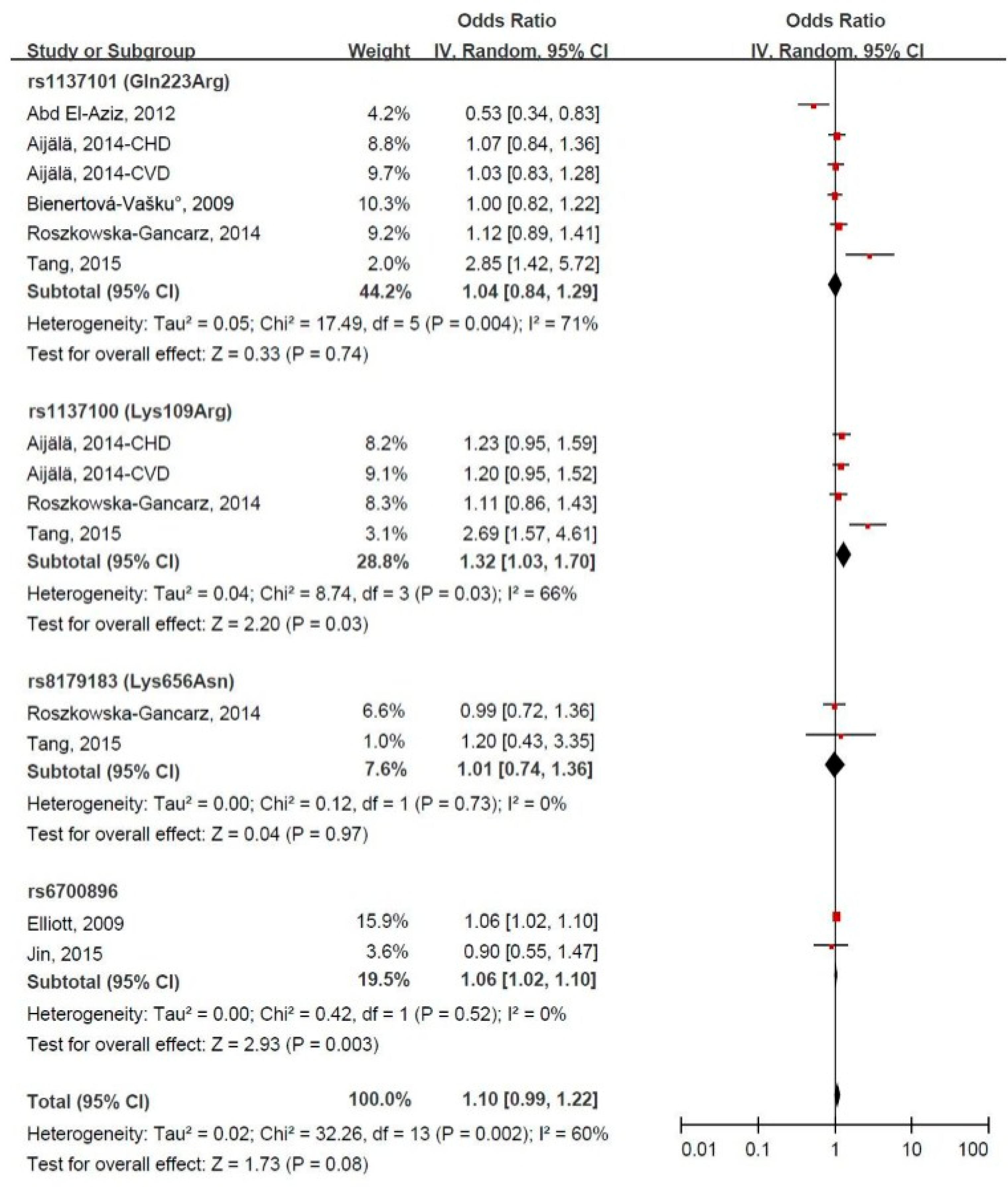
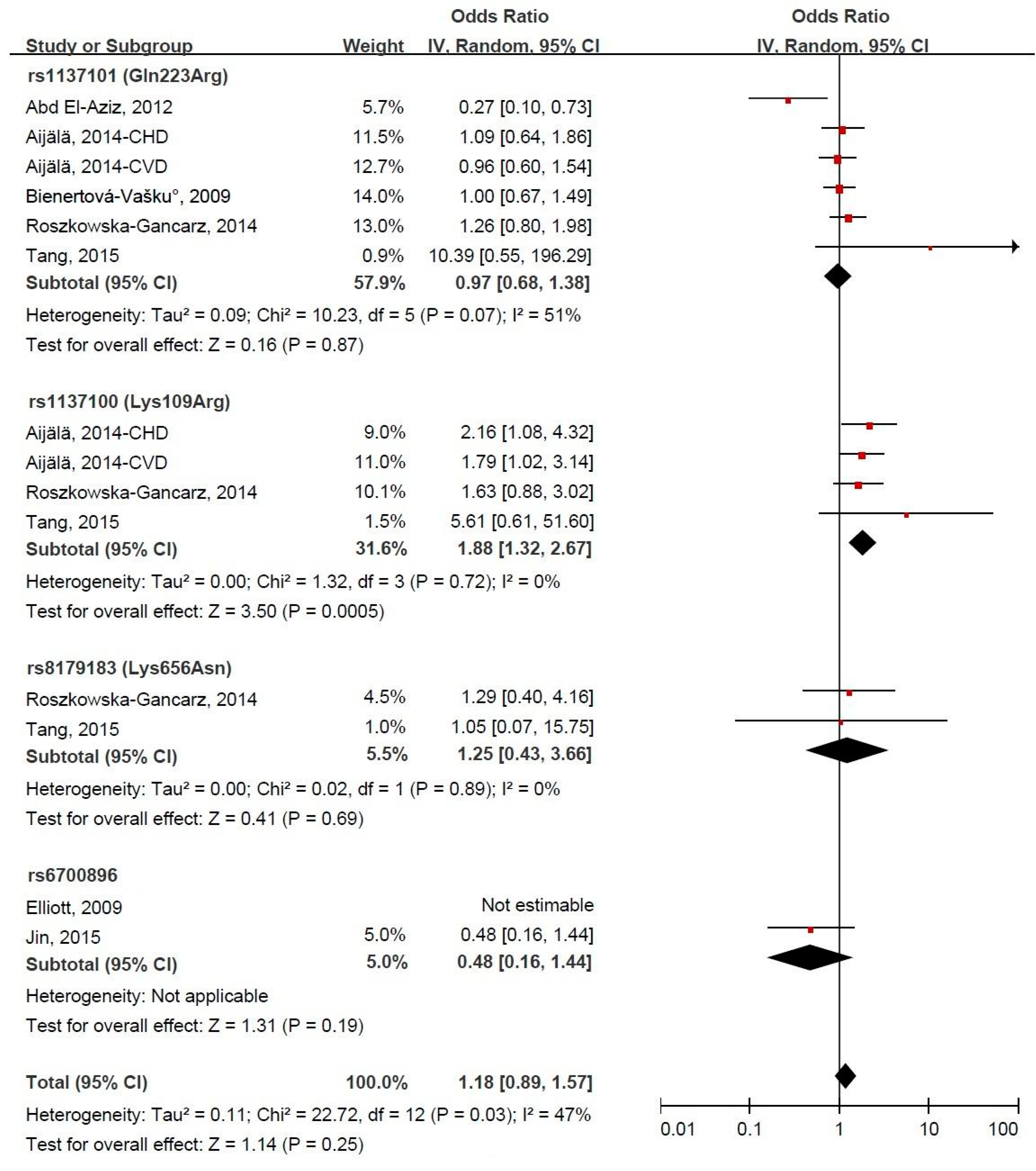
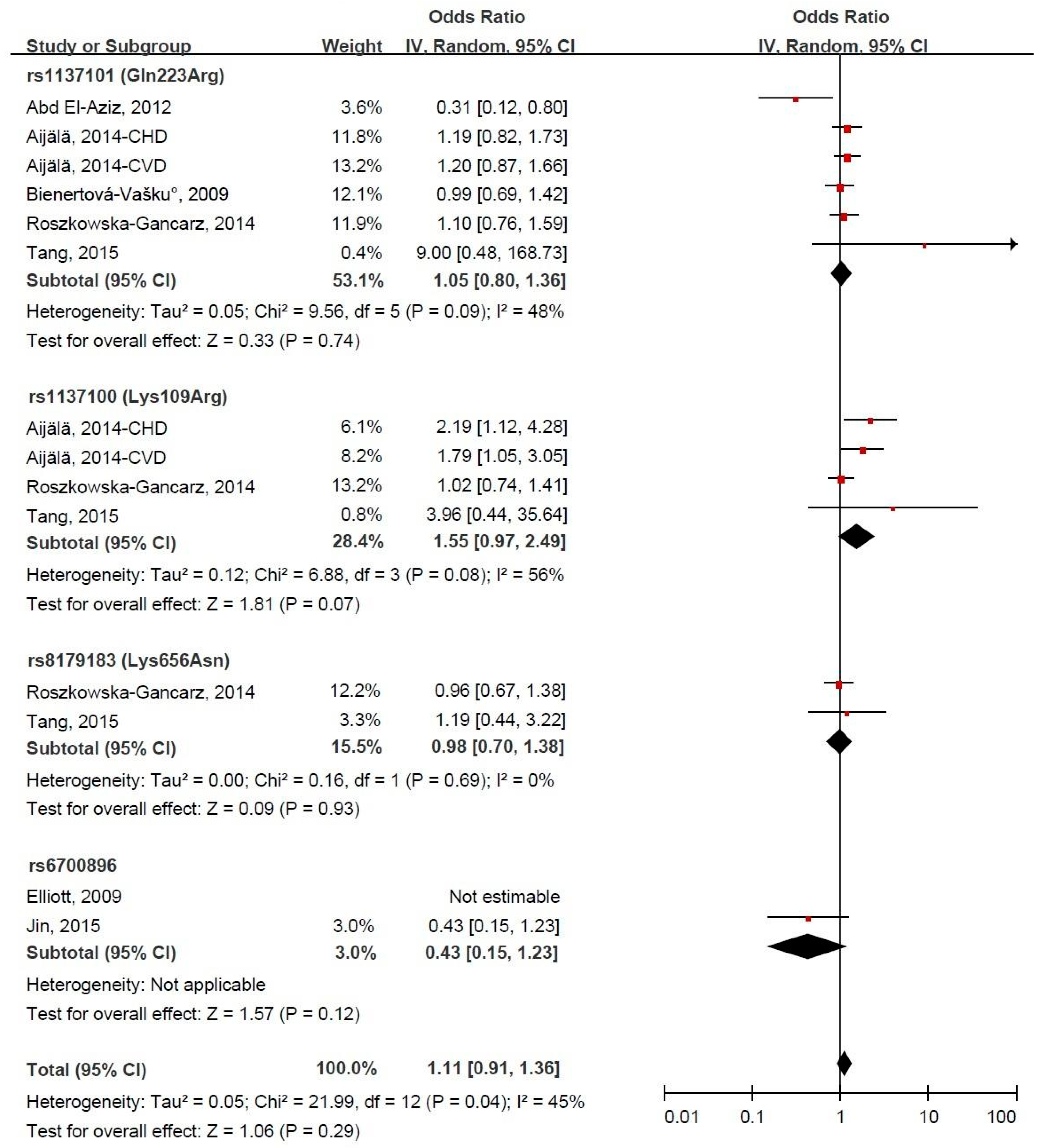
3.4. LEPR Gene Polymorphism and CVD Risk
3.5. Subgroup Meta-Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
Funding
References
- Bansilal, S.; Castellano, J.M.; Fuster, V. Global burden of CVD: Focus on secondary prevention of cardiovascular disease. Int. J. Cardiol. 2015, 201 (Suppl. 1), S1–S7. [Google Scholar] [CrossRef]
- Joseph, P.G.; Pare, G.; Anand, S.S. Exploring gene-environment relationships in cardiovascular disease. Can. J. Cardiol. 2013, 29, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Pasipoularides, A. Linking Genes to Cardiovascular Diseases: Gene Action and Gene-Environment Interactions. J. Cardiovasc. Transl. Res. 2015, 8, 506–527. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.G.; Newton-Cheh, C. Genome-wide association studies of late-onset cardiovascular disease. J. Mol. Cell. Cardiol. 2015, 83, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Schulze, P.C.; Kratzsch, J. Leptin as a new diagnostic tool in chronic heart failure. Clin. Chim. Acta 2005, 362, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, L.A.; Dembski, M.; Weng, X.; Deng, N.; Culpepper, J.; Devos, R.; Richards, G.J.; Campfield, L.A.; Clark, F.T.; Deeds, J.; et al. Identification and expression cloning of a leptin receptor, OB-R. Cell 1995, 83, 1263–1271. [Google Scholar] [CrossRef]
- Su, S.; Zhang, C.; Zhang, F.; Li, H.; Yang, X.; Tang, X. The association between leptin receptor gene polymorphisms and type 2 diabetes mellitus: A systematic review and meta-analysis. Diabetes Res. Clin. Pract. 2016, 121, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Zhang, Q.; Shang, X.M.; Li, Y.Q.; Liu, X.K.; Liu, C.Q.; Liu, X.M.; Zhang, Q.H. Association of two well-defined polymorphisms in leptin and leptin receptor genes with hypertension and circulating leptin: A meta-analysis. Arch. Med. Res. 2015, 46, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Lao, X.; Li, S.; Qin, X. Need for clarification of data in a recent meta-analysis about polymorphisms in the leptin receptor gene and risk of breast cancer. Breast Cancer Res Treat. 2013, 138, 655–656. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Aziz, T.A.; Mohamed, R.H.; Mohamed, R.H.; Pasha, H.F. Leptin, leptin gene and leptin receptor gene polymorphism in heart failure with preserved ejection fraction. Heart Vessels 2012, 27, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Bienertová-Vasků, J.A.; Spinarová, L.; Bienert, P.; Vasků, A. Association between variants in the genes for leptin, leptin receptor, and proopiomelanocortin with chronic heart failure in the Czech population. Heart Vessels 2009, 24, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Zhang, Z.; Li, Z.K.; Lin, J.; Fang, D.Z. Association of leptin receptor gene polymorphisms with genetic susceptibility to ischemic stroke. J. Stroke Cerebrovasc. Dis. 2015, 24, 2128–2133. [Google Scholar] [CrossRef] [PubMed]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B.; et al. Meta-analysis of observational studies in epidemiology: A proposal for reporting. Meta-analysis of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000, 283, 2008–2012. [Google Scholar] [CrossRef] [PubMed]
- Shamliyan, T.A.; Kane, R.L.; Ansari, M.T.; Raman, G.; Berkman, N.D.; Grant, M.; Janes, G.; Maglione, M.; Moher, D.; Nasser, M.; et al. Development qualitycriteria to evaluate nontherapeutic studies of incidence, prevalence, or risk factors of chronic diseases: Pilot study of new checklists. J. Clin. Epidemiol. 2011, 64, 637–657. [Google Scholar] [CrossRef] [PubMed]
- Quality Assessment Tool for Observational Cohort and Cross-Sectional Studies. Available online: http://www.nhlbi.nih.gov/health-pro/guidelines/in-develop/cardiovascular-risk-reduction/tools/cohort (accessed on 1 March 2017).
- Begg, C.B.; Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994, 50, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Biasin meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Aijälä, M.; Santaniemi, M.; Bloigu, R.; Kesäniemi, Y.A.; Ukkola, O. Leptin receptor Arg109 homozygotes display decreased total mortality as well as lower incidence of cardiovascular disease and related death. Gene 2014, 534, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Elliott, P.; Chambers, J.C.; Zhang, W.; Clarke, R.; Hopewell, J.C.; Peden, J.F.; Erdmann, J.; Braund, P.; Engert, J.C.; Bennett, D.; et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA 2009, 302, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.G.; Chen, G.L.; Yang, S.L.; Zhao, M.Y. Gene-gene interactions among CX3CL1, LEPR and IL-6 related to coronary artery disease in Chinese Han population. Int. J. Clin. Exp. Pathol. 2015, 8, 5968–5973. [Google Scholar] [PubMed]
- Roszkowska-Gancarz, M.; Kurylowicz, A.; Polosak, J.; Mossakowska, M.; Franek, E.; Puzianowska-Kuźnicka, M. Functional polymorphisms of the leptin and leptin receptor genes are associated with longevity and with the risk of myocardial infarction and of type 2 diabetes mellitus. Endokrynol. Pol. 2014, 65, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.R.; Maia, L.L.; Santos, M.; Peterle, G.T.; Alves, L.U.; Takamori, J.T.; Souza, R.P.; Barbosa, W.M.; Mercante, A.M.; Nunes, F.D.; et al. Leptin receptor expression and Gln223Arg polymorphism as prognostic markers in oral and oropharyngeal cancer. Genet. Mol. Res. 2015, 14, 14979–14988. [Google Scholar] [CrossRef] [PubMed]
- He, B.S.; Pan, Y.Q.; Zhang, Y.; Xu, Y.Q.; Wang, S.K. Effect of LEPR Gln223Arg polymorphism on breast cancer risk in different ethnic populations: A meta-analysis. Mol. Biol. Rep. 2012, 39, 3117–3122. [Google Scholar] [CrossRef] [PubMed]
- Duggal, P.; Guo, X.; Haque, R.; Peterson, K.M.; Ricklefs, S.; Mondal, D.; Alam, F.; Noor, Z.; Verkerke, H.P.; Marie, C.; et al. A mutation in the leptin receptor is associated with Entamoeba histolytica infectionin children. J. Clin. Investig. 2011, 121, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, S.Q.; Jing, Z.H.; Hou, X.; Chen, Y.; Song, X.J.; Lv, W.S.; Wang, R.; Wang, Y.G. Association of LEPR Gln223Arg polymorphism with T2DM: A meta-analysis. Diabetes Res. Clin. Pract. 2015, 109, e21–e26. [Google Scholar] [CrossRef] [PubMed]
- Pyrzak, B.; Wisniewska, A.; Kucharska, A.; Wasik, M.; Demkow, U. No association of LEPR Gln223Arg polymorphism with leptin, obesity or metabolic disturbances in children. Eur. J. Med. Res. 2009, 14 (Suppl. 4), S201–S204. [Google Scholar] [CrossRef]
- Ragin, C.C.; Dallal, C.; Okobia, M.; Modugno, F.; Chen, J.; Garte, S.; Taioli, E. Leptin levels and leptin receptor polymorphism frequency in healthy populations. Infect. Agent. Cancer 2009, 4, S13. [Google Scholar] [CrossRef] [PubMed]
- Fong, T.M.; Huang, R.R.; Tota, M.R.; Mao, C.; Smith, T.; Varnerin, J.; Karpitskiy, V.V.; Krause, J.E.; Van der Ploeg, L.H. Localization of leptin binding domain in the leptin receptor. Mol. Pharmacol. 1998, 53, 234–240. [Google Scholar] [PubMed]
- Zabeau, L.; Defeau, D.; Van der Heyden, J.; Iserentant, H.; Vandekerckhove, J.; Tavernier, J. Functional analysis of leptin receptor activation using a Janus kinase/signal transducer and activator of transcription complementation assay. Mol. Endocrinol. 2004, 18, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Rosmond, R.; Chagnon, Y.C.; Holm, G.; Chagnon, M.; Pérusse, L.; Lindell, K.; Carlsson, B.; Bouchard, C.; Björntorp, P. Hypertension in obesity and the leptin receptor gene locus. J. Clin. Endocrinol. Metab. 2000, 85, 3126–3131. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Shin, H.D.; Park, B.L.; Cheong, H.S.; Cho, Y.M.; Lee, H.K.; Lee, J.Y.; Lee, J.K.; Oh, B.; Kimm, K. Polymorphisms in the leptin receptor (LEPR)-putative association with obesity and T2DM. J. Hum. Genet. 2006, 51, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Becer, E.; Mehmetcik, G.; Bareke, H.; Serakıncı, N. Association of leptin receptor gene Q223R polymorphism on lipid profiles in comparison study between obese and non-obese subjects. Gene 2013, 529, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.G.; Cowley, M.A.; Munzberg, H. Mechanisms of leptin action and leptin resistance. Annu. Rev. Physiol. 2008, 70, 537–556. [Google Scholar] [CrossRef] [PubMed]
| First Author, Published Year | Ethnicity | Study Design | Male (%) | Mean Age (Years) | Case | Controls | Leptin Receptor Polymorphism | ||
|---|---|---|---|---|---|---|---|---|---|
| Disease | No. | Matching | No. | ||||||
| Abd El-Aziz, 2012 [10] | Egyptian | Case-control | 26.0 | 55.5 | Coronary artery disease | 100 | Age- and sex-matched healthy subjects | 100 | rs1137101 (Gln223Arg) |
| Aijälä, 2014 [18] | European | Cohort | 49.1 | 51 | Coronary artery disease, Cardiovascular disease | 303,423 | - | 725,605 | rs1137101 (Gln223Arg), rs1137100 (Lys109Arg) |
| Bienertová-Vašku, 2009 [11] | American | Case-control | 71.2 | 56 | Chronic heart failure | 372 | Age- and sex-matched healthy subjects | 407 | rs1137101 (Gln223Arg) |
| Elliott, 2009 [19] | European and Asian a | Cohort | Both men and women, the proportion not reported | 20–75 b | Coronary heart disease | 12,148 | - | 29,127 | rs6700896 |
| Jin, 2015 [20] | Asian | Case-control | 53.3 | 59 | Coronary artery disease | 120 | Age- and sex-matched healthy subjects | 109 | rs6700896 |
| Roszkowska-Gancarz, 2014 [21] | European | Case-control | 57.0 | 47 | Myocardial infarction | 226 | Blood donors and volunteers | 190 | rs1137101 (Gln223Arg), rs1137100 (Lys109Arg), rs8179183 (Lys656Asn) |
| Tang, 2015 [12] | Asian | Case-control | 67.5 | 61 | Ischemic stroke | 101 | Age- and sex-matched healthy subjects | 105 | rs1137101 (Gln223Arg), rs1137100 (Lys109Arg), rs8179183 (Lys656Asn) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Sun, D. Leptin Receptor Gene Polymorphism and the Risk of Cardiovascular Disease: A Systemic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2017, 14, 375. https://doi.org/10.3390/ijerph14040375
Wu L, Sun D. Leptin Receptor Gene Polymorphism and the Risk of Cardiovascular Disease: A Systemic Review and Meta-Analysis. International Journal of Environmental Research and Public Health. 2017; 14(4):375. https://doi.org/10.3390/ijerph14040375
Chicago/Turabian StyleWu, Lei, and Dali Sun. 2017. "Leptin Receptor Gene Polymorphism and the Risk of Cardiovascular Disease: A Systemic Review and Meta-Analysis" International Journal of Environmental Research and Public Health 14, no. 4: 375. https://doi.org/10.3390/ijerph14040375
APA StyleWu, L., & Sun, D. (2017). Leptin Receptor Gene Polymorphism and the Risk of Cardiovascular Disease: A Systemic Review and Meta-Analysis. International Journal of Environmental Research and Public Health, 14(4), 375. https://doi.org/10.3390/ijerph14040375




